Abstract
Sequence-specific targeting of genomic DNA by triplex-forming oligonucleotides (TFOs) is a promising strategy to modulate in vivo gene expression. Triplex formation involving G-rich oligonucleotides as third strand is, however, strongly inhibited by potassium-induced TFO self-association into G-quartet structures. We report here that G-rich TFOs with bulge insertions of (R)-1-O-[4-(1-pyrenylethynyl)-phenylmethyl] glycerol (called twisted intercalating nucleic acids, TINA) show a much lower tendency to aggregate in potassium than wild-type analogues do. We designed purine-motif TINA–TFOs for binding to a regulatory polypurine-polypyrimidine (pur/pyr) motif present in the promoter of the KRAS proto-oncogene. The binding of TINA–TFOs to the KRAS target has been analysed by electrophoresis mobility shift assays and DNase I footprinting experiments. We discovered that in the presence of potassium the wild-type TFOs did not bind to the KRAS target, differently from the TINA analogues, whose binding was observed up to 140 mM KCl. The designed TINA–TFOs were found to abrogate the formation of a DNA–protein complex at the pur/pyr site and to down-regulate the transcription of CAT driven by the murine KRAS promoter. Molecular modelling of the DNA/TINA–TFO triplexes are also reported. This study provides a new and promising approach to create TFOs to target in vivo the genome.
INTRODUCTION
Polypurine-polypyrimidine (pur/pyr) motifs are potential sites of triplex formation that have been identified through the genome with a relatively high frequency in the control region of the genes (1). As pur/pyr sequences are often located immediately upstream of transcription initiation sites, triplex forming oligonucleotides (TFOs) are attractive molecules to specifically modulate gene expression (2–5). TFOs are designed to bind to the major groove of the pur/pyr sequence through the formation of a triple-helical complex stabilized by hydrogen bonds in the Hoogsteen or reverse-Hoogsteen configuration, depending on the nature of the third strand. When TFOs contain C and T nucleotides, they bind to the target in the parallel orientation through the formation of Hoogsteen hydrogen bonds (CT triplex motif) (6–8). In contrast, TFOs containing G and A/T nucleotides bind to the target in an antiparallel orientation, with reverse-Hoogsteen hydrogen bonds, although GT-TFOs may in some cases prefer a parallel binding (9). Since in the parallel triplex motif the cytosines of the third strand are protonated, CT-TFOs show poor binding at neutral pH and are therefore not very useful to manipulate gene expression in culture cells or in vivo. For these applications AG-and GT-TFOs are preferred because they have no pH-limits. However, the binding of G-rich TFOs to DNA under physiological conditions can be disfavoured by intracellular potassium (10–12). High potassium concentrations induce G-rich oligonucleotides to assume intra- or inter-molecular G-quartet structures that subtract the TFO from the equilibrium of triplex formation, with the result that the triplex is not observed even at sub-physiological potassium concentrations. This is the reason why most of the literature data about in vitro triplex formation by G-rich TFOs have been obtained in the absence of potassium. One therefore wonders if the antigene activity reported by certain G-rich oligonucleotides is really mediated by triplex formation at the corresponding target site. In order to potentiate the capacity of TFOs to form stable triplexes in the cellular context, several chemical modifications at the sugar phosphate backbone have been proposed. The most promising modified TFOs are peptide nucleic acids (13–15), locked nucleic acids (16,17), 2′-aminoethyl oligoribonucleotides (18) and N3′-P5′ phosphoroamidites (19). Stabilization of triplex DNA has also been obtained by linking the 5′ or 3′ end of TFOs to an intercalator, a modification that can enhance the Tm of parallel triplexes up to 16°C (20–25). However, little is known about the effect on triplex formation of intercalators inserted as a bulge in the middle of the TFO sequence. This type of modification has been recently tested in TFOs forming CT-motif parallel triplexes (26). Bulged insertions of (R)-1-O-[4-(1-pyrenylethynyl) phenylmethyl] glycerol inside CT-TFOs (with a cytosine content up to 36%) were found to form stable parallel triplexes, whereas unmodified TFOs were not able to bind to the target (27). Molecular modelling with MacroModel 8.0 showed that both pyrene and phenyl are twisted around the triple bond with a torsion angle of about 15.3 degrees (27). This suggested that twisting helps the intercalator unit to adjust within the target duplex, so that the parallel triple helical complex is significantly stabilized. The study of Filichev et al. represents a new development in triplex technology, but the concept of pyrene bulge insertions was only applied to CT-motif parallel triplexes, as well as to non-natural target sequences. We therefore wondered what effect pyrene bulge insertions might have on triplex formation by purine-motif TFOs, capable to form antiparallel triplexes with natural and therapeutically important targets. Here, we addressed two main questions: (i) do purine twisted-intercalating TFOs form stable triple helices in the antiparallel reverse-Hoogsteen conformation?; (ii) do G-rich twisted intercalating TFOs self-associate into quadruplex structures and subtract themselves from the interaction with duplex DNA? To answer these questions we designed twisted-intercalating TFOs for two natural pur/pyr sequences present in the regulatory region of human and murine KRAS genes. The results obtained show that bulge insertions stabilize triplex DNA and provide a strategy to overcome potassium-induced self-aggregation phenomena of G-rich TFOs that severely limits triplex formation under physiological conditions. All the data collected in this study point to the potential of purine twisted-intercalating TFOs as antigene molecules in cancer therapy.
MATERIALS AND METHODS
Synthesis and purification of TINA oligonucleotides
The TINA oligonucleotides shown in Figure 1 were synthesized in a 0.2 µmol scale on an Expedite™ Nucleic Acid Synthesis System Model 8909 from Applied Biosystems using 4,5-dicyanoimidazole as an activator. Increased deprotection time (100 s) and coupling time (2 min) for 0.075 M solution of TINA phosphoramidite (28) in a 1:1 mixture of dry MeCN/CH2Cl2 was applied. To obtain phosphorthioates, a freshly prepared 0.0225 M solution of 3-amino-1,2,4-dithiazole-5-thione in dry mixture of pyridine (10%, v/v) and CH3CN was used in a sulfurization step (29), which was repeated twice for 1 min instead of oxidation step. Use of the solution, which was 3 days old, resulted in an unsuccessful synthesis. The oligonucleotides were cleaved off were cleaved off from the solid support (room temperature, 2 h) and de-protected (55°C, overnight) using 32% aqueous ammonia (1 ml). Purification of 5′-O-DMT-on ONs was accomplished using a reverse-phase semi-preparative HPLC on Waters Xterra™ MS C18 column. After evaporation of the solvent on a speed-vac, ONs were treated with 80% aq AcOH (100 µl) for 25 min, diluted with 1.5 M aq NaOAc (150 µl) and precipitated from EtOH (550 µl). Solutions were kept in a freezer for 1 h, centrifuged, decanted and the precipitate was washed with EtOH and dried. The modified ONs were confirmed by MALDI-TOF analysis on a Voyager Elite Biospectrometry Research Station from PerSeptive Biosystems [calculated and experimental MW values are: 1037 (7892.5/7892.0); 1451 (7892.5/7892.2); 1452 (7425.1/7428.2); 1453 (7876.5/7877.0); 1454 (7409.1/7410.2), 1455 (7876.52/7876.0); 1456 (7676.52/7677.5); 1457 (7209.12/7210.1); 2218 (7828.2/7829.7)]. The purity of the final TFOs was checked by ion-exchange chromatography using LaChrom system from Merck Hitachi on GenPak-Fax column (Waters) and found to be over 90%.
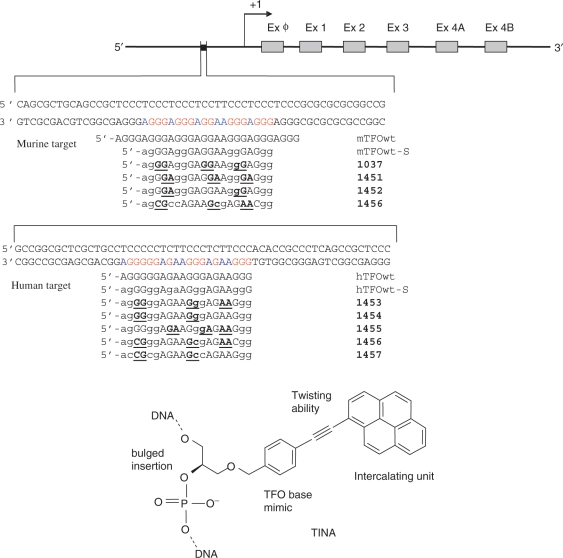
Figure 1.
Pur/pyr motifs present in a regulatory element of the mouse and human KRAS promoters. The sequences of the designed wild-type and twisted-intercalating TFOs are reported. Bulge insertions of (R)-1-O-[4-1-(1-pyrenylethynyl)-phenylmethyl]glycerol (TINA) have been placed between the underlined bases of the TFOs. Lower case letters indicate the bases that are linked with a thioate group to make the TFOs resistant to endogenous nucleases. The structure of the TINA intercalating unit is reported.
Plasmids
Plasmid pKRS-413, used for the transient transfection experiments, was kindly gifted by D. George (University of Pennsylvania). This plasmid, whose structure has been previously described, is derived from the chloramphenicol acetyltransferase (CAT) vector pSVAOcat, in which a 380-bp segment of the KRAS promoter has been cloned. This construct is able to drive CAT transcription in host cells. In addition, plasmid pTKβgal, containing the β-galactosidase gene driven by the thymidine kinase promoter, was used to evaluate the transfection efficiency. The construction of the mutant plasmid (pKRS-413-4mut) was done as follows: 75 ng of plasmid pKRS-413 were amplified in a PCR reaction with 1.25 units of AccuPrime™ Pfx DNA Polymerase, in a 25 μl volume containing ×1 AccuPrime™ Pfx reaction mix as indicated by the manufacturer (Invitrogen, Milan, Italy). The primers used are Mut 42, 5′-TGCAGCCGCTCCCTCTCTCTCTCCTTCTCTCTCTCCCGCGCG-3′, with four C-to-T substitutions (bold in Figure 9b) and Mut 25, 5′-GAGGGAGCGGCTGCAGCGCTGGGAG-3′ (GeneTailor™ Site-Directed Mutagenesis System, Invitrogen, Milan, Italy). The primers are partially overlapping and were used at a concentration of 100 nM. The PCR reaction was carried out according to the following protocol: 2′ at 95°C, 25 cycles each comprising a denaturation of 30 s at 95°C, annealing of 30 s at 68°C, elongation of 5 min at 68°C. The final cycle was performed with an elongation step of 10 min at 68°C. Competent bacteria were transformed with 7.5 μl of PCR product. The mutant plasmid was sequenced to confirm the introduction of the mutations.
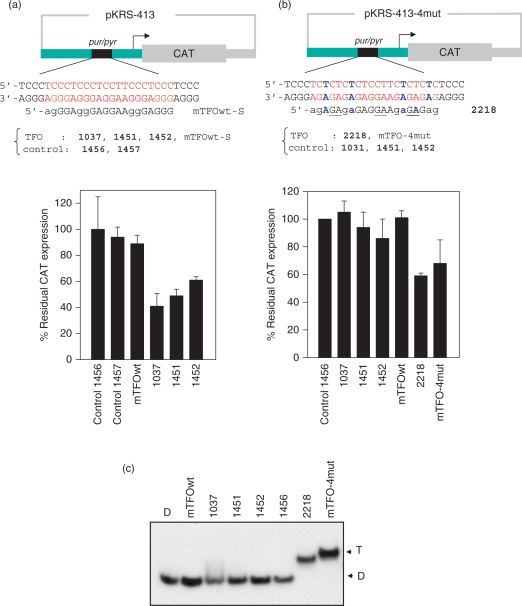
Figure 9.
(a) CAT assay to determine the effect of the designed twisted-intercalating TFOs on the activity of KRAS promoter. Plasmids pKRS-413 (2 μg), containing CAT driven by the mouse KRAS promoter, and pTK-βgal (50 ng) were co-transfected with murine TFOs 1037,1451,1452, mTFOwt or control 1456 and 1457 (1 μM), using jet-PEI. The expression of CAT, with respect to that of β-gal, was determined by an ELISA assay, 48 h after transfection. The percentage of residual CAT expression, [(CAT/β-gal)TFO/(CAT/β-gal)Control] × 100, is reported in the histograms. The values are the average of three independent experiments in duplicate. Error bar are ± SE; (b) transfection experiment as in (a) but with the mutated plasmid pKRS-413-4mut in which the KRAS promoter has been mutated at the pur/pyr target introducing 4 G-to-A mutations. The mutated target is recognized by the TINA conjugate 2218 (5′-agAGPAgaGAG-GPAAgaGPAGag; P indicates a TINA modification; the lower case letters indicate the bases linked by a phosphorothioate group) and mTFO-4mut (5′-AGGGAGAGAGAGAGGAAGAGAGAGAGGG); (c) EMSA showing that triplex formation occur between the mutated pur/pyr target and TINA–TFO 2218 and mTFO-4mut, but not with the TFOs designed for the wild-type pur/pyr sequence (mTFOwt, 1037, 1451, 1452). TFOs and target were incubated overnight at 37°C in 50 mM Tris–HCl, pH 7.4, 50 mM KCl, 10 mM MgCl2, 2 mM spermidine and run in a native 20% polyacrylamide gel. The mutated duplex was formed mixing 1:1 5′-GCTCCCTCTCTCTCTCCTTCTCTCTCTCCCG and 5′-CGGGAGAGAGAGAAGGAGAGAGAGAGGGAGC.
EMSA
Mobility-shift experiments were performed with 39-mer mouse and 32-mer human critical KRAS promoter targets. For the preparation of mouse 39-mer duplex 30 pmole of pyrimidine strand was labelled with [γ-P32]ATP and T4 polynucleotide kinase, then the duplex was prepared by annealing with the purine complementary strand and 100 mM NaCl (10 min at 95°C and overnight at room temperature). Human 32-mer duplex was prepared by labelling the pyrimidine strand in the same way as mentioned previously. A fixed amount of ∼10 nM labelled 39-mer mouse duplex or 32-mer human duplex was mixed with increasing concentrations of mouse TFOs (mTFOwt, 1037, 1451, 1452 and control 1456) or human TFOs (hTFOwt, 1453, 1454, 1455 and control 1457), in 50 mM Tris–HCl, pH 7.4, 50 mM KCl, 10 mM MgCl2 and 2 mM spermidine (binding buffer) and incubated overnight at 37°C. After incubation, the samples were immediately loaded in 15% native polyacrylamide gel prepared in TB1x and performed at 37°C with a fixed voltage of 200 V for 5 h. After running, the gel was dried and exposed to autoradiography (Hyperfilm, Amersham Biosciences) for few hours. Wild-type and twisted-intercalating TFOs (10 μM) were analysed in a 15% TBE gel at 20°C which was stained with the stains-all dye. Electrophoresis mobility shift assays (EMSA) with nuclear extract from NIH 3T3 and Panc-1 cells were performed with labelled mouse 39-mer and human 32-mer duplexes. Two protocols have been followed. In one case, the triplexes were allowed to form overnight at 37°C in binding buffer before the protein extract was added and the resulting mixtures incubated for 2 h. In the other case, TFO and protein extract were added simultaneously to the target duplex and mixtures incubated for 2 h. The protein extract was equilibrated in 20 mM Tris–HCl, pH 8, 30 mM KCl, 10 mM MgCl2, 1 mM DTT, glycerol 8%, Phosphatase Inhibitor Cocktail I (Sigma) 1%, 5 mM NaF, 1 mM Na3VO4, 2.5 nM poly dIdC. After incubation, the mixtures were immediately loaded in 5% TBE ×1 native polyacrylamide gel, with the temperature fixed at 20°C.
Calculation of the free energy change of triplex formation
The duplex-to-triplex equilibrium can be written as:
| 1 |
where D and T are the duplex and triplex species. This equilibrium is governed by a dissociation constant Kd given by:
| 2 |
We can define the fraction of triplex fT in the reaction mixture as:
| 3 |
Combining Equations (2) and (3) it is obtained that:
| 4 |
As in each reaction mixture, the TFO concentration is always in excess over the target duplex D, the equilibrium concentration [TFO] can be reasonably approximated to the total concentration [TFO]T. Equation (4) has been used to fit the experimental fT versus [TFO]T plot, using a nonlinear best-fit program (Marquardt program, Jandel Scientific). The fT values have been measured experimentally from electrophoretic titrations, quantifying bands D and T with an Image Quant TL V2003.03 apparatus (Amersham Biosciences). The free energy of reaction (1) was obtained from the general equation
Circular dichroism
CD spectra were obtained using a JASCO J-600 spectro-polarimeter equipped with a thermostatted cell holder. The oligonucleotides used for the CD experiments were at a concentration of 3 μM, in 50 mM Tris–HCl, pH 7.4 and 100 mM KCl. Spectra were recorded in 0.5 cm quartz cuvette. A thermometer placed in the cuvette holder allowed a precise measurement of the sample temperature. The spectra were calculated with J-700 Standard Analysis software (Japan Spectroscopic Co., Ltd) and are reported as ellipticity (mdeg) versus wavelength (nm). Each spectrum was recorded three times, smoothed and subtracted from the baseline.
Spectrofluorimetry measurements
Fluorescence measurements were performed on a Perkin Elmer spectrofluorimeter. The spectra of TFO or TFO-duplex solutions were recorded at room temperature in 50 mM Tris-HCl, pH 7.4, 10 mM MgCl2. TFO and duplex were used at a concentration of 0.5 μM.
DNase I footprinting
DNase I footprints were performed with DNA fragments of different length. Either the purine or the pyrimidine strand of a murine 60-mer duplex was end-labelled with [γ-32P]ATP and T4 polynucleotide kinase, PAGE purified and annealed with the complementary cold strand. The labelled target and the TFOs were pre-incubated, at different ratios, overnight in 50 mM Tris-HCl, pH 7.4, 50 mM KCl, 10 mM MgCl2 and 2 mM spermidine and digested with DNase I: 1 μl of a solution containing 0.001 mg/ml DNase I, 50 mM Tris-HCl, pH 7.4, 0.1 mg/ml BSA, 30 mM MnCl2. The reaction was conducted in a final volume of 10 μl, for 1 min at room temperature, and stopped by adding to the reaction mixture 10 μl of stop solution (90% formamide, 50 mM EDTA, bromophenol blue). Experiments with the murine target were also conducted with a DNA of 153 bp, which was obtained from plasmid pKRS-413 digested with Not I and Ava I. In this case we labelled the purine strand using [α-P32]CTP and Klenow fragment. A standard Maxam-Gilbert G reaction was carried out in order to identify the TFO binding site. DNase I footprinting with the human target were performed with a 79-mer duplex labelled at the purine strand with [γ-P32]ATP and the T4 polynucleotide kinase. The digested DNA was heated for 10 min at 97°C and loaded in 18% polyacrylamide gel in TBE ×1 (50 mM Tris base, 50 mM Boric acid, 1 mM EDTA) and 8 M urea, pre-equilibrated at 55°C in a BioRad Sequi-Gen GT nucleic acids electrophoresis apparatus, which was equipped with thermocouple allowing temperature control. After running, the gels were fixed in a solution containing 10% acetic acid, 10% methanol, dried at 80°C in a Bio-Rad dryer and exposed to autoradiography (Hyperfilm, Amersham Biosciences) at –80°C for few hours.
CAT assay
NIH 3T3 cells were transfected with mixtures containing plasmid pKRS-413 (or mutant pKRS-413-4mut), containing the CAT reporter gene driven by the murine KRAS promoter, plasmid pPTK-βgal to determine transfection efficiency, and a specific oligonucleotide. The transfections were performed using jet-PEI as transfecting agent. Forty-eight hours after transfection the cells were washed three times with PBS. The cells were then lysed with MOPS containing 0.1% Triton X-100, pH 6.5. Protein extracts were obtained and used for the CAT and β-gal assay using a commercial ELISA kit (Boehringer Manheim, Germany). A volume of 200 μl of protein extract was incubated in each well of the plate coated with the anti-CAT antibody linked to digoxygenin for 1 h. The plate was washed and incubated with anti-digoxygenin antibody conjugated to peroxidase enzyme. Finally, the peroxidase substrate was added to the reaction mixture. The reaction catalyzed by peroxidase gave a green colour that was quantified by optical density. For the β-gal procedure a similar assay was performed.
Molecular modelling
Molecular modelling was performed with MacroModel v7.5 from Schrödinger. All calculations were conducted with AMBER* force field and the GB/SA water model. The dynamic simulation was preformed with stochastic dynamics, a SHAKE algorithm to constrain bonds to hydrogen, time step of 1.5 fs and simulation temperature of 300 K. Simulation for 0.5 ns with an equilibration time of 150 ps generated 250 structures, which were all minimized using the PRCG method with convergence threshold of 0.05 kJ/mol. The minimized structures were examined with Xcluster from Schrödinger, and a representative low-energy structure was selected. The starting structure was generated from an NMR-based antiparallel triplex (downloaded from www.rcsb.org/pdb; PDB ID: 134d, 30) which was modified into a truncated 1037 antiparallel triplex.
RESULTS
Targets and design of TINA–TFOs
As target for the TFOs in this study we chose a perfect pur/pyr motif present in the promoter of the KRAS proto-oncogene. This sequence, which is conserved in human and mouse is essential for transcription, as its excision results in a strong decline of gene expression (31–33) (Figure 1). Previously we tried to knock down the human KRAS gene by TFOs conjugated to the 5′ end to polyethylene glycol, but the results we obtained were not satisfactory (34). In the present study we have introduced covalent modifications in the middle of the TFO sequence in the form of bulge insertions of (R)-1-O-[4-1-(1-pyrenylethynyl) phenylmethyl] glycerol: this is an intercalating unit composed by a pyrenylethynyl moiety attached to the end of a flexible phenyl methyl glycerol arm (Figure 1). In previous molecular modelling studies on pyrimidine motif triplexes we have shown that the intercalator, due to its flexible phenylmethyl glycerol arm, twists around the triple bond and nicely fits inside the triplex. The TFO was therefore called ‘twisted- intercalating nucleic acid’ (TINA) (27). In the present work we have extended the design of twisted-intercalating TFOs to G-rich oligonucleotides forming reverse-Hoogsteen antiparallel triplexes with C+G rich targets. It is known that triplex formation by G-rich TFOs depends on two variables: the sequence of the target and the tendency of TFOs to self-associate or fold into quadruplex structures, in particular in the presence of physiological levels of potassium (35,36). We therefore reasoned that bulge insertions of pyrene units should stabilize antiparallel triplexes, as already observed for parallel triplexes (27), but at the same time they should inhibit TFO self-association. Indeed, according to molecular modelling, TINA–TFOs with two or three bulge intercalating units are expected to bind to duplex DNA giving rise to a compact and stacked triplex, but they can hardly self-associate in an intermolecular quadruplex by accommodating 8/12 intercalators. As proof of concept we designed twisted intercalating TFOs for a regulatory pur/pyr element in the KRAS promoter. We considered the pur/pyr sequences present in both murine and human promoters and designed for these targets 20-mer TFOs with two or three pyrene bulge insertions, differently distributed along the TFO sequence. These insertions have been put either inside the guanine runs (1037, 1453, 1454) or outside/adjacent to them (1451, 1452, 1455). In addition we designed 20-mer control oligonucleotides with pyrene bulge insertions (1456 and 1457), where some guanines were substituted by cytosines. To make the designed TFOs resistant to endogenous nucleases, one oxygen was replaced by sulphur in four internucleotide phosphate groups of each conjugate (Figure 1)
Triplex formation by G-rich TINA–TFOs in the presence of potassium ions
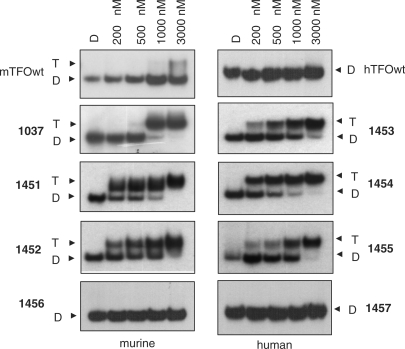
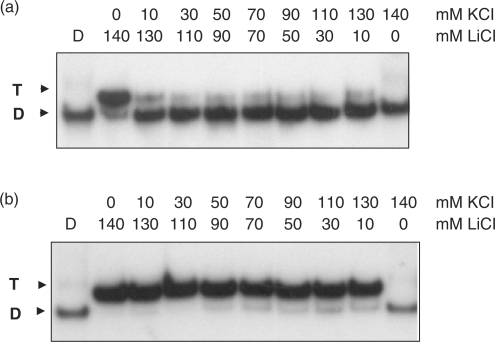
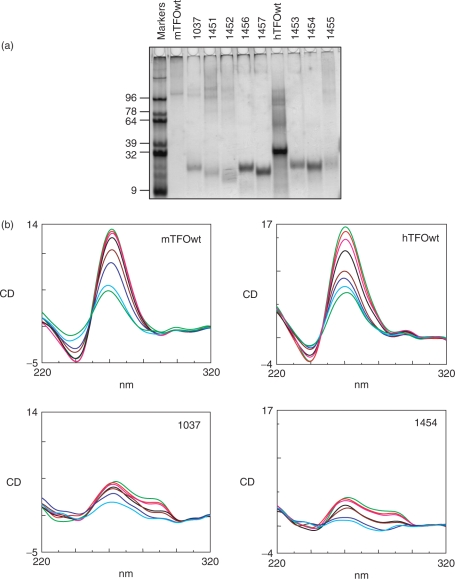
The binding to the KRAS targets of the designed wild-type and TINA TFOs was studied by EMSA. Figure 2 shows EMSA titrations performed in 50 mM Tris–HCl, pH 7.4, 50 mM KCl, 10 mM MgCl2, 2 mM spermidine (binding buffer). TFOs and target duplex were incubated overnight at 37°C in the binding buffer and the mixtures separated by 15% polyacrylamide gels, thermostated at 37°C. It can be seen that both murine and human wild-type TFOs did not practically bind to the corresponding targets, whereas TINA–TFOs showed binding, although the oligonucleotides have a G content as high as 65–70%. From electrophoresis titrations we determined for each TFO the Kd as described in Materials and Methods. The values obtained (Table 1) are of the order of 10–7 M, except for 1452 which showed a higher Kd. There seems to be a correlation between triplex formation and the number of TINA insertions in the TFO: the human TFO 1454 with two bulge insertions shows a higher target affinity than 1453 which has one more bulge insertion than 1454, between an AA dinucleotide. This suggests that the accommodation of three TINA intercalating units in a 20-mer triplex may cause some distortion and destabilization to the structure. The fact that controls 1457 and 1456 (containing respectively two and three bulge insertions) do not bind to the target suggests that the TINA–TFOs maintain the sequence specificity typical of triplex DNA. To evaluate if potassium concentrations higher than 50 mM might inhibit triplex formation by the TINA–TFOs, we performed EMSA experiments in which lithium was gradually replaced by potassium, while the total ion concentration was kept constant at 140 mM. Figure 3 shows a typical experiment performed with murine 1037 and wild-type mTFOwt. It can be seen that mTFOwt binds to the target in the presence of 140 mM LiCl. By just adding 10 mM KCl, triplex formation by the wild-type sequence is almost completely abrogated. Oligonucleotide 1037 with three TINA insertions in its sequence also binds to the target in 140 LiCl, but its binding does not decrease as lithium is gradually replaced by potassium, keeping the total ionic strength of the solution fixed at 140 mM (Figure 3b). This experiment shows that twisted-intercalating TFOs, contrarily to wild-type analogues, are less prone to self-aggregation in the presence of potassium. In order to gain more evidence for this behaviour we analysed the electrophoretic mobility and the CD spectra of the wild-type and twisted-intercalating TFOs. The oligonucleotides (10 μM) were incubated overnight in 50 mM Tris–HCl, pH 7.4, 100 mM KCl, 37°C and analysed by a native 15% PAGE (Figure 4a). It can be seen that whereas mTFOwt migrated with a strongly retarded band due to self-aggregation, the TINA analogues 1037, 1451, 1452 migrated mainly as a single-stranded species. Note that TINA–TFOs also form aggregates, but to a much lower extent than mTFOwt. The aggregates of the murine TFOs are likely to be intermolecular G4-DNA structures. Indeed, DMS-footprinting assays show that in 100 mM KCl, but not in 100 mM LiCl, the guanines of mTFOwt, except those of the central GG couple, are protected from cleavage. Thus, PAGE and DMS probing suggest the formation of a concatenamer of 6–8 strands in which the runs of three guanines form the G-tetrads. In addition to the concatenamers, it is possible that some mTFOwt folds into an intramolecular G4-DNA (Supplementary Data S1A). In contrast, murine TINA TFOs do not show any footprinting in KCl, in accord with a lower aggregation. A typical DMS-footprinting for 1037 is shown in Supplementary Data S1B. As for human wild-type hTFOwt, it showed a mobility comparable to that of a 32-mer duplex, indicating that this oligonucleotide either assumes a homoduplex or, more likely, a folded bimolecular G-quadruplex (Supplementary Data S2). TINA derivatives 1453, 1454 and partly 1455 migrate as 20-base species, indicating that they do not aggregate in the potassium containing medium. The resistance to aggregation of 1453 and 1454 can be ascribed to the pyrenyl intercalators, which in these oligonucleotides have been put inside the guanine runs, disrupting the G-motif and preventing the formation of G-quartet structures. So the electrophoretic analysis corroborates the working hypothesis raised in this study according to which, contrarily to wild-type sequences, TINA–TFOs bind to the KRAS target in the presence of potassium because they are not inclined to aggregation. Further data suggesting that twisted-intercalating TFOs undergo little self-association in KCl were obtained by circular dichroism. It is known that potassium and, to a lesser extent, sodium induce G-rich oligonucleotides to form a variety of structures including GG duplexes and G-quartets (36,37). The designed TFOs were analysed by CD: the wild-type sequences exhibited a strong positive ellipticity at 260 nm and a negative ellipticity at 240 nm, which suggests either a GG homoduplex (37) or a parallel G-quadruplex (38,39). In contrast, TINA analogues 1037 and 1454 showed a significantly weaker ellipticity at 260 nm, suggesting that they are slightly associated in the potassium buffer (Figure 4b). Plotting the 260 nm ellipticity as a function of temperature we obtained cooperative curves from which we evaluated Tm's of 71°C and 55°C for the putative murine concatenamer and human bimolecular G-quadruplex, respectively.
Figure 2.
Electrophoretic mobility shift assays showing triplex formation at 37°C in 50 mM Tris–HCl, pH 7.4, 50 mM KCl, 10 mM MgCl2 and 2 mM spermidine (binding buffer). Experiments were performed with [γ-32P]ATP-labelled 39-mer murine duplex (5 nM) and 32-mer human duplex (15 nM) of the KRAS gene. Increasing concentrations of TFOs (as indicated) were mixed with the target duplex and incubated overnight in the binding buffer at 37°C. The mixtures were run at 37°C in a 15% native polyacrylamide gel.
Table 1.
Thermodynamic parameters for duplex–triplex equilibrium in 50 mM Tris-HCl, pH 7.4, 50 mM KCl, 10 mM MgCl2, 2 mM spermidine, 37°C, relative to human and murine KRAS duplexes and TINA-TFOs
| Oligonucleotide | Triplex | Kd (M) | ΔG 37 a (kcal/mol) |
|---|---|---|---|
| mTFOwt | No | – | – |
| 1037 | Yes | 8.3 × 10−7 | −8.6 ± 0.4 |
| 1451 | Yes | 3.4 × 10−7 | −9.2 ± 0.2 |
| 1452 | Yes | 2.6 × 10−6 | −7.9 ± 0.2 |
| 1456 | No | – | – |
| hTFOwt | No | – | – |
| 1453 | Yes | 6.1 × 10−7 | −8.8 ± 0.1 |
| 1454 | Yes | 1.3 × 10−7 | −9.8 ± 0.1 |
| 1455 | Yes | 8.7 × 10−7 | −8.6 ± 0.1 |
| 1457 | No | – | – |
aUncertainty estimated by the Marquardt non-linear best-fit program.
Figure 3.
Effect of lithium and potassium on triplex formation at the pur/pyr element of murine KRAS promoter. Oligonucleotides mTFOwt (panel a) and 1037 (panel b), were incubated at 37°C with murine 39-mer duplex (5 nM) in 50 mM Tris–HCl, pH 7.4, 10 mM MgCl2, 2 mM spermidine and 140 mM monovalent cations at various lithium/potassium ratios. The mixtures were incubated overnight at 37°C in the specific buffer and run (at 37°C) in a native 20% polyacrylamide gel.
Figure 4.
(a) Native polyacrylamide gel of wild-type and TINA–TFOs. TFOs (10 µM) were incubated overnight at 37°C in 50 mM Tris–HCl, pH 7.4, 100 mM Kcl and run in a native 15% PAGE. The gel was stained with ‘stains all’ dye; (b) Circular dichroism spectra of murine mTFOwt, 1037 and human hTFOwt, 1454 at increasing temperatures from 25°C to 95°C. All samples were used at the concentration of 3 µM, incubated overnight in 50 mM Tris–HCl, pH 7.4, 100 mM KCl before the analysis. The CD spectra of 1037 and 1454 in 100 mM LiCl are similar to those reported in the figure.
Footprinting studies
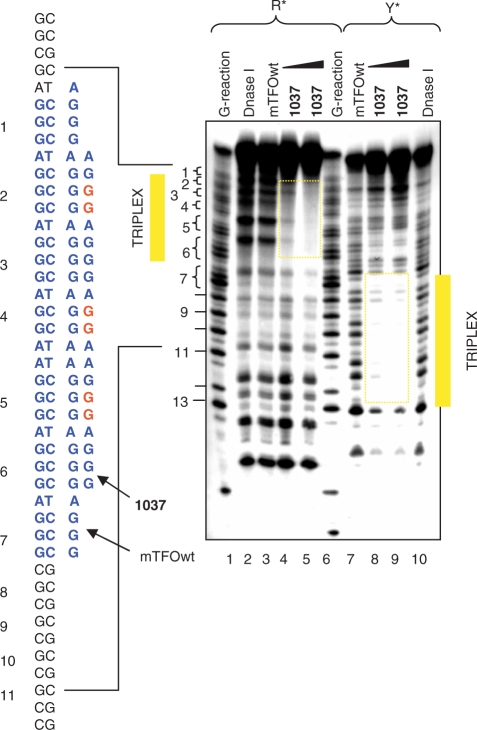
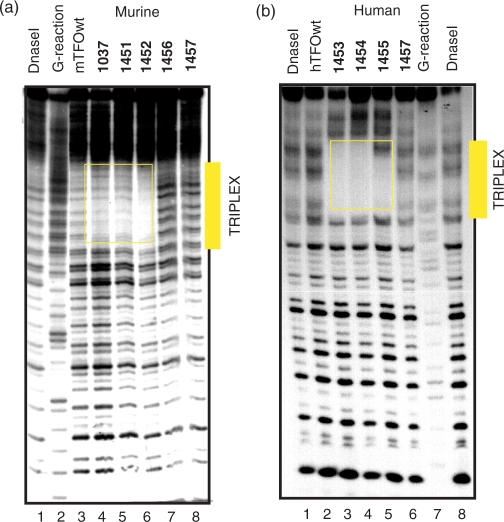
Considering that the pyrenyl units strongly favour triplex formation by the TINA–TFOs, one could argue that the stabilization may reduce the sequence binding specificity. To address this question we performed DNase I footprinting experiments under different ionic conditions. First, we analysed in detail triplex formation between the murine KRAS target, mTFOwt and TINA analogue 1037. Figure 5 shows the footprinting obtained by labelling either the purine or the pyrimidine strand of a 60-mer DNA duplex spanning over the murine pur/pyr sequence. To identify the TFO binding site, a standard Maxam-Gilbert G reaction was performed with the G strand of the target. In accord with EMSA it was found that mTFOwt (5 μM) did not bind to the target in the presence of 50 mM KCl (lane 3, R strand labelled). Instead, oligonucleotide 1037 saturated all the target sites by triple-helix formation at 2 and 5 μM (lanes 4, 5). When the pyrimidine strand of the target was end labelled with 32P, a similar result was obtained, with the difference that the target site was now observed near the 5′ end of the 60-mer duplex. In this case too, mTFOwt did not bind to duplex DNA in the presence of potassium, whereas 1037 did the binding at both concentrations. In order to test the binding specificity of all the designed murine TINA–TFOs, we performed footprinting experiments with a 153-mer duplex in the presence of 50 mM sodium or potassium ions. In NaCl we observed that both wild-type and TINA–TFOs bound to the target (Supplementary Data, S3). Instead, in 50 mM KCl the wild-type mTFOwt did not appreciably bind to the target, while the three TINA–TFOs 1037, 1451 and 1452 did (Figure 6a). As expected, control sequences 1457 and 1456 with three and two pyrene insertions, respectively, did not protect duplex DNA from DNase I even at the concentration of 5 μM. It is worth noting that 1452, despite it is characterized by a less favourable ΔG of binding, seems to protect the target more efficiently than 1037 and 1451. This is probably due to a lower tendency of 1452 to aggregate at 5 μM (Figure 4a). Footprinting studies carried out with the human target gave similar results: hTFOwt and TINA analogues 1453, 1454 and 1455 efficiently bound to duplex DNA in the presence of 50 mM NaCl (Supplementary Data S4), whereas only the TINA analogues showed strong binding in the presence of 50 mM KCl (Figure 6b). In conclusion, footprinting and EMSA demonstrate that TINA–TFOs are able to bind to duplex DNA in the presence of 50 mM KCl in a sequence-specific manner while the wild-type analogues do not have that capacity.
Figure 5.
DNase I footprinting showing the binding of murine mTFOwt and 1037 to a 60-mer target duplex (D) spanning over the pur/pyr motif (20 nM), in which either the purine or the pyrimidine strand was labelled with [γ-P32]ATP and polynucleotide kinase. Lanes 1 and 6, Maxam-Gilbert G-reaction of D; lanes 2 and 10, DNase I digestion of D; lanes 3 and 7, digestion of D in the presence of 5 µM mTFOwt; lanes 4, 5 and 8, 9, D digestion in the presence of 2 and 5 µM 1037. In lanes 2–5, D was labelled at the purine strand; in lanes 7–10, D was labelled at the pyrimidine strand. In this experiment we used a wild-type mTFOwt of 28 nt. The bases of 1037 in read indicate the position of the TINA intercalators.
Figure 6.
(a) DNase I footprinting showing the binding of murine TFOs 1037, 1451, 1452 to a 153-mer duplex obtained from the murine promoter labelled at the purine strand with [α-P32] CTP and Kleenow fragment. Target digestion in the absence of TFO (lane 1), in the presence of 5 μM mTFOwt (lane 3), 1037 (lane 4), 1451 (lane 5), 1452 (lane 6), control 1456 (lane 7), control 1457 (lane 8). A G-reaction with the target duplex is shown in lane 2; (b) DNase I footprinting showing the binding of human TFOs 1453, 1454 and 1455 to a 79-mer human duplex labelled at the purine strand with [α-P32]ATP and polynucleotide kinase. Target digestion in the presence of 3 μM hTFOwt (lane 1), 1453 (lane 2), 1454 (lane 3), 1455 (lane 4), 1457 (lane 5). Lane 6 shows the digestion of the 79-mer target duplex. The footprints have been performed in 50 mM Tris–HCl, pH 7.4, 50 mM KCl, 10 mM MgCl2, 2 mM spermine (binding buffer). The same experiment was performed with the TFOs at the concentration of 5 μM and similar results were obtained (Supplementary Data S5).
Molecular modelling studies
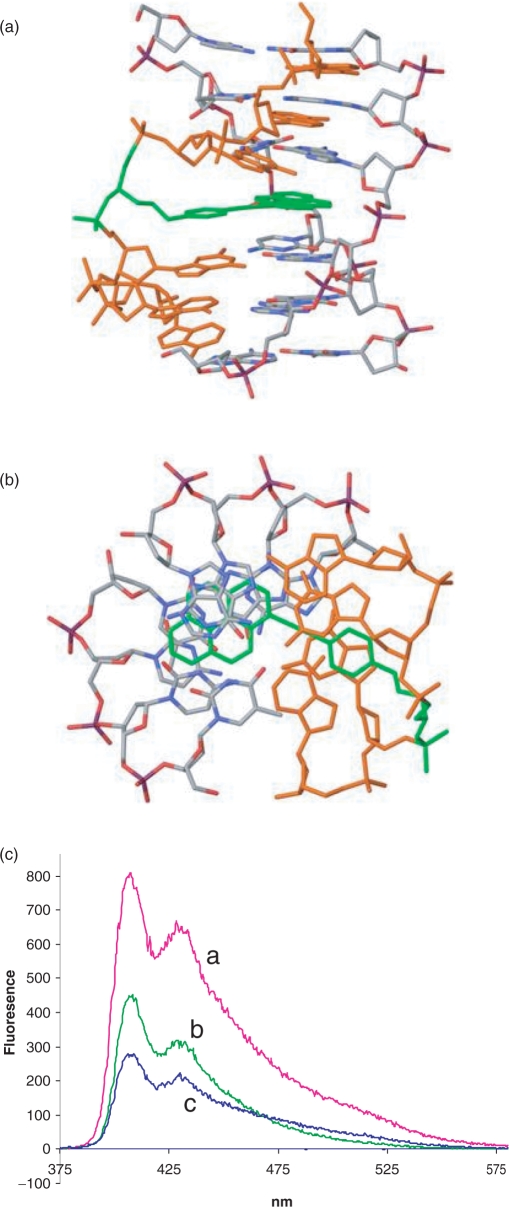
Structural calculation (MacroModel) of triplex DNA with TINA oligonucleotides as third strand shows that the pyrene moiety intercalates in the Watson–Crick duplex portion of the structure while the phenyl in the phenylmethyl glycerol arm is coaxial with the nucleobases of the third strand (Figure 7a and b). Thereby the TINA–monomer is mimicking the spatial void from a base triplet and adds to the triplex stability by stacking interactions. But as opposed to a base triplet the covalently bound pyrene moiety may anchor the TFO to the duplex resulting in a higher stability. It was also found that the intercalator twists around the triple bond by 1–10 degrees in the antiparallel triplex, depending on the structure selected from the cluster analysis, compared to 15.3 degrees for the monomeric structure (27), showing the intercalators’ ability to adjust secondary structural changes in order to attain an optimal fit. It was found that, unlike the TFO-backbone in the parallel triplex, the TFO-backbone in the antiparallel triplex was positioned in the middle of the major groove relative to the backbones of the duplex. This secondary structural property of the antiparallel triplex decreases the electrostatic repulsion between the negatively charged backbones, which is considered to be one of the main obstacles for the formation of the parallel triplex (40).
Figure 7.
(a, b) Calculated structures obtained by molecular modelling showing the intercalation of the pyrene moiety within the double helical portion of the triplex and the stacking of the phenyl between the nucleotides of the third strand (TFO); (c) Fluorescence spectra in 50 mM Tris–HCl, pH 7.4, 100 mM KCl of 500 nM 1037 (a), 500 nM 1037 + 500 nM 39-mer murine duplex (b), 500 nM 1037 + 1000 nM 39-mer murine duplex.
An indication that pyrene does intercalate into the double helix portion of the triplex was obtained by fluorescence experiments. When TINA-conjugated oligonucleotides are excited at 340 nm, a characteristic fluorescence spectrum with maxima at 400 and 421 nm is usually observed. We observed that the formation of antiparallel triplexes by guanine-rich TINA–TFOs with a 39-mer murine duplex resulted in fluorescence quenching. A typical quenching effect obtained by adding to a solution of oligonucleotide 1037 the 32-mer murine duplex is reported in Figure 7c. This is in keeping with the previous finding that the presence of guanine next to the pyrene induces fluorescence quenching (41). This result supports the conclusion from molecular modelling that pyrene is intercalated in the duplex portion of the structure.
DNA–protein interaction: competition experiments
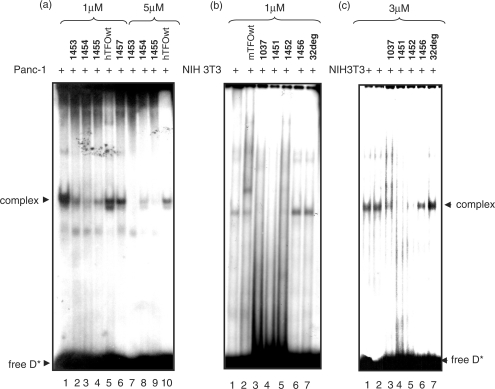
As the pur/pyr motif of KRAS used as target for the designed TINA–TFOs is a regulatory transcriptional element, we have searched for putative transcription factors binding to this critical sequence by using a computer program from Geno Matix, MatInspector (www.genomatix.de). It was found that the KRAS promoter region spanning over the pur/pyr motif should be recognized by three zing finger transcription factors: ZF5F, MZF1, ZBPF. There is indeed a high sequence homology (>90%) between the pur/pyr element and the consensus sequence of these proteins. When the 32-mer duplex corresponding to the human pur/pyr motif is incubated with nuclear extract, one main DNA–protein complex is observed by EMSA (Figure 8a). As this complex is competed away by excess unlabelled pur/pyr duplex but not by unrelated DNA duplexes (17-mer duplex with the Sp1 motif and DNA from plasmids pSVCAT, pTK-βgal), we concluded that the pur/pyr motif is bound by the nuclear proteins in a sequence-specific manner (data not shown). We have not yet identified the proteins present in this DNA–protein complex; however, they could be indeed by the abovementioned zing finger transcription factors. In order to see if the designed twisted-intercalating TFOs are able to block the binding of the nuclear factors to the regulatory pur/pyr motif, we performed competition experiments. Figure 8a shows that when the TINA–TFOs 1453, 1454 and 1455 are bound to the human pur/pyr sequence, they prevent the formation of the DNA–protein complex. As the nuclear extract contains 140 mM KCl, both wild-type hTFOwt and control TINA oligonucleotide 1457 did not abrogate protein binding. When the experiment was carried out with a higher TFO concentration (5 μM), the inhibition by the TINA TFOs was even stronger while hTFOwt was still found unable to inhibit protein binding. To rule out the possibility that the abrogation of the DNA–protein complex is due to direct binding of the TINA TFOs to the proteins, rather than being due to triplex formation, we labelled 1453, 1454 and 1455 with 32P, incubated them with crude extract and found that they did not bind to any protein (not shown). EMSA experiments were also carried out with the murine 39-mer pur/pyr duplex and crude extract from murine NIH 3T3 cells (Figure 8b). Also in this case we observed the formation of a DNA–protein complex. As happens with the human pur/pyr sequence, this complex did not form when the target was previously treated with the designed TINA TFOs 1037, 1451 and 1452. Instead, oligonucleotides hTFOwt, 1456 and 32deg did not interfere with protein binding, as is to be expected. We also investigated whether adding simultaneously TFO and extract to the DNA target, still results in the abrogation of the DNA–protein complex. Figure 8c shows that 1037, 1051 or 1052 (3 μM) are able to block the binding of the protein to the DNA target even when all the reagents are mixed in the reaction tube at the same time. Taken together these data provide strong evidence that the designed twisted-intercalating TFOs are able to block the binding of nuclear factors to a transcription regulatory pur/pyr element located in the promoter of the KRAS gene.
Figure 8.
EMSA showing the abrogation by the designed TINA TFOs of a DNA–protein complex C1 formed between the murine and human KRAS pur/pyr element and a nuclear extract. (a) DNA–protein interaction between a 32-mer human duplex (5′-AGGGCGGTGTGGGAAGAGGGAAGAGGGGGAGG; 5′-CCTCCCCCTCTTCCCTCTTCCCACACCGCCCT) and Panc-1 nuclear extract in the presence or absence of TFO. Loading: human target (10 nM) with 3 μg extract (lane 1); human target (10 nM) + 1 μM 1453, 1454, 1455, hTFOwt and 1457 incubated overnight at 37°C in binding buffer (see EMSA), then the mixtures treated with 3 μg extract for 2 h (lanes 2–6); human target (10 nM) + 5 μM 1453, 1454, 1455 and hTFOwt incubated overnight, then the mixtures treated for 2 h with 3 μg extract (lanes 7–10); (b) DNA–protein interaction between a 39-mer murine duplex (5′-CGCGCGGGAGGGAGGGAAGGAGGGAGGGAGGGAGCGGCT; 5′-AGCCGCTCCCTCCCTCCCTCCTTCC-CTCC-CTCCCGCGCG) and NIH 3T3 nuclear extract in the presence or absence of TFO. Loading: 39-mer murine duplex (10 nM) incubated with 12 μg NIH 3T3 extract for 2 h (lane 1); 32-mer duplex (10 nM) + 1 μM mTFOwt; 1037, 1451, 1452, 1456, 32deg incubated overnight in binding buffer at 37°C, then treated for 2 h with 12 μg extract (lanes 2–7); (c) 39-mer murine duplex (10 nM) added simultaneously with 12 μg NIH 3T3 extract and 3 μM 1037, 1451, 1452, 1456, 32deg and incubated for 2 h (lanes 3–7); duplex with only 12 μg NIH 3T3 extract (lanes 1 and 2). The sequence of 32deg is: 5′-GCATTCTGATTACACGTATTACCTTCACTCCA.
Transient transfection experiments
As the designed TINA–TFOs efficiently block protein binding to the regulatory pur/pyr motif of the KRAS gene which is essential for transcription, we investigated their capacity to down-regulate the activity of KRAS in NIH 3T3 cells. The cells were transfected with mixtures containing plasmid pKRS-413 (2 μg), bearing the CAT gene driven by the murine KRAS promoter, and either a TFOs or a control oligonucleotide (1 μM). In the transfection mixtures we also added plasmid PTK-βgal (50 ng) to evaluate transfection efficiency. The mixtures were transfected in NIH 3T3 cells by using a commercial polyethylenimine liposome. To quantify the expression of CAT and β-gal, we performed immunoassays with anti–CAT and anti-β-gal antibodies. The residual CAT expression in NIH 3T3 cells treated with the TFOs or control oligonucleotides is shown in Figure 9a. It can be seen that TINA–TFOs 1037, 1451 and 1452 reduced CAT expression by roughly 50%, whereas mTFOwt and control TINA-oligonucleotides 1456 and 1457 did not have any significant effect on CAT. Note that 1452 shows a lower capacity to inhibit CAT expression (40% inhibition) compared to 1037 (60%) and 1451 (50%), in keeping with a less favourable ΔG of triplex formation found for 1452 compared to 1037 and 1051 (Table 1). To check if the designed TINA TFOs promote aptameric effects, we introduced four G-to-A mutations in the murine pur/pyr sequence of plasmid pKRS-413. This brought about a change in the pur/pyr sequence but not in its polypurine character (Figure 8b). Despite these substitutions, the mutant plasmid, pKRS-413-4mut, maintained a capacity to express CAT. The mutated pur/pyr element should not be recognized by 1037, 1451 and 1452 TFOs, but by 2218 and by its unconjugated analogue mTFO-4mut, as these two oligonucleotides have been designed with a sequence specific for the mutated target (Figure 9b). Indeed, EMSA shows that 1037, 1451 and 1452 do not bind to the mutated target, whereas 2218 and mTFO-4mut do (Figure 9c). Contrary to mTFOwt, the mutant analogue mTFO-4mut binds to the target even in the presence of potassium, due to its lower G-content and therefore reduced tendency to aggregate. When pKRS-413-4mut was transfected with the designed TINA TFOs (1037, 1451 and 1452) in NIH 3T3 cells, we did not observe any CAT inhibition. But when it was transfected with 2218 (with which it forms a triplex), a CAT inhibition of about 40% was observed. This experiment indicates that the down-regulation of KRAS promoted by the designed TINA TFOs is likely mediated by triple helix formation at the promoter. These data, together with those obtained by EMSA, footprinting and DNA–protein interaction assays, suggest that TINA–TFOs, contrarily to the wild-type TFOs, have a potential as therapeutic antigene molecules.
DISCUSSION
In a recent study Vasquez and co-workers (1) reported that about 98% of human genes contain at least one high affinity TFO target in the promoter or, less frequently, in the coding region. This suggests that it is in principle possible to target most human genes with specific TFOs, which explains why triplex DNA could be an interesting approach to manipulate the genome.
However, even if pyrimidine oligonucleotides have been discovered to form triplex helical complexes at acidic conditions almost two decades ago, several problems still remain to be solved, and this limits the use of DNA triplex technology in vivo. One of the major obstacles is that triplex formation by G-rich oligonucleotides can be severely inhibited by physiological potassium concentrations (10–12). This is because TFOs containing guanine runs tend to self-associate into G-quartet structures (36,38,42), with the result that they are subtracted from the duplex-to-triplex equilibrium. Of the two types of TFOs, CT-oligonucleotides forming parallel Hoogsteen triplexes and AG/GT-oligonucleotides forming antiparallel reverse-Hoogsteen triplexes, the latter have the potential to bind to duplex DNA under physiological conditions. Indeed, several research groups have studied AG/GT oligonucleotides as antigene effector molecules in the context of anticancer therapy (43–49). But two major problems, i.e. oligonucleotide self-association and the susceptibility to endogenous nucleases, have up to now prevented TFOs from being used as therapeutic agents. There are several strategies to circumvent self-association by G-rich oligonucleotides, as reported in the literature: (i) incorporation of 6-thioguanine in a limited number of positions within purine oligonucleotides reduces G-quartet formation under physiological conditions, but it also slightly destabilizes the triplex (10); (ii) replacement of negative phosphodiester bonds with positively-charged phosphoramidite linkages may also favour triplex formation in the presence of KCl (11); (iii) hybridization of the TFO 3′ or 5′ ends to a short complementary oligonucleotide has shown to prevent aggregation without affecting triplex formation (12); (iv) replacement of guanine with 7-deazaguanine eliminates the ability of G-rich oligonucleotides to form Hoogsteen hydrogen-bond between the 2-amino and N7, which is essential for formation of G-quartets (50,51), but unfortunately the antiparallel triplex is destabilized due to a change in the atomic charge pattern (52). Recently, Filichev and Pedersen (27) have designed a new type of pyrimidine TFO containing inside its sequence one or two bulge insertions of (R)-1-O-[4-1-(1-pyrenylethynyl)-phenylmethyl] glycerol; i.e. an intercalating unit characterized by a flexible phenyl glycerol linker at the end of which a pyrene molecule is covalently attached. When bulge insertions were introduced in a parallel Hoogsteen-type CT triplex with a cytosine content of 36%, triplex formation was even observed at pH 7.2, whereas the unconjugated analogues were unable to bind to the target (27). Molecular modelling showed that the flexible phenylmethyl glycerol arm places the pyrenyl moiety into the dsDNA part of the triplex while the phenyl assumes a coaxial position with the nucleobases of the TFO. A twist around the triple helical bond allows the phenyl and pyrene to fit properly inside the triplex, promoting stabilizing stacking interactions. This is clearly illustrated in the structure shown in Figure 7. In our study this approach has been applied to antiparallel reverse-Hoogsteen triplex DNA. We discovered that G-rich TINA–TFOs form stable triple helical complexes under physiological conditions and, more surprising, that they were resistant to potassium-induced self-aggregation. We designed TINA–TFOs specific for a regulatory pur/pyr site in the murine and human KRAS genes. As this element is recognized by nuclear factors essential for transcription, it is an ideal target for TFOs. The TINA–TFOs were designed with two or three bulge insertions located either inside or adjacent to the guanine runs. Electrophoresis mobility-shift and footprinting experiments showed that twisted-intercalating TFOs, but not the wild-type analogues, bind to duplex DNA in the presence of potassium up to 140 mM. The different binding behaviour between TINA and wild-type TFOs correlate with the high tendency of the latter to self-associate into unusual G-quadruplex structures. Native electrophoresis shows that the murine mTFOwt, which has four AGGG repeats, forms a high molecular weight species in potassium: probably a quadruplex concatenamer of several strands which, according to CD, has a Tm of 71°C (Supplementary Data, S1). When however the TFO is modified with two or three bulge TINA insertions, it has a much lower tendency to aggregate, most likely because the guanine runs are disrupted by the pyrene intercalator (there should be 26 intercalating units per concatenamer of eight strands). The human hTFOwt having a less regular guanine distribution forms a structure that, according to electrophoresis, could be a dimeric parallel quadruplex with a Tm of 50°C (38). This structure is not formed by the TINA analogues 1453, 1454 and 1455 because the bulge insertions in these oligonucleotides strongly destabilize the quadruplex. It is remarkable that 1455, where the bulge insertions are placed outside or adjacent to the guanine runs, shows some aggregation. This suggests that when the pyrenyl insertions are put inside the guanine runs, they destabilize the G-quartets and reduce or abrogate self-aggregation, but when they are located adjacent to or outside the guanine runs, the formation of the G-quartet structure is only partially disturbed.
As regards the binding specificity of the designed TINA TFOs, the data presented here show that, though the TINA insertion is a non-sequence specific modification, the conjugates maintain the binding specificity typical of TFOs. The target with four G-to-A mutations is not recognized by the designed TINA–TFOs, and control TINA TFOs 1456 and 1457 do not bind to the pur/pyr sequence either. Moreover, we found that pyrimidine TINA TFOs containing two or three TINA insertions, just as the purine TFOs, did not bind to the murine and human KRAS targets at pH 7.4.
Molecular dynamic calculations show that the pyrenyl moieties intercalate into the double helix portion of the triplex promoting stacking interactions that further stabilize the structure. The conjugates 1453 and 1454 have bulge insertions between G3-G4 and G11-G12, but 1453 has one more insertion between A16-A17. EMSA shows that 1454 (ΔG = –9.8 kcal/mol) with only two insertions forms a more stable triplex than 1453 (ΔG = −8.8 kcal/mol), probably because the accommodation in the latter of three bulged 1-methylethynylpyrenes may slightly distort the structure of the triplex, thus destabilizing it. This comparison does not hold for the murine sequence, because the bulge insertions are located in different places in the various conjugates. A systematic study on the effect of the number and position of the bulge insertions on triplex formation will be necessary to gain more insight into this issue. We also synthesized for the murine KRAS pur/pyr target pyrimidine TINA–TFOs, which bind to DNA in the parallel fashion. Although TINA monomers stabilize parallel triplexes (27), the high cytosine content of the pyrimidine TINA–TFOs only allows triplex formation at pH 6, but not at pH 7, even though the TFOs contained 5-methylcytosine (53). This suggests that pur/pyr sites with a high C + G content (>50%) can be efficiently targeted only by purine TINA–TFOs.
The antigene activity of the TINA–TFOs was evaluated by co-transfecting NIH 3T3 cells with plasmid pKRS-413, which contains CAT driven by the KRAS promoter, and TINA–TFOs. Interestingly, murine TFOs 1037, 1451 and 1452 were found to inhibit roughly 50% CAT expression, compared to control 1456 and 1457. As this inhibition could be attributed to aptameric effects promoted by the TFOs against the proteins binding to the pur/pyr element, we mutated the pur/pyr sequence in pKRS-413 (by introducing four G-to-A mutations). The results obtained showed that the designed TINA–TFOs did not affect the CAT expression by the mutant plasmid, whereas CAT was significantly inhibited by the 2218, a TINA–TFO that binds to the mutant pur/pyr target. We have seen that the CAT inhibition promoted by the TINA–TFOs nicely correlates with their capacity to block protein binding to the KRAS pur/pyr site. This blocking was observed either when we added TFO and extract simultaneously to the target, and also when we allowed the triplex to be formed before the protein extract was added. Considering that the mean residence time on DNA of transcription factors should be in the order of few tens of seconds (54), the TFOs should not need to compete with existing DNA–protein complexes to inhibit transcription.
In conclusion, the present study opens new perspectives in the sequence-selective targeting of duplex DNA by using the triplex oligonucleotide technology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work has been carried out with the financial support of the Italian Ministry of Scientific Research (PRIN 2005), the Italian Association for Cancer Research (AIRC 2007), and Nucleic Acid Center, which is funded by The Danish National Research Foundation for studies on nucleic acid chemical biology. We thank Imrich Géci for the synthesis of oligonucleotide 2218. Funding to pay the Open Access publication charges for this article was provided by DSTB (University of Udine).
Conflict of interest statement. None declared.
REFERENCES
- 1.Wu Q, Gaddis SS, MacLeod MC, Walborg EF, Thames HD, DiGiovanni J, Vasquez KM. High-affinity triplex-forming oligonucleotide target sequences in mammalian genomes. Mol. Carcinog. 2007;46:15–23. doi: 10.1002/mc.20261. [DOI] [PubMed] [Google Scholar]
- 2.Belotserkovskii BP, De Silva E, Tornaletti S, Wang G, Vasquez KM, Hanawalt PC. A triplex-forming sequence from the human c-MYC promoter interferes with DNA transcription. J. Biol. Chem. 2007;282:32433–32441. doi: 10.1074/jbc.M704618200. [DOI] [PubMed] [Google Scholar]
- 3.Rogers FA, Lloyd JA, Glazer PM. Triplex-forming oligonucleotides as potential tools for modulation of gene expression. Curr. Med. Chem. Anticancer Agents. 2005;5:319–326. doi: 10.2174/1568011054222300. [DOI] [PubMed] [Google Scholar]
- 4.Majumdar A, Puri N, McCollum N, Richards S, Cuenoud B, Miller P, Seidman MM. Gene targeting by triple helix-forming oligonucleotides. Ann. NY Acad. Sci. 2003;1002:141–153. doi: 10.1196/annals.1281.013. [DOI] [PubMed] [Google Scholar]
- 5.Besch R, Giovannangeli C, Degitz K. Triplex-forming oligonucleotides – sequence-specific DNA ligands as tools for gene inhibition and for modulation of DNA-associated functions. Curr. Drug Targets. 2004;5:691–703. doi: 10.2174/1389450043345100. [DOI] [PubMed] [Google Scholar]
- 6.Moser HE, Dervan PB. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987;238:645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- 7.Manzini G, Xodo LE, Gasparotto D, Quadrifoglio F, van der Marel GA, van Boom JH. Triple helix formation by oligopurine-oligopyrimidine DNA fragments. Electrophoretic and thermodynamic behavior. J. Mol. Biol. 1990;213:833–843. doi: 10.1016/S0022-2836(05)80267-0. [DOI] [PubMed] [Google Scholar]
- 8.Francois JC, Saison-Behmoaras T, Helene C. Sequence-specific recognition of the major groove of DNA by oligodeoxynucleotides via triple helix formation. Footprinting studies. Nucleic Acids Res. 1988;16:11431–11440. doi: 10.1093/nar/16.24.11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beal PA, Dervan PB. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991;251:1360–1366. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- 10.Olivas WM, Maher L.J., 3rd Overcoming potassium-mediated triplex inhibition. Nucleic Acids Res. 1995;23:1936–1941. doi: 10.1093/nar/23.11.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dagle JM, Weeks DL. Positively charged oligonucleotides overcome potassium-mediated inhibition of triplex DNA formation. Nucleic Acids Res. 1996;24:2143–2149. doi: 10.1093/nar/24.11.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Svinarchuk F, Cherny D, Debin A, Delain E, Malvy C. A new approach to overcome potassium-mediated inhibition of triplex formation. Nucleic Acids Res. 1996;24:3858–3865. doi: 10.1093/nar/24.19.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 14.Wang G, Xu XS. Peptide nucleic acid (PNA) binding-mediated gene regulation. Cell Res. 2004;14:111–116. doi: 10.1038/sj.cr.7290209. [DOI] [PubMed] [Google Scholar]
- 15.Ziemba AJ, Zhilina ZV, Krotova-Khan Y, Stankova L, Ebbinghaus SW. Targeting and regulation of the HER-2/neu oncogene promoter with bis-peptide nucleic acids. Oligonucleotides. 2005;15:36–50. doi: 10.1089/oli.2005.15.36. [DOI] [PubMed] [Google Scholar]
- 16.Petersen M, Wengel J. LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol. 2003;21:74–81. doi: 10.1016/S0167-7799(02)00038-0. [DOI] [PubMed] [Google Scholar]
- 17.Brunet E, Alberti P, Perrouault L, Babu R, Wengel J, Giovannangeli C. Exploring cellular activity of locked nucleic acid-modified triplex-forming oligonucleotides and defining its molecular basis. J. Biol. Chem. 2005;280:20076–20085. doi: 10.1074/jbc.M500021200. [DOI] [PubMed] [Google Scholar]
- 18.Blommers MJ, Natt F, Jahnke W, Cuenoud B. Dual recognition of double-stranded DNA by 2′-aminoethoxy-modified oligonucleotides: the solution structure of an intramolecular triplex obtained by NMR spectroscopy. Biochemistry. 1998;37:17714–17725. doi: 10.1021/bi9816352. [DOI] [PubMed] [Google Scholar]
- 19.Gryaznov SM, Lloyd DH, Chen JK, Schultz RG, DeDionisio LA, Ratmeyer L, Wilson WD. Oligonucleotide N3′–>P5′ phosphoramidates. Proc. Natl Acad. Sci. USA. 1995;92:5798–5802. doi: 10.1073/pnas.92.13.5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Escudé C, Nguyen CH, Kukreti S, Janin Y, Sun JS, Bisagni E, Garestier T, Hélène C. Rational design of a triple helix-specific ligand. Proc. Natl Acad. Sci. USA. 1998;95:3591–3596. doi: 10.1073/pnas.95.7.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strekowski L, Hojjat M, Wolinska E, Parker AN, Paliakov E, Gorecki T, Tanious FA, Wilson WD. New triple-helix DNA stabilizing agents. Bioorg. Med. Chem. Lett. 2005;15:1097–1100. doi: 10.1016/j.bmcl.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 22.Puri N, Zamaratski E, Sund C, Chattopadhyaya J. Tetrahedron. 1997;53:10409–10432. [Google Scholar]
- 23.Mouscadet JF, Ketterle C, Goulaouic H, Carteau S, Subra F, Le Bret M, Auclair C. Triple helix formation with short oligonucleotide-intercalator conjugates matching the HIV-1 U3 LTR end sequence. Biochemistry. 1994;33:4187–4196. doi: 10.1021/bi00180a011. [DOI] [PubMed] [Google Scholar]
- 24.Mohammadi S, Slama-Schwok A, Leger G, el Manouni D, Shchyolkina A, Leroux Y, Taillandier E. Triple helix formation and homologous strand exchange in pyrene-labeled oligonucleotides. Biochemistry. 1997;36:14836–14844. doi: 10.1021/bi971710t. [DOI] [PubMed] [Google Scholar]
- 25.Cheng E, Asseline U. Synthesis and binding properties of perylene-oligo-2′-deoxyribonucleotide conjugates. Tetrahedron Lett. 2001;42:9005–9010. [Google Scholar]
- 26.Jessen CH, Pedersen EB. Design of an intercalating uniut linker leading to the first efficiently 5′,5′-linked alternate-strand Hoogsteen triplex with high stability and specificity. Helv. Chim. Acta. 2004;87:2465–2471. [Google Scholar]
- 27.Filichev VV, Pedersen EB. Stable and selective formation of hoogsteen-type triplexes and duplexes using twisted intercalating nucleic acids (TINA) prepared via postsynthetic Sonogashira solid-phase coupling reactions. J. Am. Chem. Soc. 2005;127:14849–14858. doi: 10.1021/ja053645d. [DOI] [PubMed] [Google Scholar]
- 28.Filichev VV, Gaber H, Olsen TR, Jorgensen PT, Jessen CH, Pedersen EB. Twisted intercalating nucleic acids – intercalator influence on parallel triplex stabilities. Eur. J. Organ. Chem. 2006;17:3960–3968. [Google Scholar]
- 29.Tang J-Y, Han Y, Tang JX, Zhang Z. Large scale synthesis of oligonucleotide phosphorothioates using 3-amino-1,2,4-dithiazole-5-thione as an efficient sulphur-transfer reagent. Organic Process Res. Dev. 2000;4:194–198. [Google Scholar]
- 30.Radhakrishnan I, Patel DJ. Solution structure of a purine·purine·pyrimidine DNA triplex containing G·GC and T·AT triples. Structure. 1993;1:135–152. doi: 10.1016/0969-2126(93)90028-f. [DOI] [PubMed] [Google Scholar]
- 31.Hoffman EK, Trusko SP, Freeman N, George DL. Structural and functional characterization of the promoter region of the mouse c-Ki-ras gene. Mol. Cell Biol. 1987;7:2592–2596. doi: 10.1128/mcb.7.7.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffman EK, Trusko SP, Murphy M, George DL. An S1 nuclease-sensitive homopurine/homopyrimidine domain in the c-Ki-ras promoter interacts with a nuclear factor. Proc. Natl Acad. Sci. USA. 1990;87:2705–2709. doi: 10.1073/pnas.87.7.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jordano J, Perucho M. Chromatin structure of the promoter region of the human c-K-ras gene. Nucleic Acids Res. 1986;14:7361–7378. doi: 10.1093/nar/14.18.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cogoi S, Ballico M, Bonora GM, Xodo LE. Antiproliferative activity of a triplex-forming oligonucleotide recognizing a Ki-ras polypurine/polypyrimidine motif correlates with protein binding. Cancer Gene Ther. 2004;11:465–476. doi: 10.1038/sj.cgt.7700722. [DOI] [PubMed] [Google Scholar]
- 35.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng AJ, Wang JC, Van Dyke MW. Self-association of G-rich oligonucleotides under conditions promoting purine-motif triplex formation. Antisense Nucleic Acid Drug Dev. 1998;8:215–225. doi: 10.1089/oli.1.1998.8.215. [DOI] [PubMed] [Google Scholar]
- 37.Kejnovska I, Kypr J, Vorlickova M. Circular dichroism spectroscopy of conformers of (guanine + adenine) repeat strands of DNA. Chirality. 2003;15:584–592. doi: 10.1002/chir.10249. [DOI] [PubMed] [Google Scholar]
- 38.Rujan IN, Meleney JC, Bolton PH. Vertebrate telomere repeat DNAs favor external loop propeller quadruplex structures in the presence of high concentrations of potassium. Nucleic Acids Res. 2005;33:2022–2031. doi: 10.1093/nar/gki345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hazel P, Huppert J, Balasubramanian S, Neidle S. Loop-length-dependent folding of G-quadruplexes. J. Am. Chem. Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 40.Vasquez KM, Glazer PM. Triplex-forming oligonucleotides: principles and applications. Q. Rev. Biophys. 2002;35:89–107. doi: 10.1017/s0033583502003773. [DOI] [PubMed] [Google Scholar]
- 41.Manoharan M, Tivel KL, Zhao M, Nafisi K, Netzel TL. Base-Sequence Dependence of Emission Lifetimes for DNA Oligomers and Duplexes Covalently Labeled with Pyrene – Relative Electron-Transfer Quenching Efficiencies of A-Nucleoside, G-Nucleoside, C-Nucleoside, and T-Nucleoside toward Pyrene. J. Phys. Chem. 1995;99:17461–17472. [Google Scholar]
- 42.Fry M. Tetraplex DNA and its interacting proteins. Front Biosci. 2007;12:336–351. doi: 10.2741/2391. [DOI] [PubMed] [Google Scholar]
- 43.Cheng K, Ye Z, Guntaka RV, Mahato RI. Biodistribution and hepatic uptake of triplex-forming oligonucleotides against type alpha1(I) collagen gene promoter in normal and fibrotic rats. Mol. Pharm. 2005;2:206–217. doi: 10.1021/mp050012x. [DOI] [PubMed] [Google Scholar]
- 44.Cheng K, Ye Z, Guntaka RV, Mahato RI. Enhanced hepatic uptake and bioactivity of type alpha1(I) collagen gene promoter-specific triplex-forming oligonucleotides after conjugation with cholesterol. J. Pharmacol. Exp. Ther. 2006;317:797–805. doi: 10.1124/jpet.105.100347. [DOI] [PubMed] [Google Scholar]
- 45.Hewett PW, Daft EL, Laughton CA, Ahmad S, Ahmed A, Murray JC. Selective inhibition of the human tie-1 promoter with triplex-forming oligonucleotides targeted to Ets binding sites. Mol. Med. 2006;12:8–16. doi: 10.2119/2005-00046.Hewett. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carbone GM, Napoli S, Valentini A, Cavalli F, Watson DK, Catapano CV. Triplex DNA-mediated downregulation of Ets2 expression results in growth inhibition and apoptosis in human prostate cancer cells. Nucleic Acids Res. 2004;32:4358–4367. doi: 10.1093/nar/gkh744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carbone GM, McGuffie EM, Collier A, Catapano CV. Selective inhibition of transcription of the Ets2 gene in prostate cancer cells by a triplex-forming oligonucleotide. Nucleic Acids Res. 2003;31:833–843. doi: 10.1093/nar/gkg198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stütz AM, Hoeck J, Natt F, Cuenoud B, Woisetschläger M. Inhibition of interleukin-4- and CD40-induced IgE germline gene promoter activity by 2′-aminoethoxy-modified triplex-forming oligonucleotides. J. Biol. Chem. 2001;276:11759–11765. doi: 10.1074/jbc.M010260200. [DOI] [PubMed] [Google Scholar]
- 49.Rapozzi V, Cogoi S, Spessotto P, Risso A, Bonora GM, Quadrifoglio F, Xodo LE. Antigene effect in K562 cells of a PEG-conjugated triplex-forming oligonucleotide targeted to the bcr/abl oncogene. Biochemistry. 2002;41:502–510. doi: 10.1021/bi011314h. [DOI] [PubMed] [Google Scholar]
- 50.Murchie AIH, Lilley DMJ. Retinoblastoma susceptibility genes contain 5′ sequences with a high propensity to form guanine-tetrad structures. Nucleic Acids Res. 1992;20:49–53. doi: 10.1093/nar/20.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seela F, Mersmann K. 7-Deazaguanosne: synthesis of an oligoribonucleotide building block and disaggregation of the U-G-G-G-G-U G4 structure by the modified base. Helv. Chim. Acta. 1993;76:1435–1449. [Google Scholar]
- 52.Milligan JF, Krawczyk SH, Wadwani S, Matteucci MD. An anti-parallel triple helix motif with oligodeoxynucleotides containing 2′-deoxyguanosine and 7-deaza-2′-deoxy-xanthosine. Nucleic Acids Res. 1993;21:327–333. doi: 10.1093/nar/21.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xodo LE, Manzini G, Quadrifoglio F, Vandermarel GA, Vanboom JH. Effect of 5-methylcytosine on the stability of triple-stranded dna - a thermodynamic study. Nucleic Acids Res. 1991;19:5625–5631. doi: 10.1093/nar/19.20.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paroni G, Fontanini A, Cernotta N, Foti C, Gupta MP, Yang X.-J, Fasino D, Brancolini C. Dephosphorilation and caspase processing generate distinct nuclear pools of histone deacetylase. Mol. Cell Biol. 2007;27:6718–6732. doi: 10.1128/MCB.00853-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.