Abstract
Somatic genome assembly in the ciliate Paramecium involves the precise excision of thousands of short internal eliminated sequences (IESs) that are scattered throughout the germline genome and often interrupt open reading frames. Excision is initiated by double-strand breaks centered on the TA dinucleotides that are conserved at each IES boundary, but the factors that drive cleavage site recognition remain unknown. A degenerate consensus was identified previously at IES ends and genetic analyses confirmed the participation of their nucleotide sequence in efficient excision. Even for wild-type IESs, however, variant excision patterns (excised or nonexcised) may be inherited maternally through sexual events, in a homology-dependent manner. We show here that this maternal epigenetic control interferes with the targeting of DNA breaks at IES ends. Furthermore, we demonstrate that a mutation in the TA at one end of an IES impairs DNA cleavage not only at the mutant end but also at the wild-type end. We conclude that crosstalk between both ends takes place prior to their cleavage and propose that the ability of an IES to adopt an excision-prone conformation depends on the combination of its nucleotide sequence and of additional determinants.
INTRODUCTION
Developmentally programmed DNA elimination has been documented in a variety of organisms ranging from bacteria to humans. In a few cases, such elimination events have been associated with the assembly of functional reading frames. In vertebrates, for instance, the large repertoire of immunoglobulin genes is generated by excision of noncoding intervening sequences, followed by imprecise closure of chromosomal excision sites (1). Ciliates exhibit the unique property of rearranging their whole genome at each sexual cycle (2,3). This stems from the coexistence, in the cytoplasm of these unicellular organisms, of two different types of nuclei. The diploid micronucleus, transcriptionally silent during vegetative growth, undergoes meiosis at each sexual cycle and, following fertilization and karyogamy, transmits the germline genome to the zygotic nucleus of the next generation. In contrast, the highly polyploid (800–1000 n) somatic macronucleus, although responsible for all gene transcription in the cell, is destroyed during these processes and a new macronucleus differentiates from a mitotic product of the zygotic nucleus. Macronuclear development is characterized by extensive DNA amplification. Most notably, genome-wide and programmed DNA rearrangements take place in the developing macronucleus to yield the mature somatic genome. In Paramecium tetraurelia, two modes of DNA elimination have been reported (4). Imprecise elimination of sequences ranging up to several kilobases in length can lead either to internal deletions between variable boundaries or to chromosome fragmentation followed by addition of telomeric repeats to the heterogeneous new chromosome ends (5). Such eliminated sequences often contain repeated elements, such as transposons or minisatellites (5). Moreover, imprecise elimination of unique sequences can be induced experimentally and maintained through maternal inheritance in the following sexual generations (6). The second type of DNA rearrangement is the precise excision of single-copy, short (26–882 bp) noncoding sequences, also called IESs (internal eliminated sequences). Although only the rearranged version of the genome present in the macronucleus has been sequenced (7), analysis of a few known micronuclear loci indicates that IESs are extremely numerous (an estimated ∼60 000 per haploid genome), scattered throughout the genome—both in coding and noncoding regions—and probably interrupt most open reading frames in the micronucleus (8).
Paramecium IESs are A+T-rich and flanked by two TA dinucleotides, a single copy of which is retained at their excision site in the macronuclear genome. Their programmed excision takes place after 3–4 rounds of DNA replication have taken place in the developing macronucleus (9). Excision starts with a 4-bp staggered double-strand DNA cleavage centered on the conserved 5′-TA-3′ at each end (10). We proposed that precise closure of excision sites is achieved through highly controlled joining of broken chromosome ends, which involves pairing of the TA dinucleotides embedded within the 5′ overhangs generated by DNA breakage (Figure 1). Limited end processing is thought to take place in this paired-end intermediate, with removal of the 5′ terminal residue and polymerization of one nucleotide to the 3′ recessive end prior to a final ligation step. The enzyme(s) involved in DNA cutting have not been identified and the way IES ends are targeted for cleavage still awaits further elucidation. Statistical analysis of the nucleotide sequence of ∼80 IESs led to an 8-bp degenerate consensus sequence 5′-TA(C/T)AG(C/T)N(A/G)-3′, which includes the flanking TA dinucleotide and defines loosely conserved terminal inverted repeats at IES ends (8,11). Genetic studies indicated that base substitutions in the TA or at the fifth internal position of the consensus can abolish excision (12–16). However, the nucleotide sequence may not be the sole determinant for IES elimination, since several lines of evidence have pointed to epigenetic maternal control of this process in Paramecium (17,18). This was documented for a subset of IESs, the excision of which is inhibited in the new macronucleus by the presence of homologous nonexcised copies in the old macronucleus. Maternal control can eventually lead to the emergence of stable macronuclear variant cell lines, in which a given IES is not excised, although its nucleotide sequence is fully wild-type and while all other IESs are excised normally. Maternally controlled IESs may represent up to one-third of all IESs but the specific features that distinguish them from other Paramecium IESs are still unclear (18).
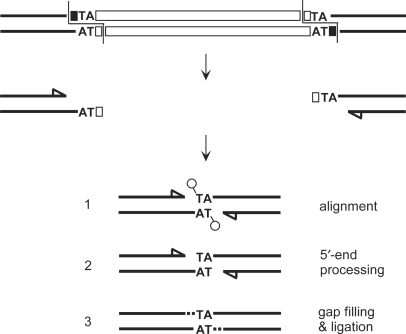
Figure 1.
Model for the precise repair of IES excision sites in Paramecium. IES excision involves the introduction of 4-base staggered double-strand breaks, one at each IES end (10). IES sequences are represented by white boxes and flanking DNA is shown in black. 5′ terminal nucleotides are drawn as circles, recessive 3′ ends as arrowheads. Broken chromosome ends are thought to align within a junction repair intermediate through the pairing of their central TA dinucleotides (Step 1). Macronuclear junction assembly results from processing of the 5′ flapped nucleotides (Step 2), fill-in of the 3′ recessed ends (dotted lines in Step 3) and ligation.
In this study, we focused on the hypothesis that an additional feature, such as DNA conformation, may participate in IES recognition. To investigate whether both ends of an IES interact early during excision, we examined whether they are cut in a concerted manner. For this purpose, we followed IES end cleavage during macronuclear development in three available mutant strains, each of which carries a point mutation within the TA dinucleotide at one end of a particular IES: 51A1835 (28 bp) in strain AIM-5 (15), 51A6649 (370 bp) in AIM-2 (14) and sm19-576 (66 bp) in d4-2 sm19-1 (16). Previous work done by others had established that each mutant IES is retained in the macronucleus. Here, using ligation-mediated PCR (LMPCR), we show that these mutations do not only inhibit DNA cleavage at the mutant end, but also severely impair that of the wild-type end. This study leads us to speculate on the relative contributions of nucleotide sequence, DNA structure and maternal control to the precise targeting of IES end cleavage.
MATERIALS AND METHODS
Paramecium tetraurelia strains and growth conditions
Paramecium tetraurelia stock 51 new was described in (10) and carries the wild-type A51 surface antigen gene. Stock d4-2 is largely isogenic but carries the A29 allele. A derivative of d4-2 harboring the sm19-1 mutation (16) was kindly provided by F. Ruiz. An IES+ macronuclear variant of 51 new was obtained as described in (17), after transformation of the vegetative macronucleus with plasmid p851 and selection of postautogamous cells, in which IES 51G4404 was retained in all macronuclear copies of the surface antigen G51 gene. Homozygous strains AIM-2 and AIM-5 were obtained from a cross between 51 new and 51 stocks carrying the AIM-2 (14) or AIM-5 (15) mutations in A51, respectively. Macronuclear variants with a deletion of the A gene were obtained as described in (6). Approximately 5 pl of a 5 mg/ml solution of linearized plasmid p29A-SX carrying the A29 gene on a SalI–XhoI fragment were injected into the vegetative macronucleus of 51 new or AIM-2 cells. The same procedure was applied to strain AIM-5 with plasmid p51A-SB carrying the A51 gene on a SalI–BbsI fragment. Following autogamy of injected cells, deletion of the A gene from the zygotic macronucleus was checked by PCR amplification of the region encompassing different IESs (Supplementary Figures S1 and S2). This led to stocks 51ΔA, AIM-2ΔA and AIM-5ΔA, in which the macronuclear deletion is maintained by maternal inheritance through autogamy.
For time-course analyses, a ∼3.6-l culture of each strain was grown at 27°C (9) and autogamy of starved cells was monitored by DAPI staining. The T0 time-point was arbitrarily chosen as the time when the percentage of cells with fragmented maternal macronucleus reached ∼50%. More details about the cytological states of the cultures at each time-point can be found in Supplementary Figure S3. For molecular studies, total genomic DNA was extracted from 200 to 400 ml aliquots of the cultures as described in ref. (10).
PCR and detection of broken DNA ends
All enzymes were purchased from New England Biolabs, Massachussetts, USA, except for Taq DNA polymerase Sequencing Grade (Promega, Wisconsin, USA) and DyNAzyme II DNA polymerase (Finnzymes, Finland). For specific applications, the Expand Long Template PCR System (Roche, Switzerland) was used following the supplier's recommendations, with buffer 2 and final concentrations of 750 µM dATP, 750 µM dTTP, 250 µM dGTP and 250 µM dCTP.
Oligonucleotides were purchased from MWG Biotech, Germany, Genaxis, France or Sigma–Aldrich, France. Paramecium specific primers are listed in Supplementary Table 1.
Poly(dG)-tailing of free 3′OH ends was as described in ref. (10). So was LMPCR detection of double-strand breaks, except that oligonucleotide 5′-GAATTCGGATCCGCTCGGACCGTGGC-3′ (PCRhaut) was used in combination with a Paramecium specific primer for the amplification of ligation products. This modification was found to reduce nonspecific background.
Sequencing templates were amplified from agarose gel slices or, for some primer extension products, from dried sequencing gels (10). Samples and primers were sent to MWG Biotech AG for automated Value Read sequencing. For the generation of size ladders, DNA sequencing reactions were performed manually using the fmol DNA Cycle Sequencing System (Promega) and 33P-labeled primers.
RESULTS
Detection of single-end cleaved molecules for wild-type IESs
We concentrated on IESs 51A1835 and 51A6649 located in surface antigen A51 gene (Figure 2) to examine whether wild-type IESs could be cleaved at one end only during macronuclear development, while remaining attached to flanking chromosomal DNA at their opposite end. For this purpose, we slightly modified our original LMPCR procedure for the detection of transient DNA double-strand breaks at IES ends (10 and Figure 3A). Specific LMPCR products could be detected in P. tetraurelia cells undergoing a self-fertilization process called autogamy (Figure 3B), with exactly the same time course as the chromosomal double-strand breaks generated at each excision site (see 51 new in Figure 6). The structure of these molecules was confirmed by DNA sequencing of the bands cut from the gels and amplified by PCR. Only one sequence was recovered from each band, which corresponded to a single-end cleaved product carrying a full-size 4-base 5′-overhang (not shown). Thus, in contrast to the double-strand breaks generated at chromosomal excision sites and at the ends of excised IESs, for which both 4-base and 3-base 5′ overhangs are observed (10), single end-cleaved molecules do not exhibit 5′-end processing. Although the exact nature of these products is unclear (they could be excision intermediates on their way to second end cleavage or abortive excision products), our data indicate that the two ends of an IES may not be cut simultaneously. This does not necessarily imply, however, that each end is recognized and cleaved independently from the other. Furthermore, our experiments do not reveal any substantial difference in the relative cleavage order of the two ends.
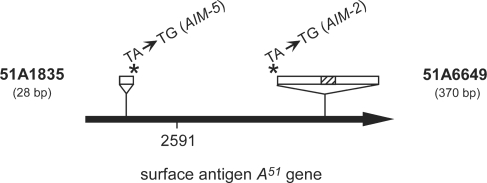
Figure 2.
Schematic map of P. tetraurelia gene A51. IESs 51A1835 and 51A6649 (white rectangles) are displayed on top (not to scale), with the hatched box representing a 29-bp internal IES embedded within IES 51A6649. The position of each point mutation is marked by an asterisk and the name of the corresponding mutant strain is indicated between brackets. The position of IES 51A2591, used as a wild-type control in this work, is also indicated.
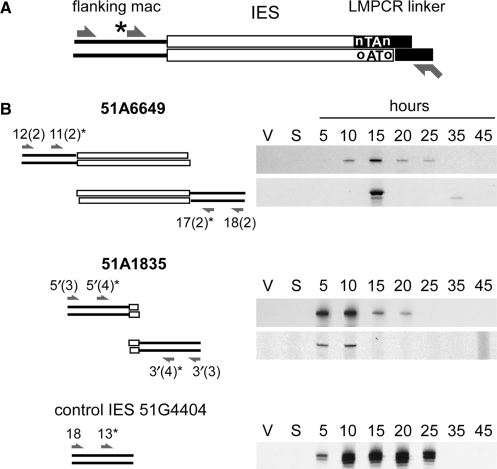
Figure 3.
Detection of single-end cleaved IES molecules during macronuclear development. (A) Experimental strategy. A standard LMPCR linker (black boxes with 5′-nTAn overhang) is ligated to the complementary broken end of an IES (white boxes with 5′-oTAo overhang). Ligation products corresponding to single-end cleaved molecules are selectively amplified with linker-specific primer PCRhaut (shown at the bottom) and a second primer (arrow on top) hybridizing within the macronuclear-destined sequences (black lines) that flank the opposite IES boundary. Primer extension is performed with a nested specific primer (marked with an asterisk). (B) LMPCR detection of single-end cleaved molecules for IESs 51A6649 and 51A1835 during autogamy of strain 51 new. The molecules detected in each assay are drawn on the left of each panel, with IESs represented by white boxes and their flanking DNA by black lines. For clarity, the linkers used in the first ligation step are not represented: I′/(GTAG)J′ was used for both ends of IES 51A6649, and I′/(ATAT)J′ and I′/(ATAA)J′ were used for the detection of molecules cleaved at the right or left end of IES 51A1835, respectively. Oligonucleotides used for PCR and primer extension are indicated by arrows. Bottom panel: detection of broken chromosome ends at the left boundary of control IES 51G4404 with linker I′/(GTAT)J′.
Figure 6.
Inhibition of DNA double-strand cleavage at the wild-type end of mutant IESs. Linkers I′/(CTAC)J′ and I′/(TTAT)J′ were used for LMPCR analysis of IES 51A6649 right end (top) and IES 51A1835 left end (bottom), respectively. Strain names and autogamy time-points are indicated on top of each panel, together with the oligonucleotides used for PCR and primer extension (asterisk). Broken chromosome ends at the left end of control IES 51A2591 were detected as described in Figures 3 and 4.
Maternal control of IES end cleavage
The observation that IESs can be cleaved at one end only justifies our strategy to examine whether the wild-type end of an IES can be cleaved in the presence of a point mutation at the other end. Formally, as an additional difficulty to our study, inhibition of IES excision during macronuclear development in mutant cell lines could be due to the superimposition of two factors: cis-effect of the mutated TA and maternal control due to the presence of homologous IES copies in the old macronucleus. The effect of maternal control on the excision reaction has not been described at the molecular level, although it was clearly shown to be independent of the presence of any point mutation in the IES (17,18). Inhibition could act early, during recognition and cleavage of IES ends, or later, during closure of the excision site, to restore the germline sequence in the new macronucleus. We tested the hypothesis that maternal control interferes with the earliest steps of the reaction (e.g. recognition or cleavage).
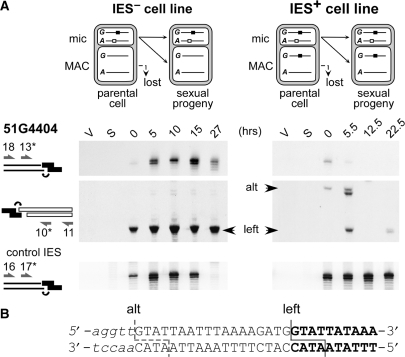
To compare DNA cleavage patterns in a wild-type (IES−) cell line and an IES+ macronuclear variant, we chose to study IES 51G4404 from the surface antigen G51 gene because of its high sensitivity to maternal control, which allows the establishment of a stable IES+ cell line (17). During macronuclear development in the IES+ variant, far fewer double-strand breaks were detected by LMPCR at 51G4404 left end than in the IES− cell line (Figure 4A), while similar levels of DNA cutting were observed in both lines at the left end of a control IES from surface antigen A51 gene (bottom panels). In a sporadic manner, we could detect faint bands of higher molecular weights in the IES+ cell line, the sequencing of one of which (‘alt’) suggested that ligation of the linker had occurred at an alternative position, 18-bp upstream of the left end (Figure 4B). At the right end of IES 51G4404, no cleavage at all was detected in the IES+ cell line (data not shown). These results indicate that the presence of a maternally controlled IES in the parental macronucleus strongly impinges on the efficiency, and perhaps also on the accuracy, of DNA cleavage at the ends of the homologous IES in the new macronucleus, even in the absence of any mutation in its nucleotide sequence.
Figure 4.
Maternal control of IES end cleavage in an IES+ cell line. (A) LMPCR detection of DNA double-strand breaks at the left boundary of IES 51G4404 during autogamy of a wild-type IES− cell line (left) and its IES+ macronuclear variant (right). Top panels: broken chromosome ends revealed following ligation of linker I′/(GTAT)J′. Middle panels: broken IES ends analyzed with linker I′/(ATAC)J′. Flanking chromosomal sequences are drawn as thin lines, IES sequences as white rectangles and each linker is shown in black. Oligonucleotides are represented by arrows. Bottom panels: broken chromosome ends at the left boundary of control IES 51A2591 analyzed with linker I′/(GTAT)J′. (B) Sequence analysis of band ‘alt’ identified at the left end of IES 51G4404. Bold letters represent the sequence ligated to the linker in the wild-type LMPCR product (‘left’), additional nucleotides found in the ‘alt’ product are in uppercase and upstream flanking sequences in lower case. The positions of the deduced double-strand breaks are shown.
A point mutation within a flanking TA impairs DNA cleavage at the mutant end
Coming back to our initial question, and because IES 51A6649 excision is maternally controlled (18), we removed mutant IES 51A6649 from the macronucleus of stock AIM-2 by injecting high-copy numbers of an A transgene into the macronucleus of vegetative cells, as described previously (6). Following autogamy of injected cells, we recovered a deletion of the entire A gene in the zygotic macronucleus, while the mutant allele was still present in the micronucleus. The induced macronuclear deletion was inherited through autogamy, leading to variant cell line AIM-2ΔA (see Materials and methods section). A 51ΔA cell line was derived from wild-type stock 51 new using the same procedure. We then examined the effect of a TA to TG mutation on the cleavage of IES 51A6649 mutant left end in these four strains, using wild-type IES 51A2591 as a positive control of excision in the macronuclear variants. As shown in Figure 5A (bottom), no significant difference was observed for the control IES between the LMPCR patterns obtained in strains 51 new or AIM-2 and in their respective variants deleted for the A gene in their macronucleus. This indicates that the lack of a pre-existing rearranged version of the A gene in the parental macronucleus does not prevent double-strand cleavages from being targeted precisely at the ends of a wild-type IES during development of a new macronucleus. In cell lines deleted for the macronuclear A gene, we could even detect the precise closure of IES excision junctions at the rearranged locus prior to maternally inherited complete elimination of the A gene (Supplementary Figure S2).
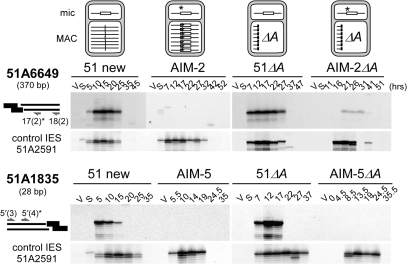
Figure 5.
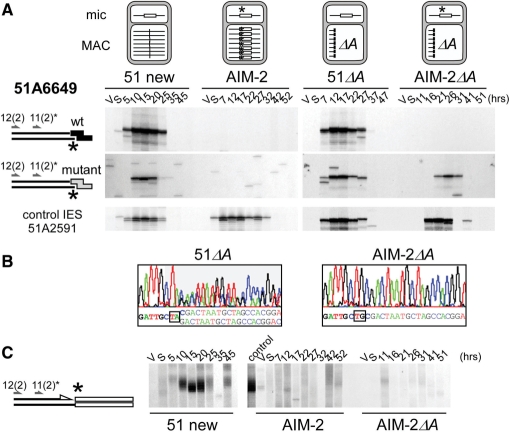
Effect of a germline mutation at the left end of IES 51A6649 on the cleavage of the mutant end. (A) LMPCR detection of broken chromosome ends in strains 51 new, AIM-2, 51ΔA and AIM-2ΔA, using linkers I′/(CTAC)J′ (wt: top panels) or I′/(CTGC)J′ (mutant: middle panels). The structure of the A locus in the maternal macronucleus of each strain is shown on top of the figure, with IES 51A6649 drawn as a white box and its mutant end marked with an asterisk. Bottom panels: broken chromosome ends at the left end of control IES 51A2591 analyzed as in Figure 3. Autogamy time-points are indicated on top of the figure. (B) En masse sequencing of the re-amplified LMPCR products obtained following ligation of mutant linker I′/(CTAC)J′ to the T17 and T26 genomic DNA samples from 51ΔA and AIM-2ΔA, respectively. Oligonucleotide 51A6649-11(2) was used as a sequencing primer. (C) Terminal transferase-mediated poly(dG) tailing of the free 3′ hydroxyl ends on the macronucleus-destined side of IES 51A6649 left boundary, during autogamy of strains 51 new, AIM-2 and AIM-2ΔA. As a control, the T20 sample from 51 new was loaded next to the AIM-2 samples. Oligonucleotides used for PCR and primer extension are indicated by arrows.
In a first set of experiments, we used double-stranded LMPCR linkers with 5′-overhangs complementary to IES 51A6649 wild-type left end. As expected, similar amounts of double-strand breaks accumulate transiently in wild-type stocks 51 new and 51ΔA (Figure 5A top), with the same timing as for control IES 51A2591. In contrast, no specific LMPCR band was detected at IES 51A6649 mutant end in strains AIM-2 or AIM-2ΔA. In a second step, taking into consideration that a linker with a wild-type 5′-CTAC overhang would hybridize less efficiently to an IES mutant end, we repeated the experiment with an LMPCR linker carrying a 5′-CTGC extension fully complementary to the putative mutant broken end (Figure 5A, middle). Use of the mutant linker still revealed developmental double-strand breaks at the left end of wild-type IES 51A6649 in strains 51 new and 51ΔA. For the mutant IES, while no specific LMPCR band was apparent in strain AIM-2, double-strand breaks were detected at the mutant end in the AIM-2ΔA variant, albeit with an ∼5 h delay relative to control IES 51A2591 (Figure 5A, middle and bottom right). LMPCR products obtained in strains 51ΔA and AIM-2ΔA were cut from the gels and PCR amplified prior to DNA sequencing. A double sequence indicative of efficient 5′-end processing of the broken end was recovered from the 51ΔA sample (Figure 5B, left). In contrast, although DNA cleavage at the broken left end of mutant IES 51A6649 in strain AIM-2ΔA had occurred at the same position as for the wild-type end, no evidence for processing was obtained (Figure 5B, right).
As no LMPCR primer is appropriate to compare directly the efficiency of DNA cleavage at IES 51A6649 mutant and wild-type left ends, we followed the formation of free 3′-hydroxyl DNA ends at the left boundary of IES 51A6649 by terminal transferase-mediated poly(dG) tailing (10), an approach which should be biased in favor of neither the mutant nor the wild-type end. Specific free 3′ ends accumulated transiently during autogamy in stock 51 new (Figure 5C, left), with the same time course as the double-strand breaks detected by LMPCR (Figure 5A, top left). In contrast, little or no signal was observed in mutant strains AIM-2 or AIM-2ΔA (Figure 5C, middle and right). Thus, although delayed double-strand cleavage at the mutant end of IES 51A6649 can be detected in a sensitive LMPCR assay, the TA to TG mutation severely impinges on cleavage efficiency. As judged by the failure to detect any LMPCR product in strain AIM-2, the presence of the mutant IES in the maternal macronucleus strengthens the inhibitory effect of the point mutation. For IES 51A1835, the excision of which is not submitted to maternal control, cleavage of the mutant end was abolished in strain AIM-5 to the same extent as in its macronuclear variant AIM-5ΔA (Supplementary Figure S4). These observations confirm the essential role of the conserved TA dinucleotide on the efficiency of DNA cleavage at IES ends.
A point mutation at one IES end inhibits cleavage at the wild-type end
We then examined whether a mutation at one IES end could interfere at a distance with the cleavage of the other (wild-type) end of the same IES. Here again, strains AIM-5 and AIM-2 and their variants deleted from the A51 gene in their macronucleus were used in parallel for the study of IESs 51A1835 and 51A6649, respectively. No specific cleavage of the right (wild-type) end of IES 51A6649 could be detected in mutant strain AIM-2, while some cutting was revealed during macronuclear development in the AIM-2ΔA variant (Figure 6). In these experiments, no hybridization bias of the LMPCR linker is expected at the right end of the mutant or the wild-type IES, because the AIM-2 mutation lies within the left end. We can therefore conclude that specific cleavage of the right, wild-type, end of IES 51A6649 is reduced in strain AIM-2ΔA and completely abolished in strain AIM-2. Furthermore, the same 5 h delay relative to control IES 51A2591 is observed in the timing of residual double-strand cleavage at both ends of the mutant IES in strain AIM-2ΔA (Figures 5A and 6). DNA sequencing of LMPCR products indicates that no 5′ end processing takes place following cleavage of either end of mutant IES 51A6649 (Figure 5B and data not shown).
For nonmaternally controlled IES 51A1835, cleavage of the wild-type left end is completely abolished by the AIM-5 mutation at the right end, no matter the presence or absence of the mutant IES in the macronucleus of the previous sexual generation (compare AIM-5 and AIM-5ΔA in Figure 6). Similar experiments were carried out for IES sm19-576 in mutant strain d4-2 sm19-1 (Supplementary Figure S5): a TA to CA mutation at the right end was found to interfere with the cleavage of both the mutant and the wild-type ends. Taken together, our data support the idea that the two ends of each mutant IES interact at an early stage during the excision reaction. Such an interaction may take place in cis, with the two IES ends located on the same molecule, or in trans, between homologous IES copies carried by distinct chromosomes (e.g. sister chromatids or homologs). To address this question, we used PCR to monitor the excision pattern of IES sm19-576 in heterozygous cells harboring the wild-type and sm19-1 mutant alleles in their germline genome (Supplementary Figure S6). Similar amounts of IES+ and IES− macronuclear chromosomes were observed, which indicate that wild-type IES copies are excised normally in the presence of mutant IES copies on homologous chromosomes.
DISCUSSION
Crosstalk between IES ends
For the three IESs examined in this study, a germline mutation within one of their flanking TAs results in a severe decrease in the level of detectable double-strand breaks, not only at their mutant end but also at their wild-type end. This could be attributed either to inhibition of DNA cleavage at both ends of the mutant IESs or to normal cleavage followed by more efficient resealing of the double-strand break, which would reduce the transient amount of free broken IES ends. We do not favor the latter hypothesis because, in contrast to a wild-type IES, unrepaired broken ends of the mutant IESs do not accumulate in cells deficient for double-strand break repair at IES excision sites (A. Kapusta, S.M., M.B., unpublished data). Because both ends must be wild-type to be cleaved efficiently, a strong implication of our data is, therefore, that a crosstalk between the two boundaries of an IES is established before DNA cleavage.
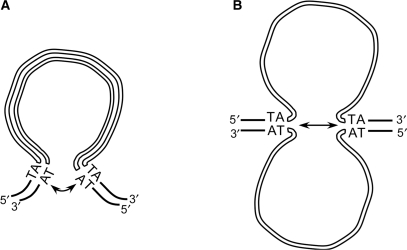
No trans-inhibitory effect was observed between IES copies carried on homologous chromosomes. Thus, even though we cannot rule out that crosstalk takes place between newly replicated sister chromosomes [an estimated 16–32 copies of each chromosome are present in the developing macronucleus by the time IES excision starts (9)], we propose that this interaction takes place in cis, during assembly of a precleavage complex in which the two ends of an IES would be brought together. Such a conformation could facilitate IES recognition by the DNA cutting machinery or stimulate its catalytic activity once it is bound to DNA. This would be reminiscent of site-specific recombination or DNA transposition systems (19), in which assembly of a synaptic complex including a recombination protein and its cognate substrate DNA sites is a prerequisite to DNA cleavage. Formation of a double-stranded DNA loop could allow the two ends of a given IES to come together prior to initial cleavage (Figure 7A). The observation that covalently closed circular IES molecules accumulate transiently during macronuclear development supports the notion that some IESs may accommodate significant curvature of their DNA double helix (9). Strikingly, however, >50% of known IESs are shorter than 80 bp and the ability of such short DNA fragments to circularize, even in the presence of DNA bending proteins, has been the subject of recent controversy (20,21). Circular excised molecules could not be detected for short IESs (8) and we proposed that the primary products of excision are linear molecules, which, if flexible enough, could be converted to circles in a subsequent end-joining step. The sequences studied here are representative of Paramecium IES size range (28, 66 and 370 bp) and similar observations were made for all three. Thus, assuming that the same geometry shapes the assembly of the excision initiation complex for all IESs, we speculate that crosstalk between ends is perhaps not mediated by looping out of double-stranded DNA but by local melting of the two DNA strands (Figure 7B). Strand separation may be favored by the A+T richness of IES DNA and by ongoing replication or transcription, both of which were revealed by pulse-chase labeling of the developing macronucleus (22). Single-stranded DNA was recently shown to be the substrate for DNA cleavage in other site-specific recombination systems (23–26). For Paramecium IESs, the formation of secondary structures on either strand, e.g. through the pairing of terminal inverted repeats at IES ends, could further stabilize extruded DNA bubbles or hairpin-loops and provide appropriate substrates to the excision machinery.
Figure 7.
Possible models for crosstalk between IES ends. (A) Formation of a double-stranded DNA loop. (B) Opening of the DNA double helix. IES strands are drawn as thick white lines, flanking DNA is represented by thin black lines.
Precise targeting of DNA cleavage at IES ends: a combination of sequence and epigenetic determinants
Our data point to a role of cis determinants (nucleotide sequence or secondary structure) in the targeting of DNA cleavages to Paramecium IES ends during developmental excision of these elements. The comparison of two IESs from the A51 surface antigen gene suggests that differences may exist between maternally controlled IESs and those whose excision is insensitive to the presence of homologous copies in the maternal macronucleus. IES 51A1835 belongs to the latter category (18) and a point mutation within its left TA is sufficient to abolish cleavage at both ends. Thus, the nucleotide sequence of its ends appears to be the key determinant for DNA cleavage. In contrast, DNA cutting at both ends of maternally controlled IES 51A6649 is strongly reduced by the AIM-2 mutation but not completely knocked out, as judged by detection of correctly positioned residual cleavage at both ends in the variant mutant strain AIM-2ΔA. Complete inhibition was observed only in strain AIM-2, which harbors the mutant IES in its macronucleus. These results indicate that, for IES 51A6649, another factor than simply the nucleotide sequence contributes to the determination of DNA cleavage sites. Consistent with an interaction between ends, we observed that residual cleavage at both ends of mutant IES 51A6649 is delayed to the same extent relative to a control wild-type IES in strain AIM-2ΔA. Interestingly, these residual double-strand breaks do not undergo 5′ processing: with regards to our current model for IES excision in Paramecium (10), this suggests that the absence of a central TA dinucleotide on the 5′ overhang generated at the mutant broken chromosome end may prevent correct alignment of the two chromosome arms at the excision site and, consequently, inhibit their controlled processing and repair. Residual double-strand breaks, therefore, would not lead to productive excision of the mutant IES.
Epigenetic programming of genome rearrangements has been documented in various ciliates (27). Current models involve homology-dependent trans-nuclear comparison of the maternal germline and somatic genomes mediated by noncoding RNAs (27,28). Short RNA molecules (scnRNAs), produced during meiosis from the micronuclear genome by an RNAi-related pathway, play an essential role in the search for germline-specific sequences that are absent from the maternal macronucleus (29–31). Germline scnRNAs selected by this scanning process are thought to be exported to the developing new macronucleus to participate in depositing an epigenetic mark on chromatin (32–34). This would trigger the elimination of homologous sequences from the new macronuclear genome. Long noncoding transcripts produced from the maternal macronucleus were also reported to participate in the regulation of genome rearrangements, but different roles have been proposed for these molecules (35 and G.L., M.B., E.M. and S.D., Genes and Development, in press). While they are thought to guide gene unscrambling in the developing macronucleus of Oxytricha, the fact that IESs are correctly excised from the A gene in the absence of a prerearranged version of the locus in the old macronucleus suggests that maternal transcripts may act differently in Paramecium. Rather, we have proposed that they are used as hybridization targets for the scnRNAs during the scanning for germline-specific sequences. Maternal control was reported for a subset of IESs (17,18), the excision of which depends on the NOWA1p RNA-binding protein, which may transport scnRNAs from the maternal to the new macronucleus (36). Intriguingly, the other IESs are excised efficiently from the new macronucleus, independently of the presence or absence of homologous sequences in the maternal somatic genome. Moreover, we show here that IESs are correctly excised even if they are embedded within a larger, imprecisely eliminated germline region. In the context of the scanning model, this suggests that some additional feature participates in the precise targeting of double-strand breaks at IES ends. The present evidence for crosstalk between ends prior to DNA cleavage suggests that DNA conformation may be a cis-acting determinant for excision. ScnRNAs might assist the excision of maternally controlled IESs by guiding the folding of separated DNA strands in order to bring IES ends close together, while they would not be necessary for the other IESs to adopt an excision-prone conformation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful to Aurélie Kapusta, Céline Baudry, Khaled Bouhouch and Anne Le Mouël for their support at different stages of this work. Warm thanks also to Jean Cohen, Bénédicte Michel, François-Xavier Barre and Frédéric Boccard labs for stimulating discussions, and to Linda Sperling for critical reading of the article. This work was funded by a CNRS ATIP grant to M.B. and by grants from the Ligue contre le cancer (RS06/75-32), the Fondation pour la Recherche Médicale (INE20061108404), the ACI BCMS 2004 (BCMS287), the Association pour la Recherche sur le Cancer (3608) and the Agence Nationale de la Recherche (NT05-2_41522). A.G. and G.L. were doctoral fellows of the French Ministère de l’Enseignement Supérieur et de la Recherche and of the Association pour la Recherche sur le Cancer. Funding to pay the Open Access publication charges for this article was provided by CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dudley DD, Chaudhuri J, Bassing CH, Alt FW. Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences. Adv. Immunol. 2005;86:43–112. doi: 10.1016/S0065-2776(04)86002-4. [DOI] [PubMed] [Google Scholar]
- 2.Jahn CL, Klobutcher LA. Genome remodeling in ciliated protozoa. Annu. Rev. Microbiol. 2002;56:489–520. doi: 10.1146/annurev.micro.56.012302.160916. [DOI] [PubMed] [Google Scholar]
- 3.Yao MC, Duharcourt S, Chalker DL. Genome-wide rearrangements of DNA in ciliates. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 730–758. [Google Scholar]
- 4.Bétermier M. Large-scale genome remodelling by the developmentally programmed elimination of germ line sequences in the ciliate Paramecium. Res. Microbiol. 2004;155:399–408. doi: 10.1016/j.resmic.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 5.Le Mouël A, Butler A, Caron F, Meyer E. Developmentally regulated chromosome fragmentation linked to imprecise elimination of repeated sequences in Paramecium. Eukaryot. Cell. 2003;2:1076–1090. doi: 10.1128/EC.2.5.1076-1090.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garnier O, Serrano V, Duharcourt S, Meyer E. RNA-mediated programming of developmental genome rearrangements in Paramecium tetraurelia. Mol. Cell. Biol. 2004;24:7370–7379. doi: 10.1128/MCB.24.17.7370-7379.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Segurens B, Daubin V, Anthouard V, Aiach N, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- 8.Gratias A, Bétermier M. Developmentally programmed excision of internal DNA sequences in Paramecium aurelia. Biochimie. 2001;83:1009–1022. doi: 10.1016/s0300-9084(01)01349-9. [DOI] [PubMed] [Google Scholar]
- 9.Bétermier M, Duharcourt S, Seitz H, Meyer E. Timing of developmentally programmed excision and circularization of Paramecium internal eliminated sequences. Mol. Cell. Biol. 2000;20:1553–1561. doi: 10.1128/mcb.20.5.1553-1561.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gratias A, Bétermier M. Processing of double-strand breaks is involved in the precise excision of Paramecium IESs. Mol. Cell. Biol. 2003;23:7152–7162. doi: 10.1128/MCB.23.20.7152-7162.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klobutcher LA, Herrick G. Consensus inverted terminal repeat sequence of Paramecium IESs: resemblance to termini of Tc1-related and Euplotes Tec transposons. Nucleic Acids Res. 1995;23:2006–2013. doi: 10.1093/nar/23.11.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haynes WJ, Ling KY, Preston RR, Saimi Y, Kung C. The cloning and molecular analysis of pawn-B in Paramecium tetraurelia. Genetics. 2000;155:1105–1117. doi: 10.1093/genetics/155.3.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayer KM, Mikami K, Forney JD. A mutation in Paramecium tetraurelia reveals function and structural features of developmentally excised DNA elements. Genetics. 1998;148:139–149. doi: 10.1093/genetics/148.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayer KM, Forney JD. A mutation in the flanking 5′-TA-3′ dinucleotide prevents excision of an internal eliminated sequence from the Paramecium tetraurelia genome. Genetics. 1999;151:597–604. doi: 10.1093/genetics/151.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuda A, Mayer KM, Forney JD. Identification of single nucleotide mutations that prevent developmentally programmed DNA elimination in Paramecium tetraurelia. J. Eukaryot. Microbiol. 2004;51:664–669. doi: 10.1111/j.1550-7408.2004.tb00606.x. [DOI] [PubMed] [Google Scholar]
- 16.Ruiz F, Krzywicka A, Klotz C, Keller A, Cohen J, Koll F, Balavoine G, Beisson J. The SM19 gene, required for duplication of basal bodies in Paramecium, encodes a novel tubulin, eta-tubulin. Curr. Biol. 2000;10:1451–1454. doi: 10.1016/s0960-9822(00)00804-6. [DOI] [PubMed] [Google Scholar]
- 17.Duharcourt S, Butler A, Meyer E. Epigenetic self-regulation of developmental excision of an internal eliminated sequence in Paramecium tetraurelia. Genes Dev. 1995;9:2065–2077. doi: 10.1101/gad.9.16.2065. [DOI] [PubMed] [Google Scholar]
- 18.Duharcourt S, Keller AM, Meyer E. Homology-dependent maternal inhibition of developmental excision of internal eliminated sequences in Paramecium tetraurelia. Mol. Cell. Biol. 1998;18:7075–7085. doi: 10.1128/mcb.18.12.7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Craig NL, Craigie R, Gellert M, Lambowitz AM. Mobile DNA II. Washington DC: ASM Press; 2002. [Google Scholar]
- 20.Cloutier TE, Widom J. Spontaneous sharp bending of double-stranded DNA. Mol. Cell. 2004;14:355–362. doi: 10.1016/s1097-2765(04)00210-2. [DOI] [PubMed] [Google Scholar]
- 21.Du Q, Smith C, Shiffeldrim N, Vologodskaia M, Vologodskii A. Cyclization of short DNA fragments and bending fluctuations of the double helix. Proc. Natl Acad. Sci. USA. 2005;102:5397–5402. doi: 10.1073/pnas.0500983102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berger JD. Nuclear differentiation and nucleic acid synthesis in well-fed exconjugants of Paramecium aurelia. Chromosoma. 1973;42:247–268. doi: 10.1007/BF00284774. [DOI] [PubMed] [Google Scholar]
- 23.Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 24.Bouvier M, Demarre G, Mazel D. Integron cassette insertion: a recombination process involving a folded single strand substrate. EMBO J. 2005;24:4356–4367. doi: 10.1038/sj.emboj.7600898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ton-Hoang B, Guynet C, Ronning DR, Cointin-Marty B, Dyda F, Chandler M. Transposition of ISHp608, member of an unusual family of bacterial insertion sequences. EMBO J. 2005;24:3325–3338. doi: 10.1038/sj.emboj.7600787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Val ME, Bouvier M, Campos J, Sherratt D, Cornet F, Mazel D, Barre FX. The single-stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae. Mol. Cell. 2005;19:559–566. doi: 10.1016/j.molcel.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Meyer E, Chalker DL. Epigenetics of ciliates. In: Allis CD, Jenuwein T, Reinberg D, editors. Epigenetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2007. pp. 127–150. [Google Scholar]
- 28.Mochizuki K, Gorovsky MA. Small RNAs in genome rearrangement in Tetrahymena. Curr. Opin. Genet. Dev. 2004;14:181–187. doi: 10.1016/j.gde.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 29.Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- 30.Chalker DL, Fuller P, Yao MC. Communication between parental and developing genomes during Tetrahymena nuclear differentiation is likely mediated by homologous RNAs. Genetics. 2005;169:149–160. doi: 10.1534/genetics.104.032300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee SR, Collins K. Two classes of endogenous small RNAs in Tetrahymena thermophila. Genes Dev. 2006;20:28–33. doi: 10.1101/gad.1377006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taverna SD, Coyne RS, Allis CD. Methylation of histone H3 at lysine 9 targets programmed DNA elimination in Tetrahymena. Cell. 2002;110:701–711. doi: 10.1016/s0092-8674(02)00941-8. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Mochizuki K, Gorovsky MA. Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proc. Natl Acad. Sci. USA. 2004;101:1679–1684. doi: 10.1073/pnas.0305421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Taverna SD, Muratore TL, Shabanowitz J, Hunt DF, Allis CD. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev. 2007;21:1530–1545. doi: 10.1101/gad.1544207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nowacki M, Vijayan V, Zhou Y, Schotanus K, Doak TG, Landweber LF. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature. 2008;451:153–158. doi: 10.1038/nature06452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nowacki M, Zagorski-Ostoja W, Meyer E. Nowa1p and Nowa2p: novel putative RNA binding proteins involved in trans-nuclear crosstalk in Paramecium tetraurelia. Curr. Biol. 2005;15:1616–1628. doi: 10.1016/j.cub.2005.07.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.