Abstract
Different cDNA clones encoding a rat homeobox gene and the mouse homologue OG-12 were cloned from adult rat brain and mouse embryo mRNA, respectively. The predicted amino acid sequences of the proteins belong to the paired-related subfamily of homeodomain proteins (Prx homeodomains). Hence, the gene was named Prx3 and the mouse and rat genes are indicated as mPrx3 and rPrx3, respectively. In the mouse as well as in the rat, the predicted Prx3 proteins share the homeodomain but have three different N termini, a 12-aa residue variation in the C terminus, and contain a 14-aa residue motif common to a subset of homeodomain proteins, termed the “aristaless domain.” Genetic mapping of Prx3 in the mouse placed this gene on chromosome 3. In situ hybridization on whole mount 12.5-day-old mouse embryos and sections of rat embryos at 14.5 and 16.5 days postcoitum revealed marked neural expression in discrete regions in the lateral and medial geniculate complex, superior and inferior colliculus, the superficial gray layer of the superior colliculus, pontine reticular formation, and inferior olive. In rat and mouse embryos, nonneuronal structures around the oral cavity and in hip and shoulder regions also expressed the Prx3 gene. In the adult rat brain, Prx3 gene expression was restricted to thalamic, tectal, and brainstem structures that include relay nuclei of the visual and auditory systems as well as other ascending systems conveying somatosensory information. Prx3 may have a role in specifying neural systems involved in processing somatosensory information, as well as in face and body structure formation.
Keywords: embryo, transcription factors, OG-12, SHOX, limb
Homeodomain proteins constitute a superfamily of transcription factors mainly involved in gene regulatory events that underlay embryonic development and differentiation (1, 2). Many homeobox genes have been implicated in the development of body structures and organs, such as the limbs, eye, spinal cord, and brain areas, by laying out the organizational plan and controlling patterning (1, 3, 4). A common theme in the patterning of developing tissues is the interplay between inductive signaling triggered by growth factors and the expression of multiple homeobox genes. Many homeobox genes are expressed in unique spatiotemporal modes. The expression of some homeobox genes may be restricted to specific cell lineages and sometimes may extend to adulthood (5–9).
Although the role of homeobox genes in patterning of the neural axis at the level of the spinal cord and the rhombencephalon has been investigated widely in mammals (3, 4), the number of known homeobox genes expressed during development of the mid- and forebrain is relatively limited. Their contribution in patterning and differentiation of these latter structures is less clearly understood because of the complexity of developmental regionalization and highly organized integration of very diverse neuronal systems (10–12). Recent data suggest that some homeobox genes that are expressed in the brain and pituitary gland during embryonic development retain expression in adulthood, in a cell-specific mode, and play a role in cell-specific expression and regulation of downstream target genes (6, 13). We therefore aimed to characterize homeobox genes expressed in the brain of adult rodents and in embryos. Here, multiple transcripts of a paired-related homeobox gene designated Prx3 are identified that are expressed in distinct patterns in the nervous system and peripheral structures, suggesting a role in specifying neural systems involved in conveying somatosensory information.
METHODS AND MATERIALS
Cloning of Prx3 cDNA.
Poly(A)+ RNA from distinct areas dissected from adult rat brain were subjected to reverse transcription-PCR (RT-PCR) with Moloney murine leukemia virus reverse transcriptase (SuperScript, BRL) and the following primers encoding conserved amino acid motifs within the first and third helices of known homeoboxes, respectively, using International Union of Biochemistry nomenclature conventions: upstream: 5′-GMRSCGMSAVMGSACMMBCTTYAC-3′, downstream: 5′-TGGTTYMRVAAYCGYHGMGCMARRTG-3′. The annealing temperature was 40°C. PCR products of the expected size were cloned in the plasmid vector pGEM 7zf(+) and sequenced. One of the cloned PCR products that encoded a homeobox was used to screen 500,000 plaques of a 14-day rat embryo whole brain cDNA library in λ-ZAP (from J. Boulter, Salk Institute, San Diego, CA). The positive clones were rescued by standard procedures, and both strands of the inserts were sequenced manually using the T7 sequencing kit (Pharmacia Biotech) with either a standard protocol or deaza-G reactions. A 10-day mouse embryo cDNA library in λ-SHlox (Novagen) was screened with a 32P-labeled antisense RNA probe corresponding to a partial cDNA fragment that was obtained by RT-PCR from 12-day mouse embryo RNA. PCR primer sequences were: upstream: 5′-GGAAAATCAATTCACAAAG-3′, downstream: 5′-CTACGTTGACATAGGGTGC-3′, corresponding, respectively, to nucleotide residues 1080–1099 and 1464–1482 of OG-12 genomic DNA (14). Three recombinant λ-SHlox DNA clones were isolated, and recombinant plasmids were excised from the phage vector following the manufacturer’s instructions and sequenced using a Perkin–Elmer/Applied Biosystems DNA sequencer (model 373A) using Taq DNA polymerase and fluorescent dideoxynucleotides (Perkin–Elmer/Applied Biosystems).
mPrx3 Chromosomal Mapping.
mPrx3 was mapped by analysis of the cross (NFS/N × Mus spretus) × M. spretus or C58/J (15, 16). Chromosome 3 markers were typed as described (17).
Northern Analysis.
Total RNA extracted from tissues of the adult rat by Rnazol (Biotecx Laboratories, Houston) was fractionated on a formaldehyde–agarose gel and transferred onto a nylon membrane by downward capillary blotting. Blots were hybridized in 0.5 M NaHPO4, 1 mM Na2EDTA, 1% crystalline grade BSA (Sigma), and 7% SDS at 65°C overnight (18) with a randomly primed 32P-labeled EcoRI–BamHI DNA fragment corresponding to the complete coding region of the rPrx3A cDNA without the Δ region and part of the 3′-untranslated region (nucleotides 1–864 in Fig. 1). The final washes were with 40 mM NaHPO4, 1 mM Na2EDTA, and 1% SDS at 65°C for 15 min. Autoradiography was performed with a FUJIX (Tokyo) BAS1000 phosphor imager (Fuji Photo Film).
Figure 1.
Nucleotide sequences of mouse and rat Prx3 cDNAs and predicted amino acid sequences. The figure shows that all Prx3 proteins share the region starting 18 aa residues before the homeodomain (rPrx3A amino acid residue 12) up to the C terminus, except for the Δ region that was absent in several clones, due to alternative splicing. Other variations in amino acid sequence concern the N terminus. mPrx3A is the protein predicted by the longest ORF in mPrx3A cDNA whereas the ORF of rPrx3A cDNA starts at the position corresponding to mPrx3A amino acid residue 101. rPrx3B is a partial protein generated by alternative splicing at nucleotide 578 of mPrx3A cDNA. An 18-nt residue intron interrupts the coding sequence at this position in the mouse mPrx3 gene (14). mPrx3B is predicted in the ORF of mPrx3B cDNA that differs from mPrx3A cDNA before mPrx3A cDNA nucleotide 560, marking another splice point. mPrx3B is the smallest predicted protein. Additional variation in transcripts is observed in the 3′-untranslated region due to different polyadenylation signals; a potential polyadenylation signal is underlined. Alternative 5′-nucleotide sequences are boxed, triangles indicate splice sites in the corresponding gene (14), the homeodomain is shown in shadow, and Gly- and Ala-rich regions and the aristaless domain are indicated.
In Situ Hybridization.
Sections from rat brain and embryos were prepared for in situ hybridization essentially as described (19). 35S-Labeled RNA probes were synthesized from the sense and antisense strands of the rPrx3 homeobox fragment originally isolated by RT-PCR. Sections were exposed on Betamax films (Amersham) for 3–7 days and then subjected to autoradiography under Hypercoat LM-1 liquid emulsion (Amersham). Whole mount in situ hybridization with 12-day mouse embryos was performed essentially as described (20) using digoxigenin-labeled antisense cRNA probes synthesized from a linearized mPrx3B cDNA plasmid, according to the manufacturer (Boehringer Mannheim). Probes were hydrolyzed partially in alkaline solution (60 mM Na2CO3/40 mM NaHCO3, pH 10.2) at 60°C for 10 min. Embryos were subjected to hybridization at 65°C overnight either in the absence of probe or in the presence of digoxigenin-labeled cRNA probe with or without 50 times excess of unlabeled cRNA probe.
RESULTS
Characterization of Prx3 Gene Products.
One of the cDNA clones obtained by RT-PCR from adult rat hypothalamus poly-A+ RNA represented a homeobox. The predicted amino acid sequence of the homeodomain is identical to that of the mouse OG-12 homeodomain (14) and belongs to the paired-related family of homeodomains, which includes Phox2 (21), Prx1/Mhox1 (22, 23), and Prx2/S8 (24). Hence, the rat homeobox was named rPrx3, and the mouse counterpart, OG-12, was renamed mPrx3. A rat embryonic brain cDNA library was screened with a probe corresponding to the rPrx3 homeobox, and five independent cDNA clones were obtained all sharing the homeobox and 3′-region but some differing in the 5′-ends and in a 36-nt insertion downstream of the homeobox (Δ region; Fig. 1). rPrx3 cDNAs encode three different species of rPrx3 proteins. The rPrx3A cDNA contains an ORF of 233 aa residues, starting at an ATG within an acceptable match (50%) to the Kozak consensus nucleotide sequence (25) for the initiation of protein synthesis (Fig. 1). The alternatively spliced form, rPrx3A-Δ cDNA, encodes a protein of 211 aa residues, due to a deletion of 12 aa residues. No potential translation initiation codon was found in rPrx3B cDNA. Hence, the predicted amino acid sequence is a partial fragment of rPrx3B protein with a different N terminus than rPrx3A. Similarly, three mouse mPrx3 cDNA clones were isolated by screening a 10-day mouse embryo cDNA library with an OG-12/mPrx3 cRNA probe (Fig. 1); two overlapping cDNA clones, mPrx3A and mPrx3A′, encode the same mPrx3 protein but differ in the length of the 3′-untranslated region due to alternative polyadenylation sites. A third clone, mPrx3B cDNA, differs in the 5′-end and in a 36-nt residue insertion downstream of the homeobox (Δ region; Fig. 1). Both mPrx3A′ and mPrx3B cDNAs have ORFs that encode, respectively, 333 and 190 aa residues and that start at different ATG codons. The nucleotide sequence surrounding the putative translation initiation site for mPrx3A matches the Kozak consensus sequence by 90% whereas that surrounding the first ATG codon in mPrx3B cDNA only matches by 50%. However, the crucial G/A and G residues (25), at −3 and +1, respectively, are conserved. In mPrx3B cDNA, an in-frame termination codon is found 50-nt residues upstream of the presumptive translation initiation codon. An out-of-frame, upstream ATG that also is embedded in an acceptable nucleotide environment (50% match with the Kozak consensus) also is found in mPrx3B cDNA, which leads to a truncated peptide, 47 aa residues in length. Such upstream ATG codons may be involved in regulating translation of some cDNAs (26). Thus, two species of mPrx3 proteins are predicted, differing at both the N terminus and a 12-aa residue region (Δ region) downstream of the homeodomain that is deleted in the predicted mPrx3B protein. Inclusion and exclusion of the Δ region were observed in both mouse and rat cDNAs. However, further work is required to determine whether the Δ region is involved in a function of Prx3 proteins. The nucleotide sequences of the rPrx3A and rPrx3A-Δ cDNAs are virtually identical to those of the mPrx3A and mPrx3B cDNAs, respectively. However, longer N termini are predicted for both the mPrx3A and mPrx3B proteins, compared with the rPrx3A and rPrx3A-Δ proteins. The alignment of the nucleotide sequences of rPrx3B cDNA with mPrx3 genomic DNA (Fig. 1 and ref. 14) shows almost total identity and an intervening intron, only 18 nt residues in length. The predicted splice sites match consensus sequences for intron–exon boundaries. Southern blot analyses of restriction enzyme-digested mouse genomic DNA with a mPrx3 probe that did not include the homeobox revealed a single DNA band (data not shown), indicating that a single Prx3 gene is present in the mouse genome. Hence, alternative splicing of the mPrx3 gene results in the multiple transcripts that were cloned.
Chromosomal Localization of Prx3.
Southern blot analysis of DNA from an interspecies cross using a random primed 32P-labeled DNA probe corresponding to a 440 nt-long region within intron 2 of mPrx3 (14) identified SacI DNA fragments of 2.5 kb in NFS/N and C58/J and of 5.5 kb in M. spretus. Inheritance of the variant fragment in progeny mice was compared with that of over 800 marker loci, and Prx3 was mapped to chromosome 3 with the following gene order and recombination: Anx5-16/102 (15.7 ± 3.6); Prx3-5/115 (4.3 ± 1.9); and D3Mit22-6/116 (5.2 ± 6.5).
Expression of Prx3 During Embryonic Development.
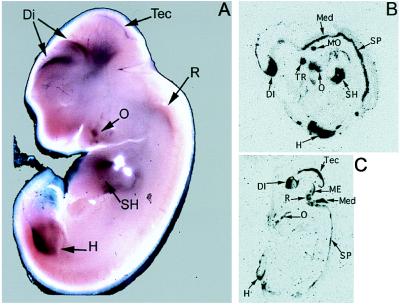
Whole mount in situ hybridization of 12-day mouse embryos with a mPrx3 cDNA probe revealed neural expression in dorsal forebrain regions, tectum, and rhombencephalon and extra-neural expression in the mesenchyme surrounding the vertebrae, hip, shoulder, and proximal parts of fore and hind limbs but not in the circular limbplates (Fig. 2A). Further detailed analyses on sections of 14.5- and 16.5-day rat embryos showed that expression in the brain was localized in the dorsal diencephalon, dorsal mesencephalon, tectum, rhombencephalon, and spinal cord, as well as in the motor trigeminal nucleus and trigeminal ganglion. Extraneural expression also was observed in tissues surrounding the oral cavity and in the hip and shoulder regions in compact structures with undefined extensions, possibly representing the developing musculature (Fig. 2 B and C).
Figure 2.
Expression of the Prx3 gene in mouse and rat embryos. In situ hybridization of Prx3 RNA with whole mount 12.5-day mouse embryo (A) and cryostat sections of rat embryos at 14.5 days (B) and 16.5 days (C). DI, diencephalon; Med, pons-medulla; H, hip region; ME, mesencephalon; MO, motor trigeminal nucleus; O, connective tissue of the oral cavity with developing teeth; R, rhombencephalon; SH, shoulder region; SP, spinal cord; Tec, tectum; TR, trigeminal ganglion.
Expression of Prx3 in Adult Rat Brain.
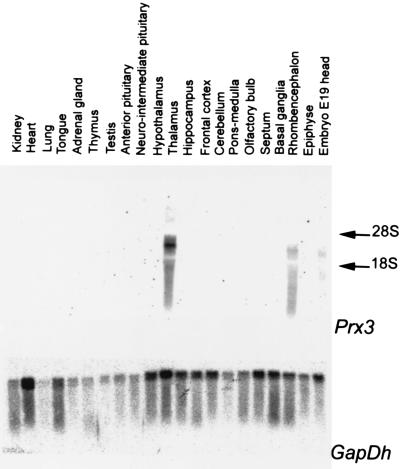
Northern analyses of total RNA from adult rat tissues and brain areas showed that Prx3 gene expression was not detected in the peripheral organs tested, including kidney, heart, lung, tongue, adrenal gland, thymus, or testis, or in the pituitary or pineal gland, but was restricted to a few regions in the brain including the thalamus and the midbrain (Fig. 3). The detected transcripts were 4–6 kb in length.
Figure 3.
Northern blot analysis of Prx3 expression in tissues and organs of the adult rat. Total RNA was analyzed by using a rPrx3 cDNA probe that hybridizes to all Prx3 transcripts detected. A glyceraldehyde phosphate dehydrogenase (GapDH) cDNA probe was used as a positive control.
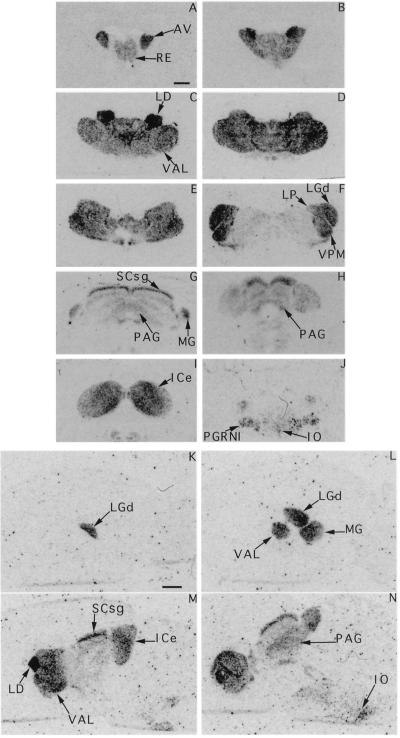
In situ hybridization of coronal and sagittal sections of adult rat brain with a rPrx3 probe common to all of the identified rPrx3 cDNAs revealed that the Prx3 gene is expressed in three defined and interconnected structures of the midbrain (Fig. 4). First, the most rostral domain of expression consists of prominent thalamic nuclei, including the ventral anterior–lateral complex, lateral dorsal, and posterior nuclei, the mediodorsal nuclei, ventral posterior–lateral, and ventral posterior–medial thalamic nuclei. High expression also was detected in the dorsal lateral geniculate complex and ventral and dorsal geniculate complexes. Second, Prx3 expression was found in the dorsal periaquaductal gray and in multiple layers of the superior colliculus with different levels of abundance. Highest expression was observed in the superficial gray layer. Third, all nuclei of the inferior colliculus showed high levels of Prx3 expression. Finally, Prx3 was expressed in the ventral hindbrain, including parts of the pontine reticular formation and the inferior olive (Fig. 4).
Figure 4.
Expression of the Prx3 gene in adult rat brain. 35S-labeled anti-sense RNA probe derived from the common region of all described Prx3 transcripts was used for in situ hybridization. (A–J) Consecutive coronal sections, from rostral to caudal. (K–N) Saggital sections, from lateral to medial. Specific signals are observed in thalamic nuclei, medial geniculate nucleus (MG), anterior–ventral (AV), reticulate nucleus (RE), the ventral anterior–lateral complex (VAL), lateral dorsal (LD), ventral posterior–medial thalamic nuclei (VPM), the dorsal lateral geniculate complex (LGd), and the dorsal periaquaductal gray (PAG) and in the superficial gray layer of the superior colliculus (SCsg); Prx3 also was expressed in the ventral hindbrain, including parts of the pontine reticular formation (PGRNI) and the inferior olive (IO).
DISCUSSION
In this report, cDNAs are described of alternatively spliced RNA transcripts of a rat homeobox gene, rPrx3, and the mouse homologue mPrx3, previously termed OG-12 (14). The mouse and rat genes display a high degree of conservation, both within and outside the homeobox, including conserved exon–intron junction sites.
Recently, a human homeobox gene, SHOX, that is mutated in individuals with idiopathic short stature and in Turner syndrome patients with the short stature phenotype was identified in the pseudoautosomal region of the human X chromosome (27). SHOX has a high degree of sequence homology to mPrx3, having an identical homeodomain and an 87% homology of the amino acid sequence of the entire protein. Hence, SHOX and mPrx3 may be human and mouse counterparts of the same gene. However, some molecular differences are present. First, the SHOX gene is expressed in two splice variants: One is closely related to the Prx3A cDNA and the other, which is expressed in low abundance and is restricted to fetal kidney, bone marrow, fibroblasts, and skeletal muscle, uses an alternative 3′-exon that has not been detected in the mouse or rat. Furthermore, the Δ-region of Prx3 was not detected in any of the human DNA libraries tested (27), suggesting that the SHOX gene or its transcripts lack the Δ-region. Southern analyses did not provide evidence for another highly homologous gene in the mouse. It would be interesting to determine whether multiple SHOX genes are present in the human genome.
The mouse Prx3 gene was mapped to mouse chromosome 3 whereas the SHOX gene resides on the human X chromosome. Mouse orthologs of several genes that map in or near the pseudoautosomal region of the human X chromosome have been mapped to mouse autosomes. However, mPrx3 is the only ortholog of a gene on the human X chromosome that has been found thus far on mouse chromosome 3. Ptx-2, a bicoid related homeobox gene (refs. 28 and 29 and M.P.S., unpublished work) also resides on mouse chromosome 3 (30).
Like mPrx3 (14), SHOX is expressed in skeletal muscle, brain, heart, and other tissues. Relatively abundant Prx3 mRNA expression was observed by in situ hybridization in limbs of both mouse and rat embryos and in mesenchyme surrounding the vertebrae of mice embryos. It is tempting to speculate that expression of mPrx3 in skeletal muscle and limb paravertebral mesenchyme might be related to skeletal development and length of the organism. Although expression data indicate that both Prx3 and SHOX are expressed in fetal and adult brain, no neural phenotype was reported for the individuals with a mutated SHOX gene (27). The mutation detected in the individuals with the idiopathic short stature phenotype and some family members is expected to result in a truncated SHOX protein that still has an intact N terminus, a homeodomain, and part of the C terminus. Data from rodents and humans demonstrate the presence of multiple species of mRNA, and hence, different protein isoforms are expected to be synthesized through tissue-specific alternative splicing. The effects of mutations at other positions of the SHOX gene have not been described.
The amino acid sequence of the Prx3 homeodomain is most similar to the rodent Phox2 homeodomain (21), and Pax3 (8) and Pax7 (31) are close relatives. A 14-aa residue region near the C terminus of Prx3 is present that is conserved in a limited number of homeodomain proteins, including Prx1, Prx2, and Phox2, but also other homeodomain proteins not directly related through homeodomain homology. This region was noted first in Otp (32), and more recently was described in solurshin, the protein encoded by the human RIEG homeobox gene (28) that represents the human homolog of Ptx2 in Rax/Rx (33, 34), a homeodomain protein involved in eye and brain development, and in Arx (35). It has therefore been addressed by different names in the literature. The only known invertebrate homeodomain protein that has this motif is the Drosophila aristaless protein. We therefore named this domain the “aristaless domain.” Homeodomain proteins containing the aristaless domain are generally expressed at high levels in craniofacial, eye, and/or brain regions (28), which also applies to Prx3. Of interest, a splice variant of Prx1 has been reported that lacks the aristaless domain (22, 23). Both the Chou–Fasman and the Garnier–Osguthorpe–Robson algorithms predict an α-helical conformation for the aristaless domain of Prx3. The conservation of amino acid sequence of the aristaless domain suggests that this domain may function as a site for molecular interaction.
The Prx3 gene was shown to be expressed in both embryonic and adult brains. Although the embryonic rodent brain expresses Prx3 in broad areas that develop into the dorsal thalamus, pretectum, and tectum, this pattern of expression becomes restricted in the adult animal. Most notably, Prx3 expression is present in nuclei that are part of the subcortical visual system. These include nuclei that receive and relay axonal input from the retina, e.g., the thalamic dorsal lateral geniculate complex and the superior colliculus (35). Prx3 is also expressed in thalamic nuclei, such as the mediodorsal nucleus and lateral dorsal nucleus, that are involved in integration of visual information with other sensory inputs, such as auditory and gustatory inputs (36). Relay nuclei along the auditory pathway, including the olivary complex in the ventral hindbrain, the medial geniculate complex in the thalamus, and the inferior colliculus, express Prx3 (36). Other thalamic nuclei expressing Prx3, i.e., the ventral posterior lateral and ventral posterior medial nuclei, play roles in conveying and integrating somatic sensory information from body and face. Thus, the expression pattern of Prx3 is consistent with functional units in the brain that coordinate and integrate sensory information being sent to the sensory cortex. However, Prx3 also is expressed in relay stations, such as the pontine reticular formation, along pathways that are important in the control of posture, via integration of sensory information with descending inputs from the motor cortex. Thalamic motor nuclei, such as the anterior ventral nucleus, which receives fibers from the globus pallidus and sends fibers to the motor cortex and the anterior ventral lateral nucleus, which represents a relay station of the cerebello-cortical pathway, also express Prx3.
Homeobox genes have been implicated in the integration of neuronal cell groups into functional circuitry during development and maintenance of phenotype and function in the adult brain. For example, Phox2 is required for the expression of an aminergic neurotransmitter phenotype in peripheral ganglia and brain (37, 38) and may also affect axonal pathfinding (39). The role of Prx3 in the formation and maintenance of specific brain nuclei that work in concert to transfer and process sensory and motor information deserves further investigation. Finally, expression data also suggest that Prx3 may play a role in the development of nonneuronal structures, particularly in the developing appendages. The relationship of this finding with the observation that a putative human homologue of Prx3, SHOX, is mutated in individuals with idiopathic short stature warrants further investigation.
Acknowledgments
We are grateful to Drs. Jim Boulter for the gift of rat cDNA libraries, Jacqueline Deschamps for help with the interpretation of embryonic expression patterns, and Frits Meijlink for providing the idea for the nomenclature of the “aristaless domain.” We thank Joke Cox-van Put for providing expertise on in situ hybridization. This work was supported by grants of Netherlands Organization for Scientific Research-MW (nr 900-546-109) and the Korczak Foundation for Autism and Related Disorders. rPrx3 cDNA cloning and characterization was performed at the RMI, Utrecht, the Netherlands; mPrx3 cloning and characterization was performed at the National Institutes of Health, Bethesda, MD.
ABBREVIATIONS
- RT-PCR
reverse transcription-PCR
- rPrx3
rat paired-related homeobox 3
- mPrx3
mouse paired-related homeobox 3 formerly termed OG-12
Footnotes
References
- 1.Krumlauf R. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 2.De Robertis E M. In: The Homeobox in Cell Differentiation and Evolution. Duboule D, editor. Oxford: Oxford Univ. Press; 1994. pp. 11–23. [Google Scholar]
- 3.Lumsden A, Krumlauf R. Science. 1996;274:1109–1115. doi: 10.1126/science.274.5290.1109. [DOI] [PubMed] [Google Scholar]
- 4.Tanabe Y, Jessell T M. Science. 1996;274:1115–1123. doi: 10.1126/science.274.5290.1115. [DOI] [PubMed] [Google Scholar]
- 5.Alvarez-Bolado G, Rosenfeld M G, Swanson L W. J Comp Neurol. 1995;355:237–295. doi: 10.1002/cne.903550207. [DOI] [PubMed] [Google Scholar]
- 6.Rosenfeld M G, Bach I, Erkman L, Li P, Lin C, Lin S, McEvilly R, Ryan A, Rhodes S, Schonnemann M D, Scully K. Recent Prog Horm Res. 1996;51:217–238. [PubMed] [Google Scholar]
- 7.Lamonerie T, Tremblay J J, Lanctot C H, Therrien M, Gauthier Y, Drouin J. Genes Dev. 1996;10:1284–1295. doi: 10.1101/gad.10.10.1284. [DOI] [PubMed] [Google Scholar]
- 8.Stoykova A, Gruss P. J Neurosci. 1994;14:1395–1412. doi: 10.1523/JNEUROSCI.14-03-01395.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thor S, Ericson J, Bannström T, Edlund T. Neuron. 1991;7:881–889. doi: 10.1016/0896-6273(91)90334-v. [DOI] [PubMed] [Google Scholar]
- 10.Puelles L, Rubenstein L R. Trends Neurosci. 1993;16:472–479. doi: 10.1016/0166-2236(93)90080-6. [DOI] [PubMed] [Google Scholar]
- 11.Figdor M C, Stern C D. Nature (London) 1993;363:630–634. doi: 10.1038/363630a0. [DOI] [PubMed] [Google Scholar]
- 12.Shimamura K, Hartigan D, Martinez S, Puelles L, Rubinstein J. Development. 1995;121:3923–3933. doi: 10.1242/dev.121.12.3923. [DOI] [PubMed] [Google Scholar]
- 13.Nakai S, Kawano H, Yudate T, Nishi M, Kuno J, Nagata A, Jishage K, Hamada H, Fujii H, Kawamura K, Shiba K, Noda T. Genes Dev. 1995;9:3109–3121. doi: 10.1101/gad.9.24.3109. [DOI] [PubMed] [Google Scholar]
- 14.Rovescalli A C, Asoh S, Nirenberg M. Proc Natl Acad Sci USA. 1996;93:10691–10696. doi: 10.1073/pnas.93.20.10691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adamson M C, Silver J, Kozak C A. Virology. 1991;183:778–781. doi: 10.1016/0042-6822(91)91010-e. [DOI] [PubMed] [Google Scholar]
- 16.Dietrich W, Katz H, Lincoln S E, Shin H-S, Friedman J, Dracopoli N C, Lander E S. Genetics. 1992;131:423–447. doi: 10.1093/genetics/131.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez-Garcia M I, Kozak C A, Morgan R O, Fernandez M P. Genomics. 1996;31:151–157. doi: 10.1006/geno.1996.0026. [DOI] [PubMed] [Google Scholar]
- 18.Church G M, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopes da Silva S, Horssen A M, van Chang C, Burbach J P H. Endocrinology. 1995;136:2276–2283. doi: 10.1210/endo.136.5.7720676. [DOI] [PubMed] [Google Scholar]
- 20.Wilkinson D G. In: In Situ Hybridization: A Practical Approach. Wilkinson D G, editor. New York: IRL; 1992. [Google Scholar]
- 21.Valarche I, Tissier-Seta J P, Hirsch M R, Martinez S, Goridis C, Brunet J F. Development. 1993;119:881–896. doi: 10.1242/dev.119.3.881. [DOI] [PubMed] [Google Scholar]
- 22.Kern M J, Witte D P, Valerius M T, Aronow B J, Potter S S. Nucleic Acids Res. 1992;20:5189–5195. doi: 10.1093/nar/20.19.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson D, Sheng G, Lecuit T, Dostatni N, Deplan C. Genes Dev. 1993;7:2120–2134. doi: 10.1101/gad.7.11.2120. [DOI] [PubMed] [Google Scholar]
- 24.Kongsuwan K, Webb E, Housiaux P, Adams J M. EMBO J. 1988;7:2131–2138. doi: 10.1002/j.1460-2075.1988.tb03052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozak M. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geballe A P, Morris D R. Trends Biochem Sci. 1994;19:159–164. doi: 10.1016/0968-0004(94)90277-1. [DOI] [PubMed] [Google Scholar]
- 27.Rao E, Weiss B, Fukami M, Rump A, Niesler B, Mertz A, Muroya K, Binder G, Kirsch S, Winkelmann M, Nordsiek G, Heinrich U, Breuning M H, Ranke M B, Rosenthal A, Ogata T, Rappold G A. Nat Genet. 1997;16:54–63. doi: 10.1038/ng0597-54. [DOI] [PubMed] [Google Scholar]
- 28.Semina E V, Reiter R, Leysens N J, Alward W L M, Small K W, Datson N A, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel B U, Carey J C, Murray J C. Nat Genet. 1996;14:392–299. doi: 10.1038/ng1296-392. [DOI] [PubMed] [Google Scholar]
- 29.Mucchielli M-L, Martinez S, Pattyn A, Goridis C, Brunet J-F. Mol Cell Neurosci. 1997;8:258–271. doi: 10.1006/mcne.1996.0062. [DOI] [PubMed] [Google Scholar]
- 30.Gage P J, Camper S A. Hum Mol Genet. 1997;6:457–464. doi: 10.1093/hmg/6.3.457. [DOI] [PubMed] [Google Scholar]
- 31.Simeone A, Acampora D, Gulisano M, Stornaiuolo A, Boncinelli E. Nature (London) 1992;358:687–690. doi: 10.1038/358687a0. [DOI] [PubMed] [Google Scholar]
- 32.Simeone A, D’Apie M R, Nigro V, Casanova J, Grazani F, Acampora D, Avantaggiato V. Neuron. 1994;13:83–101. doi: 10.1016/0896-6273(94)90461-8. [DOI] [PubMed] [Google Scholar]
- 33.Furukawa T, Kozak C A, Cepko C L. Proc Natl Acad Sci USA. 1997;94:3088–3093. doi: 10.1073/pnas.94.7.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathers P H, Grinberg A, Mahon K A, Jamrich M. Nature (London) 1997;387:603–607. doi: 10.1038/42475. [DOI] [PubMed] [Google Scholar]
- 35.Miura H, Yanazawa M, Kato K, Kitamura K. Mech Dev. 1997;65:99–109. doi: 10.1016/s0925-4773(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 36.Paxinos G. The Rat Nervous System. 2nd Ed. San Diego: Academic; 1995. [Google Scholar]
- 37.Zellmer E, Zhang Z, Greco D, Rhodes J, Cassel S, Lewis E J. J Neurosci. 1995;15:8109–8120. doi: 10.1523/JNEUROSCI.15-12-08109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morin X, Cremer H, Hirsch M-R, Kapur R P, Goridis C, Brunet J-F. Neuron. 1997;18:411–423. doi: 10.1016/s0896-6273(00)81242-8. [DOI] [PubMed] [Google Scholar]
- 39.Groves A K, George K M, Tissier-Seta J P, Engel J D, Brunet J F, Anderson D J. Genes Dev. 1995;12:887–901. doi: 10.1242/dev.121.3.887. [DOI] [PubMed] [Google Scholar]