Abstract
Drosophila Numb is a membrane associated protein of 557 amino acids (aa) that localizes asymmetrically into a cortical crescent in mitotic neural precursor cells and segregates into one of the daughter cells, where it is required for correct cell fate specification. We demonstrate here that asymmetric localization but not membrane localization of Numb in Drosophila embryos is inhibited by latrunculin A, an inhibitor of actin assembly. We also show that deletion of either the first 41 aa or aa 41–118 of Numb eliminates both localization to the cell membrane and asymmetric localization during mitosis, whereas C-terminal deletions or deletions of central portions of Numb do not affect its subcellular localization. Fusion of the first 76 or the first 119 aa of Numb to β-galactosidase results in a fusion protein that localizes to the cell membrane, but fails to localize asymmetrically during mitosis. In contrast, a fusion protein containing the first 227 aa of Numb and β-galactosidase localizes asymmetrically during mitosis and segregates into the same daughter cell as the endogenous Numb protein, demonstrating that the first 227 aa of the Numb protein are sufficient for asymmetric localization.
Asymmetric cell divisions in which a mother cell segregates a determinant into one of its daughter cells to make this cell different from its sister cell contribute to generating different cell types in a multicellular organism (1, 2). In Drosophila, the four cells that form an external sensory (ES) organ–one type of sensory structure found in the peripheral nervous system–arise during development from a single sensory organ precursor (SOP) cell that undergoes a series of asymmetric cell divisions (3–5). During these asymmetric divisions, the protein Numb acts as a segregating determinant. In wild-type animals, the SOP cell divides into a IIA cell and a IIB cell. The IIA cell gives rise to the two outer cells (hair and socket cell) whereas the IIB cell forms the two inner cells (neuron and sheath cell). In a numb mutant, the SOP cell divides into two IIA cells that give rise to four outer cells and no inner cells (6). Conversely, when the numb gene is overexpressed, the SOP cell divides into two IIB cells that form four inner cells and no outer cells (7). numb is required for correct cell fate specification not only during SOP cell division, but also during the division of the IIA and IIB cells (6–8). numb encodes a membrane associated protein that contains an N-terminal phosphotyrosine binding domain (9). In interphase cells, the protein is uniformly distributed around the circumference of the cell. In mitotic SOP cells, however, Numb concentrates in the membrane area that overlies one of the two centrosomes and preferentially enters one of the two daughter cells (7, 10). Several experiments indicate that Numb acts by inhibiting signaling via the Notch transmembrane receptor in this daughter cell (11–13).
Asymmetric localization of Numb is not restricted to SOP cells. During stage 9 of Drosophila embryonic development, epithelial cells in the procephalic neurogenic region of the ectoderm divide perpendicularly to the epithelial surface (14) and each give rise to a large apical daughter cell and a smaller basal daughter cell. Numb is preferentially segregated into the basal daughter cell during this division (15), but the function of Numb in this case has not been characterized. Neuroblasts, the progenitor cells of the central nervous system, each divide into a large apical daughter cell that retains neuroblast characteristics and a smaller basal ganglion mother cell (GMC) that later divides into two neurons (16). Numb is segregated into the GMC during neuroblast division. In the absence of Numb, however, GMCs are not transformed into additional neuroblasts, even though defects in some central nervous system lineages are observed in numb mutants (17).
Like Numb, the Drosophila protein Prospero localizes asymmetrically in dividing neuroblasts and SOP cells. Prospero is a nuclear transcription factor, which is required for correct cell fate specification in the central nervous system and for neuronal differentiation in the peripheral nervous system (18, 19). During mitosis, the protein transiently translocates to the cell membrane and colocalizes with Numb (10, 20, 21). Numb and Prospero enter the same daughter cell, where Numb stays at the membrane whereas Prospero enters the nucleus. Numb and Prospero localization are independent events (10, 21), but localization of both proteins requires the protein Inscuteable. Inscuteable also localizes asymmetrically in dividing neural precursor cells, but it is localized to the side opposite to that of Numb and Prospero localization prior to Numb and Prospero asymmetric localization (15, 22). In inscuteable mutants, Numb and Prospero either do not localize asymmetrically or form crescents at random positions around the cell (15). Inscuteable is also involved in the orientation of the mitotic spindle. In inscuteable mutants, the mitotic spindle in neuroblasts is oriented randomly, and ectopic expression of inscuteable in epithelial cells causes a reorientation of their mitotic spindle (15). Thus, Inscuteable directs and coordinates several processes during asymmetric cell division.
Localization of Numb, Prospero, and Inscuteable does not require microtubules, but disruption of actin filaments causes defects in asymmetric localization (10, 15). After treatment with cytochalasin D, Inscuteable is no longer asymmetrically localized (15). Numb and Prospero still localize asymmetrically, but their crescents are frequently misoriented (10). We show here that treatment with latrunculin A, a more potent inhibitor of actin polymerization, completely abolishes Numb and Prospero asymmetric localization, suggesting that crescent formation requires actin filaments. We also demonstrate that a small region at the N terminus of the Numb protein is required for both membrane association and asymmetric localization. When fused to β-gal, this region is sufficient for membrane localization, but not for asymmetric localization. In contrast, a β-gal fusion protein containing a much larger N-terminal fragment that includes the PTB domain is asymmetrically localized in the dividing precursor cell and then segregated into one of the two daughter cells.
METHODS
numb Deletion and numb-lacZ Fusion Constructs.
The plasmid pSKnb-myc and the deletion constructs numb-Δ1, numb-Δ2, numb-Δ3, numb-Δ4, and numb-ΔPTB have been described before (11). For RNA injection, a vector was used that contains an SP6 promoter and β-globin 5′- and 3′-untranslated sequences (described in ref. 23, will be called pXBG below). For cloning into this vector, an NcoI site was introduced at the translation initiation site of pSKnb-myc by PCR, yielding pSKnb-myc-NcoI. The deletion constructs were cloned into pSKnb-myc-NcoI using unique restriction sites flanking the deletions. The complete ORF was then inserted into pXBG using NcoI and XbaI generating pXBG-nb-myc-NcoI and derivatives.
numb-Δmyr was generated in a PCR reaction that changed the second codon of the numb coding region from GGA to GCC and introduced an NcoI site at the translation start. numb-ΔN was generated in a PCR reaction that introduced an NcoI site at the second ATG codon (nucleotides 915–917 of the published cDNA, ref. 6). The PCR fragments were cloned into pSKnb-myc-NcoI using NcoI and SphI and the resulting cDNAs were inserted into pXBG using NcoI and XbaI.
numb-04-lacZ was generated by replacing a BamHI/XbaI fragment of pXBG-nb-myc-NcoI containing the Numb C terminus (from nucleotide 1472) with a XmaI/SpeI fragment containing the lacZ gene from pPD8.
02 (24). The other numb-lacZ fusions were generated by PCR from pSK-numb-myc-NcoI using primers that introduced BamHI sites after nucleotides 914 (numb-01-lacZ), 1019 (numb-02-lacZ), or 1145 (numb-03-lacZ). PCR fragments were cloned into numb-04-lacZ via NcoI/BamHI.
To generate transgenic flies, numb-ΔN was cloned into pUAST (25) using EcoRI/XbaI. The numb-lacZ fusion constructs were cloned into pSKnumb-myc-NcoI via NcoI/StuI, excised with NotI and cloned into pUAST. Transgenic flies were generated using a w; Oregon-R stock following standard protocols.
RNA Injection Experiments.
Capped RNA was generated from the various pXBG-constructs using the SP6 Message-Machine kit (Ambion) and injected into stage 4 Drosophila embryos. The embryos were aged to stage 9 at 18°C, transferred to test tubes with heptane, fixed, and stained for immunofluorescence.
Immunofluorescence and Confocal Microscopy.
Drosophila embryos were fixed and processed for immunofluorescence essentially as described before (7), except that 5% paraformaldehyde was used as fixative and 2% normal donkey serum was used as blocking reagent. Rabbit anti-Numb (7), rabbit anti-Prospero (19), and rabbit anti-β-gal (Cappel) were used as primary antibodies, DTAF donkey-anti-rabbit (The Jackson Laboratory) was used as secondary antibody, and propidium iodide (26) was used to stain DNA. Embryos were mounted in Slow Fade mounting medium (Molecular Probes) and analyzed on a Bio-Rad MRC 600 confocal microscope.
Drug Treatment Experiments.
For treatment with latrunculin A, Drosophila embryos at the desired stage were dechorionated for 3 min with 50% household bleach and transferred to test tubes in embryo wash buffer (0.7% NaCl/0.07% Triton-X100). They were overlayed with a 1:1 mixture of n-heptane and Drosophila Schneider’s medium (GIBCO) containing latrunculin A (Molecular Probes) dissolved in dimethyl sulfide or just dimethyl sulfide as a control. After incubation with gentle shaking at room temperature for the desired time, the Schneider’s medium was replaced with 5% paraformaldehyde and the embryos were fixed for 20 min. Fifty percent of the embryos were devitellinized with methanol and used for antibody staining according to standard protocols (7). The rest of the embryos were devitellinized by hand and stained with rhodamine-phalloidin (Molecular Probes, 1:50) for 15 min.
RESULTS
Latrunculin A Disrupts Asymmetric Localization of Numb and Prospero.
Disrupting the actin cytoskeleton with cytochalasin D does not abolish the asymmetric localization of Numb and Prospero (10) even in cells where the drug treatment leads to a defect in cytokinesis. Recent experiments have shown that the drug latrunculin A is more potent than cytochalasin D in disrupting the actin cytoskeleton (27, 28). We have tested the effect of latrunculin A on the asymmetric localization of Numb and Prospero (Fig. 1). Rhodamine-phalloidin staining reveals that treatment of Drosophila embryos with 200 μM latrunculin A for 20 min results in almost complete depolymerization of F-actin (Fig. 1B), though some very faint residual staining at the cell cortex can still be detected at higher amplification using the confocal microscope (data not shown). Both Numb and Prospero are no longer asymmetrically localized but remain membrane associated in embryos treated with latrunculin A (Fig. 1 D and F), whereas their localization is not affected in embryos processed in parallel but not exposed to the drug (Fig. 1 C and E). Even when the drug concentration is increased to 400 μM and the treatment extended to 40 min, Numb and Prospero remain associated with the cell membrane (data not shown). We conclude that the asymmetric localization of Numb and Prospero is dependent on actin filaments.
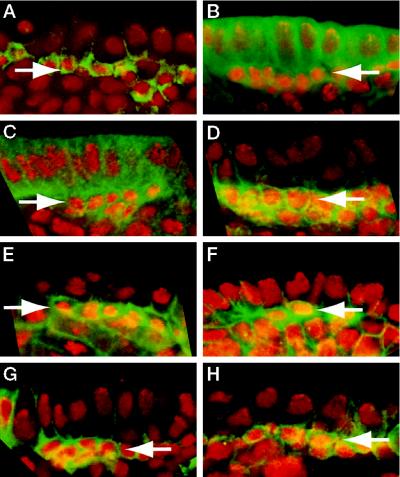
Figure 1.
Latrunculin A inhibits asymmetric localization of Numb and Prospero. Drosophila embryos were permeabilized and incubated for 20 min in Schneider’s medium with (B, D, and F) or without (A, C, and E) 200 μM latrunculin A. (A and B) Surface views of embryos stained with rhodamine-phalloidin show that filamentous actin is almost completely absent after this treatment (B), while actin fibers are unaffected in the controls (A). The images in A and B were taken with the same settings on the confocal microscope and processed identically. (C–F) Optical cross sections of embryos stained for DNA (red) and Numb (green, C and D) or Prospero (green, E and F). Two mitotic neuroblasts from different embryos are shown in each panel, apical is up and basal is down. Numb and Prospero crescents are marked by arrowheads. In control embryos, Numb and Prospero are membrane associated and asymmetrically localized (C and E). After latrunculin A treatment, Numb and Prospero are still localized to the cell membrane, but fail to localize asymmetrically (D and F). Note that the mitotic spindle frequently was not correctly oriented along the apical-basal axis after latrunculin A treatment (asterisk in F). Arrows point to the nuclei of GMCs that contain high concentrations of Prospero and appear yellow because they stain both with the anti-Prospero antibody and propidium iodide. These GMCs were generated in cell divisions that presumably occurred before the latrunculin treatment and consequently exhibited normal segregation of Prospero.
The Numb N Terminus, but Not the N-Myristoylation Signal, Is Required for Asymmetric Localization.
To identify domains in the Numb protein that are essential for membrane localization and asymmetric localization, we generated a series of deletion constructs (Fig. 2) and analyzed the subcellular localization of these mutant Numb proteins in embryos injected with RNA generated from these deletion constructs. This analysis is limited to early stages of Drosophila development, because the mRNA has to be injected before cellularization and will be degraded with time. We therefore analyzed the asymmetric localization of the modified Numb proteins in the cells of the procephalic neurogenic region of the ectoderm, the first embryonic cells that localize Numb asymmetrically. Stage 4 embryos were injected with capped mRNA, then aged to stage 9 of development, fixed and analyzed by immunofluorescence using an anti-Numb antibody (7) (Fig. 3). The tagged wild-type Numb-myc and the mutant proteins with C-terminal or central deletions (Numb-ΔPTB, Numb-Δ2, Numb-Δ3, and Numb-Δ4) exhibited the same distribution as the endogenous Numb protein (Fig. 3 A and D–G). The proteins were found at the cell membrane in all cell types, and in the cells of the procephalic neurogenic region, they segregated into the basal daughter cell during mitosis. In contrast, the two most N-terminal deletions (Numb-ΔN, Numb-Δ1) caused reproducible abnormalities in localization (Fig. 3 B and C). In all cell types, the proteins with N-terminal deletions were found mostly in the cytoplasm and failed to segregate preferentially into one of the two daughter cells during mitosis. We conclude that the N terminus of the Numb protein is required for both asymmetric localization and localization to the cell membrane.
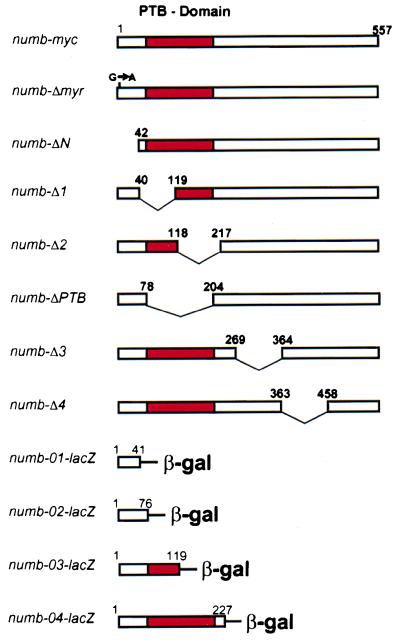
Figure 2.
Map of the Numb deletion and Numb-β-gal fusion constructs used in this study. The numbers refer to aa positions in the published sequence (6).
Figure 3.
Asymmetric segregation of Numb deletion constructs in RNA injection experiments. Stage 4 Drosophila embryos were injected with capped mRNA generated from Numb deletion constructs, aged to stage 9, then fixed and stained with an anti-Numb antibody (green) and propidium-iodide (DNA, red). Optical cross sections (apical up, basal down) through the procephalic neurogenic region of stage 9 embryos are shown. Numb-myc (A), Numb-Δ2 (D), Numb-Δ3 (E), Numb-Δ4 (F), Numb-ΔPTB (G), and Numb-Δmyr (H) are localized to the cell membrane and segregate normally into the basal daughter cell (arrow), whereas Numb-ΔN (B) and Numb-Δ1 (C) are localized in the cytoplasm and segregate equally into both daughter cells.
The N terminus of the Numb protein contains a consensus site for N-myristoylation (6). To test whether this site is required for localizing the Numb protein to the cell membrane, we substituted the glycine at position 2 of the Numb protein with alanine, a mutation that has been shown to completely abolish N-myristoylation in other proteins (29). No defects in membrane association and asymmetric localization were found when mRNA from this mutant construct (numb-Δmyr) was injected into embryos (Fig. 3H). Whether Numb is N-myristoylated in vivo is not known. If N-myristoylation of Numb does take place, it does not appear to be required for association with the cell membrane or for asymmetric localization (see Discussion).
Different Requirements for Asymmetric Localization and Downstream Function of Numb in Overexpression Experiments.
The overexpression of Numb in SOP cells leads to a cell fate transformation of the IIA cell into a second IIB cell, resulting in ES organs with four inner cells and no outer cells (7). Occasionally, only the second cell division is affected, resulting in ES organs with two hairs, but no socket. To identify the regions of Numb necessary for this overexpression function, we generated transgenic flies carrying several of the Numb deletion constructs. The results obtained for Numb-Δ2 and Numb-ΔPTB have been described (11). Consistent with our RNA injection results, no defects in asymmetric localization and membrane localization were observed when these constructs were expressed in mitotic neuroblasts (11). The same results were obtained when the localization of Numb-Δ2 and Numb-ΔPTB was analyzed in numb1 protein null-mutant embryos (data not shown). When overexpressed in SOP cells of adult ES organs, however, both proteins were completely nonfunctional (11).
Transgenic flies were also generated for numb-ΔN and the UAS/GAL4 system was used to express numb-ΔN under the control of the hairy promoter (25) in both epidermal cells and neuroblasts of Drosophila embryos. In agreement with our RNA injection results, the Numb-ΔN protein was mostly cytoplasmic in both epidermal cells and neuroblasts and no signs of asymmetric segregation were detected in dividing neuroblasts (data not shown). However, when the numb-ΔN transgene was expressed under the control of the GAL4-line GAL4109–68 in SOP cells of the adult ES organs, it caused the same overexpression phenotype as wild-type Numb (Fig. 4 B and C). Many of the sensory bristles were not visible (see arrowheads in Fig. 4B), suggesting that both external cells of the ES organs had been transformed into inner cells. Occasionally, ES organs with two hairs and no socket were formed (Fig. 4C), indicating a cell fate transformation in the second division. Thus, whereas the N terminus of Numb is required for asymmetric localization in the precursor cell, it is not necessary for the Numb protein to specify the fate of Numb-containing daughter cells.
Figure 4.
numb-ΔN causes cell fate transformations when overexpressed in SOP cells. (A) Notum of a control fly raised at 25°C. Arrowheads mark the posterior scutellar bristles. (B) Notum of a fly raised at 25°C carrying the UAS-numb-ΔN transgene and the GAL4109–68 transgene, which cause overexpression of numb-ΔN in SOP cells of adult sensory organs (11). Most macrochaetae (arrowheads mark the position of the posterior scutellars) and many microchaetae are missing. A weakly expressing line was used, because flies carrying GAL4109–68 and a strongly expressing UAS-numb-ΔN transgene were not viable. (C) High magnification view of the area boxed in B showing a duplicated microchaete bristle (arrowheads) without socket (arrow). This phenotype is caused by a cell fate transformation between the daughter cells of the IIA cell and is characteristic for Numb overexpression.
The N-Terminal Half of the Numb Protein Is Sufficient for Asymmetric Localization.
Asymmetric localization and membrane localization of Numb require the N terminus. To test whether the N terminus of the Numb protein is sufficient for asymmetric localization, we generated constructs encoding N-terminal fragments of Numb fused with the Escherichia coli β-gal protein (Fig. 2). numb-01-lacZ contains the first 41 aa, numb-02-lacZ the first 76 aa, numb-03-lacZ the first 119 aa, and numb-04-lacZ the first 227 aa of Numb. We generated transgenic flies carrying these fusion constructs and used the UAS/GAL4 system to express the constructs under the control of the scabrous promoter (25). The subcellular localization of the fusion proteins was analyzed in neuroblasts of stage 10 embryos by immunofluorescence using an anti-β-gal antibody and propidium iodide to stain DNA (Fig. 5). Numb-01-β-gal was found in the cytoplasm with no signs of asymmetric localization during mitosis (Fig. 5A). Numb-02-β-gal and Numb-03-β-gal were localized to the cell membrane, but failed to localize asymmetrically in mitotic neuroblasts. The proteins remained uniformly distributed around the cell cortex instead and segregated into both daughter cells (Fig. 5 B and C). In contrast, Numb-04-β-gal was localized to the cell membrane in interphase cells, became localized asymmetrically in mitotic neuroblasts and then segregated into the ganglion mother cell (Fig. 5 D). The same results were obtained when the localization of the Numb-β-gal fusion proteins was analyzed in numb1 protein null mutant embryos (data not shown). We conclude that the first 76 aa of Numb are sufficient to direct membrane localization but not asymmetric localization of β-gal, while the first 227 aa of Numb can direct asymmetric localization and segregation of β-gal preferentially into one of the two daughter cells.
Figure 5.
Subcellular localization of Numb-β-gal fusion proteins expressed in transgenic Drosophila embryos. Mating of flies carrying the transgenes UAS-numb-01-lacZ (A), UAS-numb-02-lacZ (B), UAS-numb-03-lacZ (C), or UAS-numb-04-lacZ (D) with flies carrying the sca-GAL4 transgene (25) generated embryos carrying both transgenes that were identified by their β-gal expression. Optical cross sections of 4–6 hr old embryos stained with anti-β-gal (green) and propidium-iodide (DNA, red) are shown. Each panel shows a mitotic neuroblast (Left, arrowhead marks the basal cell cortex) and a neuroblast/GMC pair just after cell division (Right, arrow marks the GMC). (A) Numb-01-β-gal is cytoplasmic and does not preferentially segregate into the GMC. (B and C) Numb-02-β-gal (B) and Numb-03-β-gal (C) are localized to the cell membrane but do not localize asymmetrically during mitosis and consequently segregate into both daughter cells. (D) Numb-04-β-gal first localizes to the cell membrane in interphase cells (not shown), then forms a basal cortical crescent during mitosis (arrowhead) and preferentially segregates into the GMC after cell division (arrow).
DISCUSSION
During mitosis of Drosophila neural precursor cells, the proteins Numb and Prospero localize asymmetrically and segregate into one of the two daughter cells. We show here that asymmetric localization of Numb and Prospero, but not localization to the cell membrane, can be eliminated by the drug latrunculin A that disrupts the actin cytoskeleton. Furthermore, we demonstrate that the first 119 aa of Numb are essential for asymmetric localization and for localization to the cell membrane, while most of the central and C-terminal portions of Numb are not required. When the first 119 or the first 76 aa of Numb are fused to β-gal, however, they can direct localization to the cell membrane but not asymmetric localization. In contrast, a larger N-terminal fragment containing the first 227 aa, which include the PTB domain, is sufficient for asymmetric localization and segregation into one daughter cell.
Numb and Prospero Localization Are Actin Dependent Processes.
In many organisms, asymmetric segregation of determinants during mitosis is thought to be mediated by actin-dependent mechanisms. In Caenorhabditis elegans, the first cell division of the zygote is asymmetric and establishment of asymmetry during this division requires actin but not microtubules (30, 31). In Saccharomyces cerevisiae, ASH1 mRNA segregates into one of the two daughter cells during mitosis (32, 33), where it is translated to generate the transcriptional repressor Ash1p (34, 35). The process of Ash1 segregation requires a functional actin cytoskeleton, but not microtubules (32).
In Drosophila, the asymmetric segregation of Numb and Prospero during mitosis has been shown to be microtubule independent (10) and experiments using the actin drug cytochalasin D have suggested that actin is also not required for Numb and Prospero localization. After cytochalasin D treatment, Numb and Prospero are still asymmetrically localized in cells that have defects in other actin dependent processes, such as cytokinesis (10). However, the crescents frequently form at incorrect positions and are no longer strictly correlated with the position of one of the two spindle poles. We have reinvestigated the actin dependence of Numb and Prospero localization using the drug latrunculin A (28, 36), which has recently become available. In tissue culture cells, latrunculin A has been shown to be a more potent actin inhibitor than cytochalasin D (28). Indeed, we find that after treatment of Drosophila embryos with latrunculin A, actin filaments are almost completely undetectable by rhodamine-phalloidin staining (Fig. 1B), whereas actin filaments are still present (though severely disorganized) after treatment with cytochalasin D (10). Furthermore, in contrast to cytochalasin D treatment, treatment with latrunculin A completely inhibits the asymmetric localization of Numb and Prospero. It is unlikely that our observations are due to nonspecific effects of the drug because the effects of latrunculin A at concentrations similar to the ones used in our experiments in yeast can be completely suppressed by several point mutations in the actin gene that all map to the same putative binding site (27).
Despite its effect on asymmetric localization, latrunculin A fails to disrupt membrane localization of Numb and Prospero, suggesting that these two proteins are anchored to the cell membrane by an actin independent process. However, we could reproducibly detect some residual rhodamine-phalloidin staining at the cell cortex after latrunculin A treatment of Drosophila embryos, even when the drug concentration was increased to 400 μM (data not shown). This could be due to nonspecific rhodamine-phalloidin staining or reflect an incomplete disruption of the actin cytoskeleton. Latrunculin A inhibits the polymerization of actin, but does not actively depolymerize actin filaments (27) and cortical actin filaments in Drosophila cells could have a low turnover rate that makes them resistant to latrunculin A. Thus, we cannot exclude that membrane localization of Numb and Prospero is mediated by actin filaments that are resistant to the drug treatment.
Domains that Direct Membrane Localization and Asymmetric Localization of Numb.
Our experiments show that the first 119 aa of the Numb protein contain a region that is both necessary and sufficient for localization to the cell membrane. Deletions of the first 42 aa or of aa 40–119 result in a cytoplasmic protein that is no longer asymmetrically localized (Fig. 3 B and C), whereas a fusion protein between the first 119 aa of Numb and β-gal is localized to the cell membrane. The N terminus of Numb contains an N-myristoylation signal, but mutation of this signal sequence in Numb-Δmyr does not disrupt membrane localization (Fig. 3H). Numb-Δmyr could be localized to the cell membrane by associating with endogenous Numb protein. Alternatively, Numb could be localized to the cell membrane by a process that does not require the N-myristoylation signal. The Numb N terminus is not predicted to form a transmembrane domain, but interaction of the N terminus with a integral or peripheral membrane protein or phosphilipid interactions are alternative possibilities that could account for membrane localization of Numb.
Deletion analysis of the Prospero protein has identified a 118 aa region that directs localization to the cell membrane and asymmetric localization of β-gal fusion proteins (20). A 13-aa motif within this region has some weak homology to aa 379–391–Numb (20) and it has been suggested that this region might be involved in asymmetric localization of both proteins. However, Numb-Δ4 that removes these 13 aa localizes asymmetrically and segregates into one daughter cell (Fig. 3F) suggesting that this region is not required for asymmetric localization. The observation that the first 227, but not the first 119 aa of Numb are sufficient for asymmetric localization might indicate that aa 119–227 contain the localization domain. However, Numb-Δ2 that deletes aa 117–216 and Numb-ΔPTB that deletes aa 79–203 still localize asymmetrically even in the absence of endogenous Numb protein. Several possibilities remain to be tested: It could be that the localization domain lies between aa 217 and 227. Alternatively, asymmetric localization could be mediated by a region within the first 78 aa, but this region is unable to function correctly when fused to β-gal due to incorrect folding or other steric problems. Finally, asymmetric localization could be mediated by two partially redundant domains. One of these domains would be located within aa 119–227, but in Numb-Δ2 and Numb-ΔPTB a second, more C-terminal domain could mediate asymmetric localization. In any case, the finding that the first 227 aa of Numb are involved in asymmetric localization suggests that proteins that bind to the N-terminal half of Numb may be involved in its asymmetric localization.
Acknowledgments
We thank Yee-Ming Chan, Salim Abdelilah, and Liqun Luo for helpful comments on the manuscript and all the members of the Jan Lab for fruitful discussions. We also thank Katja Brose for valuable help with generating the Numb deletion constructs. J.A.K. was supported by an European Molecular Biology Organization postdoctoral fellowship and by the Howard Hughes Medical Institute; L.Y.J. and Y.N.J. are Howard Hughes Investigators.
ABBREVIATIONS
- β-gal
β-galactosidase
- ES
organ external sensory organ
- PTB
phosphotyrosine binding
- SOP
sensory organ precursor
- GMC
ganglion mother cell
References
- 1.Horvitz H R, Herskowitz I. Cell. 1992;68:237–255. doi: 10.1016/0092-8674(92)90468-r. [DOI] [PubMed] [Google Scholar]
- 2.Knoblich, J. A. (1997) Curr. Opin. Cell Biol., in press. [DOI] [PubMed]
- 3.Bodmer R, Carretto R, Jan Y N. Neuron. 1989;3:21–32. doi: 10.1016/0896-6273(89)90112-8. [DOI] [PubMed] [Google Scholar]
- 4.Hartenstein V, Posakony J W. Development (Cambridge, UK) 1989;107:389–405. doi: 10.1242/dev.107.2.389. [DOI] [PubMed] [Google Scholar]
- 5.Jan Y N, Jan L Y. In: The Development of Drosophila melanogaster. Bate M, Martinez Arias A, editors. Vol. 2. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 1207–1244. [Google Scholar]
- 6.Uemura T, Shepherd S, Ackerman L, Jan L Y, Jan Y N. Cell. 1989;58:349–360. doi: 10.1016/0092-8674(89)90849-0. [DOI] [PubMed] [Google Scholar]
- 7.Rhyu M S, Jan L Y, Jan Y N. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 8.Wang, S., Younger-Shepherd, S., Jan, L. Y. & Jan, Y. N. (1997) Development (Cambridge, U.K.), in press. [DOI] [PubMed]
- 9.Bork P, Margolis B. Cell. 1995;80:693–694. doi: 10.1016/0092-8674(95)90347-x. [DOI] [PubMed] [Google Scholar]
- 10.Knoblich J A, Jan L Y, Jan Y N. Nature (London) 1995;377:624–627. doi: 10.1038/377624a0. [DOI] [PubMed] [Google Scholar]
- 11.Frise E, Knoblich J A, Younger-Shepherd S, Jan L Y, Jan Y N. Proc Natl Acad Sci USA. 1996;93:11925–11932. doi: 10.1073/pnas.93.21.11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spana E P, Doe C Q. Neuron. 1996;17:21–26. doi: 10.1016/s0896-6273(00)80277-9. [DOI] [PubMed] [Google Scholar]
- 13.Guo M, Jan L Y, Jan Y N. Neuron. 1996;17:27–41. doi: 10.1016/s0896-6273(00)80278-0. [DOI] [PubMed] [Google Scholar]
- 14.Foe V E. Development (Cambridge, UK) 1989;107:1–22. [PubMed] [Google Scholar]
- 15.Kraut R, Chia W, Jan L Y, Jan Y N, Knoblich J A. Nature (London) 1996;383:50–55. doi: 10.1038/383050a0. [DOI] [PubMed] [Google Scholar]
- 16.Goodman C S, Doe C Q. In: The Development of Drosophila melanogaster. Bate M, Martinez Arias A, editors. Vol. 2. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 1131–1206. [Google Scholar]
- 17.Spana E P, Kopczynski C, Goodman C S, Doe C Q. Development (Cambridge, UK) 1995;121:3489–3494. doi: 10.1242/dev.121.11.3489. [DOI] [PubMed] [Google Scholar]
- 18.Doe C Q, Chu-LaGraff Q, Wright D M, Scott M P. Cell. 1991;65:451–464. doi: 10.1016/0092-8674(91)90463-9. [DOI] [PubMed] [Google Scholar]
- 19.Vaessin H, Grell E, Wolff E, Bier E, Jan L Y, Jan Y N. Cell. 1991;67:941–953. doi: 10.1016/0092-8674(91)90367-8. [DOI] [PubMed] [Google Scholar]
- 20.Hirata J, Nakagoshi H, Nabeshima Y, Matsuzaki F. Nature (London) 1995;377:627–630. doi: 10.1038/377627a0. [DOI] [PubMed] [Google Scholar]
- 21.Spana E P, Doe C Q. Development (Cambridge, UK) 1995;121:3187–3195. doi: 10.1242/dev.121.10.3187. [DOI] [PubMed] [Google Scholar]
- 22.Kraut R, Campos-Ortega J A. Dev Biol. 1996;174:65–81. doi: 10.1006/dbio.1996.0052. [DOI] [PubMed] [Google Scholar]
- 23.Amaya E, Musci T J, Kirschner M W. Cell. 1991;66:257–270. doi: 10.1016/0092-8674(91)90616-7. [DOI] [PubMed] [Google Scholar]
- 24.Fire A, Harrison S W, Dixon D. Gene. 1990;93:189–198. doi: 10.1016/0378-1119(90)90224-f. [DOI] [PubMed] [Google Scholar]
- 25.Brand A H, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 26.Orsulic S, Peifer M. BioTechniques. 1994;16:441–447. [PubMed] [Google Scholar]
- 27.Ayscough K R, Stryker J, Pokala N, Sanders M, Crews P, Drubin D G. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spector I, Shochet N R, Blasberger D, Kashman Y. Cell Motil Cytoskeleton. 1989;13:127–144. doi: 10.1002/cm.970130302. [DOI] [PubMed] [Google Scholar]
- 29.Kamps M P, Buss J E, Sefton B M. Proc Natl Acad Sci USA. 1985;82:4625–4628. doi: 10.1073/pnas.82.14.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strome S, Wood W B. Cell. 1983;35:15–25. doi: 10.1016/0092-8674(83)90203-9. [DOI] [PubMed] [Google Scholar]
- 31.Guo S, Kemphues K J. Nature (London) 1996;382:455–458. doi: 10.1038/382455a0. [DOI] [PubMed] [Google Scholar]
- 32.Long R M, Singer R H, Meng X, Gonzalez I, Nasmyth K, Jansen R P. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- 33.Takizawa P A, Sil A, Swedlow J R, Herskowitz I, Vale R D. Nature (London) 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- 34.Bobola N, Jansen R P, Shin T H, Nasmyth K. Cell. 1996;84:699–709. doi: 10.1016/s0092-8674(00)81048-x. [DOI] [PubMed] [Google Scholar]
- 35.Sil A, Herskowitz I. Cell. 1996;84:711–722. doi: 10.1016/s0092-8674(00)81049-1. [DOI] [PubMed] [Google Scholar]
- 36.Spector I, Shochet N R, Kashman Y, Groweiss A. Science. 1983;219:493–495. doi: 10.1126/science.6681676. [DOI] [PubMed] [Google Scholar]