Abstract
Orthology is a widely used concept in comparative and evolutionary genomics. In addition to prokaryotic orthology, delineating eukaryotic orthology has provided insight into the evolution of higher organisms. Indeed, many eukaryotic ortholog databases have been established for this purpose. However, unlike prokaryotes, alternative splicing (AS) has hampered eukaryotic orthology assignments. Therefore, existing databases likely contain ambiguous eukaryotic ortholog relationships and possibly misclassify alternatively spliced protein isoforms as in-paralogs, which are duplicated genes that arise following speciation. Here, we propose a new approach for designating eukaryotic orthology using processed transcription units, and we present an orthology database prototype using the human and mouse genomes. Currently existing programs cover less than 69% of the human reference sequences when assigning human/mouse orthologs. In contrast, our method encompasses up to 80% of the human reference sequences. Moreover, the ortholog database presented herein is more than 92% consistent with the existing databases. In addition to managing AS, this approach is capable of identifying orthologs of embedded genes and fusion genes using syntenic evidence. In summary, this new approach is sensitive, specific and can generate a more comprehensive and accurate compilation of eukaryotic orthologs.
INTRODUCTION
The rapidly growing number of available complete genome sequences attests to the insistent need for functional annotations. Orthologs are defined as genes in different species that originated from a single gene locus in the last common ancestor (1–3). Therefore, orthology is a strong indicator of functional conservation, allows genome annotation based on information available from other species, provides raw material for evolutionary analysis and comparative genomics and identifies taxonomically restricted sequences. However, computer-generated ortholog designation is complicated because it demands knowledge of the ancestral state of genes and requires knowledge of complete gene repertoires. It is particularly challenging with respect to complex eukaryotic genomes due to gene duplication, conversion to pseudogenes, and gene loss and fusion.
There are several approaches for delineating orthologous genes. The phylogenetic tree-based methods HOVERGEN (4) and TreeFam (5) provide good accuracy, but because the trees are constructed by multiple sequence alignments or are corrected by experts, they provide limited comprehensiveness, homogeneity of quality and expandability to new species. Alternatively, the Best-Reciprocal-Hits (BRHs) method clusters orthologous genes based on their whole-length protein sequence similarities and was first introduced by the Cluster of Orthologous Groups (6) database. Phylogenetically, BRHs could be interpreted as genes from different species having the shortest connecting path over the distance-based tree. The identification of BRHs is widely adopted in comparative genomics for its simplicity and feasibility of application to large-scale data. For example, Ensembl Compara (7) (http://www.ensembl.org/biomart/martview/) identifies orthologous genes by searching BRHs between paired species. This notion has also been extended and applied to other approaches. Guided by a sequence similarity-based tree, HomoloGene (http://www.ncbi.nlm.nih.gov/sites/entrez?db=homologene) uses a blastp program to group proteins from input organisms and to restrict the molecular distance to prevent unlikely orthologs from being grouped together. Inparanoid (8,9) (http://inparanoid.sbc.su.se/cgi-bin/index.cgi) assigns the orthologous pair having the shortest distance among the BRHs of a given ortholog group as the anchor, and identifies other additional co-orthologs as in-paralogs. Allowing ortholog designations among multiple genomes, OrthoMCL (10,11) utilizes the Markov Cluster algorithm, a probabilistic model that groups (putative) orthologs and paralogs.
All currently available databases mentioned above are based on protein sequence alignments and do not take alternative splicing (AS) events into full consideration. AS in eukaryotic genomes plays an important role in augmenting biological complexity (12) such that a single gene can result in the generation of multiple proteins with high similarity. However, these proteins are isoforms and should not be annotated as in-paralogs or orthologs. Unfortunately, the current eukaryotic ortholog databases all discard AS by utilizing an all-against-all protein comparison that cannot exclude isoforms, which results in aberrant assignment of orthologs and in-paralogs. For example, the gene SORBS2 has two NM entries for human and several dozen XM entries in the transitory mouse annotation of May 2006 at our initial data examination (Supplementary Table 1). Identifying the ortholog of SORBS2 between human and mouse without taking AS into account would be a challenging task. Thanking the continuous efforts of the reference gene curation at National Center for Biotechnology Information (NCBI) and of the MGI (mouse genome informatics) project, this gene now has a single NM entry in the current mouse annotation. Therefore, taking the BRH of human isoforms as the only annotated ortholog at the beginning of this study would derive incomplete ortholog annotations. This serves as an example that such issues could create assignment inconsistencies among different databases with separated update schedules. Alternatively, isoforms might be misclassified as in-paralogs by methods such as Inparanoid, which assigns an anchor pair based on the best alignment score and leaves the rest as in-paralogs. Therefore, including AS in orthologous gene identification is crucial for accuracy.
In this study, we propose a new approach for delineating orthologs among species that are somewhat related. We utilized processed transcription units rather than protein sequences as the BRHs input. This accounts for AS by recognizing gene-oriented genome regions, but it maintains the advantages provided by sequence alignment-based approaches. We report that our method assigned more than 92% of the identified human/mouse orthologs available in existing ortholog databases, and covered more than 80% of the human reference sequences, providing an improved approach for simple, clear and accurate identification of eukaryotic orthologs.
MATERIALS AND METHODS
Data sources
We used genome assemblies from the human NCBI build 36 published on March 8, 2006, and the mouse NCBI build 36 available on May 8, 2006 (13,14) (http://www.ncbi.nlm.nih.gov/RefSeq/), (http://www.ncbi.nlm.nih.gov/Genomes/). There were 40 814 coding sequences (NM_ and XM_) among the 41 677 human reference sequences and 48 797 (NM_ and XM_) coding sequences among the 50 481 mouse reference sequences. This study focused only on the coding sequences. The detailed genomic locations of human reference sequences were obtained and downloaded from the NCBI ftp site: (ftp://ftp.ncbi.nlm.nih.gov/refseq/H_sapiens/H_sapiens/mapview/). This information is used as the gold standard for further dataset comparison and examination. In comparison of various datasets, we did find some inconsistent coordinates of annotated genes from different data sources, which could be resulted from different genomic mapping processes or distinct genome assembly versions. We then performed subsequent re-alignment process to ensure the genomic transcription locations and to extract genomic sequences in order to perform orthologous gene database comparison on the identical version of genome build.
The gene-oriented ortholog database
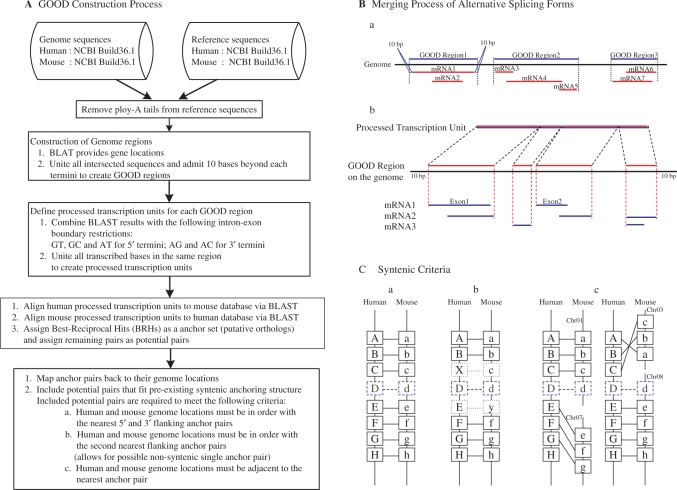
Human and mouse genomes were used to demonstrate the ortholog assignment capability of our program, and the resulting data are presented as the Gene Oriented Ortholog Database (GOOD). Figure 1A illustrates the basic methodology used to designate orthologs based on the processed transcription units of gene-oriented genomic regions. We utilized 36 018 human reference sequences and 41 815 mouse reference sequences to identify 21 544 human and 22 511 mouse transcription regions. Among these transcription regions, we identified 17 214 orthologs.
Figure 1.
Criteria involved in generating the GOOD database prototype of human and mouse orthologs. (A) Flow chart of the overall database construction process. (B) Detail representing the merging of different AS products into a processed transcription unit. The upper panel (a) illustrates how the GOOD regions are obtained from all reference sequences. The lower panel (b) illustrates that the processed transcription unit is derived from isoforms from same genomic region. (C) Detail representing the inclusion of syntenic information. Potential ortholog pairs (D/d, shown in blue) are analyzed based on the three syntenic possibilities shown in panels a, b and c. Boxes with black lines represent anchor pairs; boxes with solid lines represent anchor pairs with syntenic structure and boxes with dotted lines represent pairs that lack syntenic structure.
Identifying genomic transcription regions
To define transcription regions within genomic sequences, we used the Blast-like alignment tool (BLAT) (15) program to determine the genome locations of reference sequences. Several of the human and mouse reference sequences from NCBI have poly-A tails of varying lengths. These poly-A tails reduce the apparent sequence identities calculated by BLAT in subsequent analyses. Therefore, we removed all poly-A tails from the reference sequences to increase the accuracy of the BLAT results. Because BLAT provides all possible alignment results for each sequence, we needed to define a threshold to distinguish the optimal alignment results from all other alignment results. Therefore, only those mouse and human sequences having BLAT alignment identities greater than 0.98 were accepted to promote accurate sequence location assignments. Approximately 97% of the human sequences and 91% of the mouse sequences had an alignment identity higher than 0.98. However, there were some human (9%) and mouse (6%) sequences remaining with multiple best BLAT results above the defined threshold, allowing those sequences to be aligned to several genome positions. Because the corresponding genomic regions could not be precisely located, we excluded sequences with ambiguous locations and selected only those sequences having a unique BLAT result to define the transcription regions for the reference genes. To construct disjointed transcription regions in the genome, we integrated the transcription regions of intersecting sequences and included a 10-base extension from both termini, as shown in Figure 1B, panel a. In summary, 36 018 human reference sequences were used to define 21 544 human transcription regions, and 41 815 mouse reference sequences were used to define 22 511 mouse transcription regions.
Processed transcription units
To account for AS within the defined transcription regions, we combined exon–intron boundary rules with results from the Basic Local Alignment Search Tool (16,17). We used the BLAST program to align those reference sequences having transcription regions identified by BLAT and applied GT, GC and AT as the boundary signals of putative 5′ splice donor sites and AG and AC as the boundary signals of putative 3′ splice acceptor sites (18) to define the exon–intron boundaries. This process provided exact genomic position information for each reference transcript in its respective transcription region and resulted in the modification of 11 843 (33%) human reference sequence locations and 13 431 (32%) mouse reference sequence locations. However, there were 471 human sequences and 1012 mouse sequences that remained undefined. After manually checking these undefined reference sequence genome locations, we included 345 human BLAT results and 880 mouse BLAT results to complete the genome location information for human and mouse reference sequences in their transcription regions. Within each transcription region, we removed the purely intronic regions to obtain a concatenation of all the exons, which was defined as a processed transcription unit (Figure 1B, panel b).
Ortholog assignment
We used BLAST to align human processed transcription units to mouse processed transcription units and vice versa. We defined the BRHs of the processed transcription units (15 592 pairs) as the anchor set (see Figure 1C, boxes outlined in black), and the remaining pairs were assigned as potential pairs (see Figure 1C, boxes outlined in blue). The anchor pairs represented putative orthologs. When the anchor pairs were mapped back to their genomic locations, we discovered a pre-existing syntenic relationship between the human and mouse genomes. We therefore examined the potential pairs individually and added those pairs that fit into the syntenic anchor structure. We applied three criteria to determine fit. First, pairs in which both human and mouse genome locations were in order with the nearest 5′ and 3′ flanking anchor pairs were included (Figure 1C, panel a). Second, we included those pairs in which the genome locations were in order with the second nearest 5′ and 3′ flanking anchor pairs to allow for a possible non-syntenic single anchor pair (Figure 1C, panel b). Third, we carefully set the boundary of the syntenic blocks by including only those potential pairs in which both human and mouse genome locations were adjacent to the nearest anchor pair (Figure 1C, panel c). Using this method, we obtained 17 214 human and mouse processed transcription unit orthologous pairs.
RESULTS
Processed transcription unit is a representative of its AS-mediated isoforms
The reference sequences from the NCBI have been annotated with gene descriptions and abbreviated gene symbols. Utilizing this information, we considered distinct sequences with the same gene symbol as AS-mediated isoforms. Only 16 human genes and 24 mouse genes remained associated with different transcription regions, and the detailed locations of these regions are listed in Supplementary Tables 2 and 3. However, most of these apparent genes were the result of annotation errors. Their current annotation information had been corrected on the NCBI website (Supplementary Tables 2 and 3), and they become associated to unique transcription region instead. Although two human genes and one mouse gene remained associated with different transcription regions after correction, nearly 100% of human and mouse genes were associated with a specific transcription region. Therefore, the data indicate that processed transcription units successfully manage AS products and allow for accurate representation of individual genes.
To highlight the advantages that our algorithm offers for eukaryotic ortholog assignments, we examined the gene SORBS2. There are two NM entries for the human SORBS2. Because the currently available ortholog assignment programs do not account for AS in eukaryotes, identification of SORBS2 orthologs is often confusing and incomplete, and some isoforms are even identified as in-paralogs (8). Using our method, isoforms were associated with their transcription regions (genes) prior to ortholog delineation. This allowed a single SORBS2 ortholog assignment between human region GOODH_0040429 and mouse region GOODM_1080169 (Supplementary Table 1). Thus the GOOD database provides more accurate, straightforward and comprehensible eukaryotic ortholog assignments.
Sequence similarities of processed transcription unit pairs are much lower than that of protein pairs
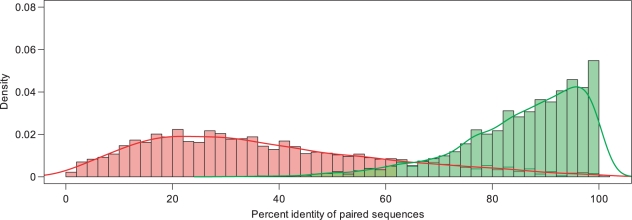
To further examine the processed transcription unit-based method of identifying eukaryotic orthology, we compared the similarities of processed transcription units with those of protein sequences. We used the UniGene database (http://www.ncbi.nlm.nih.gov/sites/entrez?db=unigene) to obtain pairs of human and mouse genes located in the same UniGene cluster and to obtain information regarding the aligned amino acid sequence similarities. To ensure that the protein sequence similarities and the processed transcription unit similarities could be analyzed in parallel, we only focused on those pairs common to the UniGene and GOOD databases. The similarity distributions of these data sets are shown in Figure 2. As expected, the protein sequence similarities of orthologs are higher than their processed transcription unit similarities, due ostensibly to decreased conservation among 5′ and 3′ untranslated regions (UTRs) and to the codon wobble hypothesis (19). For instance, the human TWSG1 transcript is 3693 bp in length whereas the mouse transcript is 3994 bp in length. Only 20% of the transcript sequence could be aligned due to long, unconserved UTRs, and the aligned bases only had 83% identity based on the codon wobble hypothesis, resulting in only 17% overall sequence similarity. In contrast, the full-length human TWSG1 protein is 223 amino acid residues in length whereas the mouse protein is 222 residues in length, allowing alignment of all residues and resulting in 93% identity. The similarities of processed transcription unit pairs are therefore much lower than that of protein pairs. In this study, we reported that the mean amino acid sequence similarity for orthologs was 86%, and about 73% of those orthologous pairs had more than 80% amino acid sequence similarity. However, the mean nucleotide sequence similarity of processed transcription units was only 37%, and only about 5% of the orthologous pairs had more than 80% nucleotide sequence similarity. Although proceeded transcription unit similarities are lower than protein similarities, eukaryotic orthology assignments between related organisms are reliable using the processed transcript unit method.
Figure 2.
Distribution of the percent identity between aligned orthologous protein and transcript pairs from the human and mouse genomes. The x axis indicates the percent identity of paired orthologous sequences, and the y axis indicates number of orthologous pairs normalized to the total number of input pairs. The aligned identities of protein sequences were obtained using UniGene and are shown in green. The aligned identities of the processed transcription units were obtained using GOOD and are shown in pink.
Comparison of GOOD with existing databases
Due to AS, genes of metazoans often have multiple sequence records. For example, SORBS2 has two AS-mediated isoforms in humans. However, the currently available databases only include one annotated isoform; the other isoform is missing. The UCSC Known Genes database has only one BRH of human NM_003603, but NM_003603 is not annotated in HomoloGene. Furthermore, the human SORBS2 protein record ENSP00000284 776 is annotated in Ensembl Compara, but the annotation in Inparanoid is ENSP00000347852. Thus although these databases contain SORBS2 orthology information, inconsistent annotations make the ortholog records confusing.
Despite these inconsistencies, we examined ortholog pairs derived by GOOD and compared them to the newest versions of the other available ortholog databases, which are derived from the same annotated genome version. Specifically, we compared GOOD with HomoloGene release 56, UCSC Known Genes hg18 and mm8 (20) (http://genome.ucsc.edu/), Ensembl Compara release 46 (7) and Inparanoid version 5.1 (9) (Table 1). We utilized the genomic locations of their orthology information and converted the results from these four available databases into processed transcription unit records for subsequent comparisons. GOOD contains 17 214 human/mouse ortholog pairs, which encompasses about 80% of the human reference genes (Figure 3), whereas the other databases each encompass less than 69% of the human reference genes. This demonstrates an 11% increased sensitivity provided by this method.
Table 1.
Comparison of 17 214 GOOD human/mouse orthologous pairs with the four existing ortholog databases
| GOOD | HomoloGene | UCSC known genes | Ensembl compara | Inparanoid | |
|---|---|---|---|---|---|
| # Reference Sequence | N/A | 16 325 HIDa | 14 692 kgIDb | 22 047 | 15 549 |
| # Region-based Orthologous Pairs | 17 214 | 14 843 | 12 111 | 12 362 | 9023 |
| # Region-based Orthologous Pairs also identified by GOOD | N/A | 14 327 | 11 889 | 11 332 | 8825 |
| Human reference gene coverage rate | ∼80% | ∼69% | ∼56% | ∼57% | ∼42% |
aHID: HomoloGene group id.
bkgID: Human/Mouse reciprocal conserved UCSC Known Genes ID pair.
N/A: not applicable.
HomoloGene: build 56
UCSC Known Genes: hg18/mm8
Ensembl Compara: release 46
Inparanoid: version 5.1
There are 21 544 human regions from the RefSeq (NCBI build 36). There might be some loss when transforming ids among different databases.
Compared to current ortholog databases, GOOD has higher consistency and also provides the highest coverage rate of the human genome.
Figure 3.
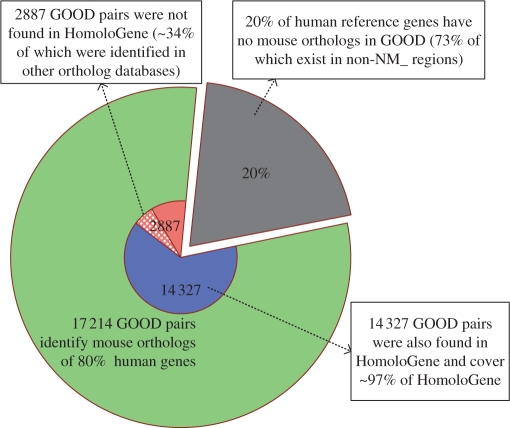
Pie chart representation of GOOD ortholog data. The whole pie reflects the total number of human reference genes (21 544 human regions) from the RefSeq (NCBI build 36). The green shading represents the percentage of human reference genes that the program was capable of considering for GOOD. The grey shading represents the percentage of human reference genes that are not represented in GOOD [73% of these reference genes are located in non-NM_regions (regions not supported by experimental evidence), where ortholog designation is difficult]. The blue shading represents those ortholog pairs identified by both GOOD and HomoloGene. GOOD identified 97% of the ortholog pairs in the HomoloGene database. The orange shading represents those ortholog pairs identified by GOOD that were not represented in HomoloGene; 34% of these ortholog pairs (light orange) were represented in one of the other three existing ortholog databases.
In addition, more than 97% of the orthologous pairs recorded in the HomoloGene and UCSC Known Genes databases and more than 92% of those recorded in the Ensembl Compara database were also identified using the processed transcription unit-based method. Furthermore, we compared the HomoloGene database—the largest dataset—with GOOD to determine specific differences between these two databases (Figure 3). There were 2887 orthologous pairs in GOOD that were not present in the HomoloGene database. However, 994 (∼34%) of these 2887 pairs were present in the other three databases (see Supplementary Table 4). Because GOOD is highly consistent with the four currently available databases, we are confident that it provides ortholog specificity.
DISCUSSION
Although AS is a formative aspect of eukaryotic genomes, it often has been disregarded in eukaryotic orthology studies due to its complexity. In this study, we propose a new method for generating an ortholog database that takes into account the AS of mRNAs. Eukaryotic ortholog assignments were made based on processed transcription units generated by revising gene transcript sequences to exclude pure intronic sequences, poly-A tails and capping signals. Here, we defined processed transcription units, which are pre-mature mRNA like sequences. They are products transcribed from a genome without pure intronic sequences and right before undergoing capping and adding poly-A tails. Therefore, the processed transcription unit represents all isoforms generated via AS and is associated with one specific gene locus. Although the similarities of transcription unit pairs were much lower than that of protein pairs, our approach still provided higher sensitivity and specificity than the other four databases with respect to human and mouse ortholog assignments.
In addition to AS, many events including pseudogenes; exon shuffling; gene duplication; gene fusion and gene loss, could further complicate eukaryotic orthology assignments. Our method not only manages AS, but also takes into account embedded genes and gene fusions. For instance, PELO is annotated as an embedded gene of ITGA1 in human region GOODH_1050094, and this embedded gene structure is conserved in mouse GOODM_0130455, which contains ITGA1 and PELO (21). Application of our algorithm allowed identification of the embedded gene ortholog using the BRH approach, demonstrating that even complex exon/intron structures such as ITGA1-PELO-ITGA2 can be represented by the processed transcription unit. However, because processed transcription units represent all sequences derived from the same transcription region, only using the BRH approach cannot completely delineate orthologies for embedded and fused genes that are not conserved. We therefore extended GOOD to include orthologs having syntenic properties. This strategy permitted inclusion of potential orthologs that maintained the syntenic structure formed by the anchor orthologs, yielding a program that accommodates gene fusion events. For example, human PALM2-AKAP2 (NM_007203 and NM_147150) in GOODH_00200210 is a fusion gene of PALM2 (NM_053016 and NM_001037293) and AKAP2 (NM_001004065) in GOODH_1090214 (22). According to the merging algorithm, these five transcripts would form just one processed transcription unit. However, these two genes are not fused in mice, and they are located at two different juxtaposed mouse regions, PALM2 in GOODM_1040154 and AKAP2 in GOODM_1040155. The BRH of GOODH_1090214 must be a single hit, specifically GOODM_1040154 in the anchor set. This indicates that some of the information of the mouse ortholog of PALM2-AKAP2 is missing. Using the syntenic evidence, the program could complement the BRH information and delineate the orthologs of PALM2 and AKAP2 in human and mouse by assigning two orthologous pairs, GOODH_00200210/GOODM_1040154 and GOODH_00200210/GOODM_1040155. Human TRIM6-TRIM34 is also an annotated fusion gene (23), and the extended GOOD program identified its mouse orthologs as TRIM6 (GOODM_1070647) and TRIM34 (GOODM_1070647). These examples demonstrate that this approach can accommodate complicated genes locations even though processed transcription units represent a complete genomic locus.
Although this method provides improved ortholog assignments between human and mouse, some conceivable drawbacks exist. First of all, the completeness of genome assemblies and transcript annotations affect the applicability of this approach. This is the primary reason that we chose to analyze the most comprehensive genomes, human and mouse, as the prototype. The exon–intron structure of an incomplete genome locus is transient because transcripts are likely to be added and removed, making the processed transcription unit less stable and thereby complicating ortholog assignments. For example, the phylogenic distance between humans and mice is approximately the same as between humans and rats. It is reasonable to expect similarly complete orthology assignments between humans and rats using the GOOD method. However, only 13 938 ortholog pairs could be designated between humans and rats (NCBI RGSC v3.4). This number is much lower than that determined for humans and mice (17 214 pairs), and the coverage of the human genome dropped from 80% using the mouse genome to 65% using the rat genome. This may result from incomplete rat genome assemblies and transcript annotations. As genome sequencing technology keeps improving, GOOD would likely provide higher quality data for every genome in the future.
Even though humans and mice belong to different orders, mice are considered closer to humans than are other metazoans. Therefore, we applied our algorithm to humans and zebrafish (NCBI v.6), which resulted in assignment of only 5166 anchor pairs. The number of human and zebrafish orthologs is too small to be acceptable and likely results not only from an imperfect zebrafish genome assembly but also from the phylogenic distance between humans and zebrafish. Nonetheless, even if genomic data are nearly complete, processed transcription units may not be sensitive enough to provide sufficient ortholog assignments between distantly related species. However, because distantly related species result from the accumulation of sequential biological evolution, we propose that ortholog assignments between distantly related species may be achieved by application of GOOD to all relative species between two phylogenetically distant species, such that the orthology between the two distantly related species would be represented as a compilation of orthology assignments from more closely related species.
In conclusion, the GOOD strategy presented in this study provides substantial improvements over the four currently available ortholog databases. Specifically, our method utilizes processed transcription units and synteny to accommodate complications often encountered in eukaryotic genomes such as AS, gene fusions and imbedded genes. Although complete GOOD orthology assignments are dependent on comprehensive genome assemblies and transcript annotations, our algorithm provides a simple, highly sensitive and specific method for producing orthology assignments and will therefore be beneficial for researchers across many fields of study.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This study was supported in part by an Academia Sinica thematic program project. Funding to pay the Open Access publication charges for this article was provided by Academia Sinica.
Conflict of interest statement. None declared.
REFERENCES
- 1.Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Gen. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- 2.Sonnhammer EL, Koonin EV. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 2002;18:619–620. doi: 10.1016/s0168-9525(02)02793-2. [DOI] [PubMed] [Google Scholar]
- 3.Fitch WM. Homology: a personal view on some of the problems. Trends Genet. 2000;16:227–231. doi: 10.1016/s0168-9525(00)02005-9. [DOI] [PubMed] [Google Scholar]
- 4.Duret L, Mouchiroud D, Gouy M. HOVERGEN: a database of homologous vertebrate genes. Nucleic Acids Res. 1994;22:2360–2365. doi: 10.1093/nar/22.12.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H, Coghlan A, Ruan J, Coin LJ, Heriche JK, Osmotherly L, Li R, Liu T, Zhang Z, Bolund L, et al. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006;34:D572–D580. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Remm M, Storm C.EV, Sonnhammer E.LL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 9.O’Brien KP, Remm M, Sonnhammer EL. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005;33:D476–D480. doi: 10.1093/nar/gki107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L, Stoeckert C.J., Jr., Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen F, Mackey AJ, Stoeckert C.J., Jr., Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brett D, Pospisil H, Valcarcel J, Reich J, Bork P. Alternative splicing and genome complexity. Nat. Genet. 2002;30:29–30. doi: 10.1038/ng803. [DOI] [PubMed] [Google Scholar]
- 13.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 18.Burset M, Seledtsov IA, Solovyev VV. SpliceDB: database of canonical and non-canonical mammalian splice sites. Nucleic Acids Res. 2001;29:255–259. doi: 10.1093/nar/29.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crick F. Codon—anticodon pairing: the wobble hypothesis. J. Mol. Biol. 1966;19:8. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- 20.Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, Sugnet CW, Stanke M, Smith KE, Siepel A, et al. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheli Y, Kanaji S, Jacquelin B, Chang M, Nugent DJ, Kunicki TJ. Transcriptional and epigenetic regulation of the integrin collagen receptor locus ITGA1-PELO-ITGA2. Biochim. Biophys. Acta. 2007;1769:546–558. doi: 10.1016/j.bbaexp.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu B, Copeland NG, Gilbert DJ, Jenkins NA, Kilimann MW. The paralemmin protein family: identification of paralemmin-2, an isoform differentially spliced to AKAP2/AKAP-KL, and of palmdelphin, a more distant cytosolic relative. Biochem. Biophys. Res. Commun. 2001;285:1369–1376. doi: 10.1006/bbrc.2001.5329. [DOI] [PubMed] [Google Scholar]
- 23.Orimo A, Tominaga N, Yoshimura K, Yamauchi Y, Nomura M, Sato M, Nogi Y, Suzuki M, Suzuki H, Ikeda K, et al. Molecular cloning of ring finger protein 21 (RNF21)/interferon-responsive finger protein (ifp1), which possesses two RING-B box-coiled coil domains in tandem. Genomics. 2000;69:143–149. doi: 10.1006/geno.2000.6318. [DOI] [PubMed] [Google Scholar]