Abstract
The human immunodeficiency virus type 1 (HIV-1) is a primate lentivirus that causes the acquired immunodeficiency syndrome (AIDS). In addition to the virion structural proteins and enzyme precursors, that are Gag, Env and Pol, HIV-1 encodes several regulatory proteins, notably a small nuclear transcriptional activator named Tat. The Tat protein is absolutely required for virus replication since it controls proviral DNA transcription to generate the full-length viral mRNA. Tat can also regulate mRNA capping and splicing and was recently found to interfere with the cellular mi- and siRNA machinery. Because of its extensive interplay with nucleic acids, and its basic and disordered nature we speculated that Tat had nucleic acid-chaperoning properties. This prompted us to examine in vitro the nucleic acid-chaperoning activities of Tat and Tat peptides made by chemical synthesis. Here we report that Tat has potent nucleic acid-chaperoning activities according to the standard DNA annealing, DNA and RNA strand exchange, RNA ribozyme cleavage and trans-splicing assays. The active Tat(44–61) peptide identified here corresponds to the smallest known sequence with DNA/RNA chaperoning properties.
INTRODUCTION
The human immunodeficiency virus type 1, HIV-1, is a member of the lentivirus family that is widespread in primates, and is the causative agent of the acquired immunodeficiency syndrome known as AIDS (1). HIV-1 codes for the three canonical retroviral polyprotein precursors, which are needed for virus formation and infectivity, namely Gag, Pol and Env (2). As a complex retrovirus, HIV-1 also encodes six regulatory proteins, namely Tat, Rev, Vpr, Vpu, Vif and Nef, that carry out important roles in virus replication, either acting at the level of viral RNA synthesis and its nuclear export in the case of Tat and Rev, or suppressing virus restriction by the host factor APOBEC 3G in the case of Vif (3,4). At the end of the early steps of HIV-1 replication, the newly made proviral DNA is integrated into the host genome via the concerted action of the viral integrase enzyme, the nucleocapsid (NC) protein, and the cellular cofactor LEDGF (5–7). In infected cells, transcription of the integrated proviral DNA requires the viral Tat protein for the synthesis of the full-length viral mRNA. As extensively documented, Tat is a small nuclear transcriptional activator which controls productive proviral DNA transcription from the 5′ long terminal repeat (LTR) of HIV-1, via its interaction with the transactivation response (TAR) RNA element (2,8,9). As for any transcriptional activator, Tat could facilitate either of two reactions taking place at the beginning of DNA transcription, namely recruitment of an RNA polymerase II (RNAP II) complex to the promoter, and the escape of that initial complex from the promoter into productive RNA synthesis (8,10,11). In fact, the most likely scenario by which Tat activates by several 100-fold transcription from the HIV-1 5′ LTR, is that Tat promotes an efficient transcription elongation. The conversion of non-processive RNAP II complexes into productive ones by Tat has been the matter of a large number of studies, which led to the identification of many Tat-associated host transcription factors and regulatory proteins (12–15). A highly simplified view proposes that Tat contacts the nascent viral TAR RNA and recruits various transcription factors such as the TATA-binding protein (TBP), the general transcription factor TFIIB and the positive transcription elongation factor B (P-TEFB), itself composed of cyclin T1 and the cyclin-dependent kinase 9 (CDK9). This leads to the formation of very active elongating transcription complexes (16,17). The hyperphosphorylation of RNAP II by CDK9 will also ensure the rapid cyclic reuse of elongation complexes to achieve a continuous and rapid reinitiation of transcription to the benefit of the viral promoter strength (18,19). This scenario favors the notion that Tat acts by interacting with multiple viral and cellular partners at a time. In accordance with the view that Tat is multi-functional it has been shown that Tat also regulates mRNA capping (20) and splicing (21), and interferes with the cellular RNA interference (RNAi) machinery through interactions with DICER and RNA (22,23).
Tat is a small basic protein conserved in all primate lentiviruses (24), and is poorly structured as a free protein in solution (25). The basic and intrinsically disordered nature of the HIV-1 Tat protein may well account for its ability to interact with several proteins and nucleic acids in a manner similar to RNA chaperones such as the HIV-1 NC protein, Flaviviridae core proteins, or the cellular fragile X mental retardation protein FMRP (26–28). These unique features of Tat made us speculate that it had general nucleic acid-chaperoning activities resembling those of HIV-1 NCp7, as suggested by recent publications (29,30). To examine this possibility, Tat and Tat peptides were synthesized and their nucleic acid-chaperoning activities were extensively studied in vitro. Here we show that Tat has potent nucleic acid-chaperoning activities, as evidenced by a number of standard RNA chaperoning assays (RCA) (31,32). We also find that the basic Tat(44–61) peptide encompassing the Tat basic domain is the smallest reported peptide with nucleic acid-chaperone activities.
| Name | Sense | Sequence (from 5′ to 3′) | Description |
|---|---|---|---|
| TAR(+) | + | GGTCTCTCTTGTTAGACCAGGTCGAGCCCGGGAGCTCTCTGGCTAG CAAGGAACCC | Nucleotides 1–56 of HIV-1 MAL strain genomic RNA |
| TAR(−) | − | GGGTTCCTTGCTAGCCAGAGAGCTCCCGGGCTCGACCTGGTCTAAC AAGAGAGACC | Nucleotides 1–56 of HIV-1 MAL strain genomic RNA |
| R(+) wt | + | GGTCTCTCTTGTTAGACCAGGTCGAGCCCGGGAGCTCTCTGGCTAG CAAGGAACCCACTGCTTAAGCCTCAATAAAGCTTGCCTTGAGTGCC TCCC | Nucleotides 1–96 of HIV-1 MAL strain genomic RNA |
| R(−) wt | − | GGGAGGCACTCAAGGCAAGCTTTATTGAGGCTTAAGCAGTGGGTTC CTTGCTAGCCAGAGAGCTCCCGGGCTCGACCTGGTCTAACAAGAGA GACC | Nucleotides 1–96 of HIV-1 MAL strain genomic RNA |
| R(−).3′-modified | − | GGGAGGCACTCAAGGCAAGCTTTATTGAGGCTTAAGCAGTGGGTTC CTTGCTAGCCAGAGAGCTCCCGGGCTCGACCTGGTCTAACATCAGT CTCTA | Complementary to R(+), but with 3′ terminal mismatches (underlined) |
| ΔU3-R(+) | + | GCAGCTGCTTTTCGCCTGTACTGGGTCTCTCTTGTTAGACCAGGTCG AGCCCGGGAGCTCTCTGGCTAGCAAGG | Corresponds to part of the LTR. The underlined sequence corresponds to the RNA sequence used in the DNA/RNA exchange assay. |
| ΔU3-R(−) | − | CCTTGCTAGCCAGAGAGCTCCCGGGCTCGACCTGGTCTAACAAGAG AGACCCAGTACAGGCGAAAAGCAGCTGC | Corresponds to part of the LTR Complementary to the ΔU3-R(+) |
MATERIALS AND METHODS
DNA substrates
The HIV-1 LTR DNA corresponding to the NY5 strain was generated by PCR, 5′ end 32P- labelled and purified by electrophoresis on a 1% agarose gel followed by QIAEX II extraction (Qiagen).
Oligodeoxynucleotides (ODNs) used for DNA binding, annealing and strand transfer assays corresponded to the HIV-1 TAR and the repeated R sequences, in the sense and anti-sense orientations, respectively. ODNs used for the HIV-1 RNA displacement assays (ΔU3-R), in the sense or anti-sense orientation, corresponded to part of the LTR. ODNs were purchased from Eurogentec (Belgium) and IBA GmbH Nucleic Acids Product Supply (Göttingen, Germany). TAR, R and ΔU3-R ODNs are 56, 96 and 74 nt in length, respectively. ODNS ΔU3-R(+) and (–) were used to mimic the start of transcription and release of the nascent RNA.
TAR(−), R(+) and ΔU3-R(−) ODNs were 32P-labelled with 50 µCi of [α-32P]ATP using T4 polynucleotide kinase. 32P-labelled ODNs were purified by 10% PAGE, 7 M urea in 50 mM Tris–borate, 1 mM EDTA, pH 8.3 (0.5× TBE). They were recovered, ethanol precipitated and dissolved in sterile H2O before use.
Fluorescently labelled TAR(−) derivatives were labelled at their 3′ termini with 5(and6)-carboxyfluorescein (Fl) using a special solid support with the dye already attached. In doubly labelled TAR(−) derivatives, the 5′ end was additionally labelled with carboxytetramethylrhodamine (TMR) via an amino-linker with a six carbon spacer arm. Labelled oligonucleotides were purified by reverse-phase HPLC and polyacrylamide gel electrophoresis.
In vitro generated RNAs
RNAs used for the ribozyme assay were synthesized by in vitro transcription with T7 RNA polymerase as previously published (31). For the trans-splicing assay (33), template DNAs were generated by digesting H1 DNA (exon 1 + 5′intron) and H2 DNA (3′intron + exon 2) with SalI (H1) or BamHI (H2), respectively. Construct H1 consists of 549 nt of exon 1 and 131 nt of the 5′ part of the intron of the T4 phage thymidylate synthase (td) gene pre-mRNA. Construct H2 consists of 147 nt of the 3′ half of the intron and 23 nt of exon 2 of td pre-mRNA. RNAs H1 and H2 were synthesized by in vitro transcription with T7 RNA polymerase as above during which they were labelled by incorporation of [35S]-UMP.
For the viral RNA displacement assay, plasmid pHIVCG4 was cut with NheI and used for in vitro synthesis of HIV-1 RNA (position 1–46).
All RNAs were purified by 5% PAGE containing 7 M urea and 50 mM Tris–borate pH 8.3, 1 mM EDTA (0.5× TBE). RNAs were recovered by elution in 0.3 M sodium acetate, 0.1% SDS, for 4 h at 37°C and ethanol precipitated. RNAs recovered by centrifugation were dissolved in sterile H2O and quantified by UV at 254 nm. RNA integrity was verified by PAGE in 7 M urea.
Tat and NC proteins
Biologically active full-length Tat protein and Tat peptides were chemically synthesized using optimized Fmoc chemistry protocols, and were purified by reverse-phase HPLC (34). Their purity was monitored by analytical RP-HPLC on a Beckman instrument (Gagny, France) with a Nucleosil C18 5-μm column (150 × 4.6 mm) and a Nucleosil C4 5-μm column (150 × 4.6 mm), respectively, using a linear gradient of 0.1% TFA in water and acetonitrile containing 0.08% TFA at a flow rate of 1.2 ml/min. Finally, integrity of each peptide was assessed by matrix-assisted laser desorption and ionization time-of-flight spectrometry on a Protein TOF™ mass spectrometer (Bruker, Wissembourg, France). The mass of Tat protein was assessed by LC/MS using a Thermofinigan LCQ. NCp7 and mutant NCp7(12–53) were chemically synthesized and purified by RP-HPLC as previously published (29,35). Peptides were dissolved at 1 mg/ml in a freshly prepared oxygen-free buffer containing 30 mM HEPES pH 6.5, 30 mM NaCl and 0.1 mM ZnCl2. Extinction coefficients of 1300, 5690 and 8250 M−1 cm−1 were used to determine the concentrations of Tat(21–43), Tat(44–61), Tat(1–20) and Tat(1–86) peptides at 280 nm. Since no aromatic residue was present in Tat(48–86), its concentration was determined at 214 nm, using an extinction coefficient of 57 600 M−1 cm−1, calculated using Equation (2) of Kuipers and Gruppen (36).
| Name | aa sequence | Mr (Da) |
|---|---|---|
| Tat(1–86) | EPVDPRLEPWKHPGSQPKTACTTCYCKKCCFHCQVCFTTKALGISYGRKKRRQRRRP PQGSQTHQVSLSKQPTSQPRGDPTGPKE | 9837 |
| Tat(1–20) | MEPVDPRLEPWKHPGSQPKT | 2311 |
| Tat(21–43) | ACTTCYCKKCCFHCQVCFTTKAL | 2605 |
| Tat(44–61) | GISYGRKKRRQRRRPPQG | 2196 |
| Tat(44–57) | GISYGRKKRRQRRR | 1817 |
| Tat(48–61) | GRKKRRQRRRPPQG | 1847 |
| Tat(48–86) | GRKKRRQRRRPPQGSQTHQASLSKQPTSQPRGDPTGPKE | 4448 |
| NCp7(1–72) | MQRGNFRNQRKNVKCFNCGKEGHTARNCRAPRKKGCWKCGKEGHQMKDCTERQ ANFLGKIWPSYKGRPGNFL | 8489 |
| NCp7(12–53) | NVKCFNCGKEGHTARNCRA PRKKGCWKCGKEGHQMKDCTERQ | 4824 |
UV-visible absorption and steady-state fluorescence measurements
Absorption spectra were recorded on a Cary 400 spectrophotometer. Fluorescence emission spectra and kinetic traces were recorded on a FluoroMax spectrofluorometer (Jobin Yvon) equipped with a thermostated cell compartment. Binding experiments were performed by adding increasing peptide concentrations to 10 nM TAR(−)-3′-Fl and monitoring the steady-state fluorescence anisotropy using a T-format SLM 8000 spectrofluorometer at 20°C. The emitted light of TAR(−)-3′-Fl was monitored through high-pass filters (520 nm) (Kodak). A home-built device ensured the automatic rotation of the excitation polarizer. The apparent binding constant Kapp was recovered by fitting the fluorescence anisotropy data using:
 |
1 |
where S0 and St are the initial and the final anisotropy, respectively; n − number of binding sites; and Pt, Nt concentrations of protein and oligonucleotide, respectively. The numbers of binding sites were determined from titrations in the absence of salt. Binding experiments were performed in 25 mM Tris (pH 7.5), 30 mM NaCl, 0.2 mM MgCl2, with the exception of the binding experiments with Tat(21–43) where 50 mM HEPES (pH 6.5), 30 mM NaCl, 0.2 mM MgCl2 was used to limit the aggregation of the peptide.
Real-time TAR(+)/TAR(−) anneling kinetics were monitored in pseudo first-order conditions by using 300 nM TAR(+) and 10 nM of the doubly labelled TMR-5′-TAR(−)-3′-Fl sequence. Tat peptides were added to the TAR sequences at a molar ratio of 1–3 peptides per oligonucleotide. The annealing reaction was triggered by mixing the TAR(+)/peptide and TMR-5′-TAR(−)-3′-Fl/peptide solutions. Excitation and emission wavelengths were 480 nm and 520 nm, respectively, to monitor the Fl fluorescence. At a concentration of 300 nM TAR(+), the kinetic traces were adequately fitted with a bi-exponential function:
| 2 |
where t0 is the dead time, kobs1,2 are the observed kinetic rate constants, x is the amplitude of the fast component, and I0 and If are the fluorescence intensities of the stem–loop and extended duplex structures, respectively. All fitting procedures were carried out with the Microcal Origin 6.1 software based on the nonlinear, least-squares method and the Levenberg–Marquardt algorithm.
Electrophoresis-based nucleic acid binding and chaperoning assays
The DNA-binding assay
The 32P-labelled TAR(−) DNA (0.03 pmol) was incubated with an increasing concentration of Tat or Tat peptide for 5 min at 37°C in buffer A (20 mM Tris–HCl pH 7.0, 30 mM NaCl, 0.1 mM MgCl2, 5 mM DTT and 10 µM ZnCl2). Next, we added 5 µl of a solution containing 20% glycerol, 20 mM EDTA pH 8.0 and 0.25% bromophenol blue. Nucleoprotein complexes were analysed by 1% agarose gel electrophoresis in stringent conditions (0.5× TB pH 8.3, 1 mM EDTA at 20°C). The agarose gel was dried and then submitted to autoradiography.
The TAR(−)/TAR(+) DNA annealing assay
TAR(+) and 32P-TAR(−) ODNs were incubated (0.03 pmol each) in 10 μl of buffer A at increasing protein concentrations as indicated in the figure legends. Reactions were performed at 37°C for 5 min except for the positive control that was incubated at 65°C. To stop the reaction and denature the protein in order to release it from the 32P-ODN, we added 5 µl of a solution containing 20% glycerol, 20 mM EDTA pH 8.0, 2% SDS, 0.25% bromophenol blue and 0.4 mg/ml calf liver tRNA. Samples were resolved by 8% native PAGE in 50 mM Tris–borate pH 8.3, 1 mM EDTA at 4°C. Subsequently, gels were autoradiographed and the amounts of labelled single-stranded and double-stranded DNA were assessed by phosphorimaging.
The DNA strand exchange assay
32P-labelled R(+) wt, R(−).3′-modified and R(−) wt were separately heat denatured for 2 min at 95°C and chilled on ice. All components were kept at 4°C. 0.03 pmol each of 32P-labelled R(+) wt and R(−).3′-modified, at a concentration of 6 × 10−8 M, were mixed with the reaction buffer to a final concentration of 20 mM Tris–HCl, pH 7.0, 30 mM NaCl, 0.1 mM MgCl2, 10 µM ZnCl2 and 5 mM DTT in 5 µl final volume, incubated for 30 min at 62°C under oil and chilled on ice. Then, 0.03 pmol of R(−) wt was added together with the protein using a protein to nucleotide molar ratio as indicated in the figure legends. Assays were conducted for 5 min at 37°C, then chilled on ice and stopped with 2.5 µl of 20% glycerol, 20 mM EDTA pH 8.0, 0.2% SDS, 0.25% bromophenol blue and 0.4 mg/ml calf liver tRNA. Samples were resolved by 6% native PAGE in 0.5× TBE at 4°C and gels were subsequently autoradiographed. Levels of DNA strand exchange were assessed by phosphorimaging.
The hammerhead ribozyme cleavage assay
Ribozyme and substrate RNA were independently heated for 1 min at 90°C in sterile H2O. The reaction buffer was added to yield final concentrations of 5 mM MgCl2, 100 mM NaCl, 20 mM Tris–HCl, pH 7.5. After slow cooling to 37°C, RNAs were incubated for 5 min at 20°C. Ribozyme measuring 0.02 pmol and 0.08 pmol of RNA substrate were then combined in a final volume of 10 µl. Each protein was added at final protein to nucleotide molar ratios as indicated in the legend of the figures. Incubations were for 20–30 min at 37°C. Reactions were stopped by adding 20 µl of the stop solution (0.3% SDS, 15 mM EDTA), and extracted with 30 µl of phenol and 15 µl of chloroform. The aqueous phase was precipitated with ethanol and the pellet re-suspended in 45% formamide, 0.5× TBE and 0.1% dyes. 32P-labelled RNAs were analysed on 8% PAGE in 7 M urea and 0.5× TBE. After electrophoresis, gels were autoradiographed and levels of RNA cleavage were assessed by phosphorimaging.
The RNA trans-splicing assay
In vitro generated RNAs (0.2 pmol of each 35S-labelled RNA) (2 × 10−8 M) were incubated for 1 min at 95°C in sterile H2O. After slow cooling down to 37°C, the splicing buffer (40 mM Tris–HCl at pH 7.4, 3 mM MgCl2, 0.4 mM spermidine, 4 mM DTT), 8 U RNasin and 0.33 pmol 32P-GTP were added. Protein was added to final protein to RNA molar ratios as indicated in the figure legends (typically at concentrations of 2 × 10−8 to 10−7 M). Incubation was for 30 min at 37°C in 10 µl final volume. Reactions were stopped by adding a stop solution (40 mM EDTA, 5 µg of tRNA). Proteins were removed by phenol–chloroform extractions, and samples processed as above. Precipitated RNAs were re-suspended in 85% formamide, 0.5× TBE and 0.1% dyes (xylene cyanol and bromophenol blue), heat denatured and loaded on a denaturing 5% PAGE in 7 M urea and 0.5× TBE. 5′ 32P-labelled FX174 DNA HinfI markers (Promega) were used for size determination (not shown). After electrophoresis, gels were autoradiographed and levels of trans-splicing were assessed by phosphorimaging.
The DNA/RNA exchange assay
The ODN ΔU3-R(−) and viral 32P-RNA (nt 1–46) (0.1 pmol each) were heat denatured for 2 min at 90°C and annealed at 60°C for 30 min in buffer A, in 10 µl final volume. Then, ODN ΔU3-R(+) (0.1 pmol) was added together with Tat or Tat peptide, followed by incubation for 5 min at 30°C. Reactions were stopped and proteins removed as indicated above. ODNs and RNA were separated on a 400 Spreadex gel under native conditions (Euromedex). Quantitations were by phosphorimaging as above.
RESULTS
Binding of Tat and Tat peptides to TAR DNA
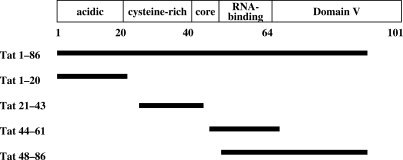
The HIV-1 transactivator Tat is a basic protein of about 11.5 kDa, which tightly binds a short nascent stem–loop RNA structure, TAR (transactivation response element), for its activity (37,38). Tat is encoded by two exons, namely exon 1 for residues 1 to 72 and exon 2 for residues 73–86 in the case of laboratory strains and 73–101 found in most clinical isolates (39). Tat contains five domains that are the N-terminal domain I (aa 1–20), the cysteine-rich domain II (aa 21–40), the hydrophobic core domain III (aa 41–48), the RNA-binding domain IV encompassing the basic RKKRRQRRR motif and a glutamine-rich region (aa 49–72) necessary for the transactivation, and the C-terminal domain V of variable size (73–86 or 73–101) (Figure 1). Tat contains a large number of conserved arginine and lysine residues, notably between positions 45 and 60 in the core and RNA-binding domains. In good agreement with recently published NMR data (25), a computer prediction of Tat folding indicated that Tat alone is mostly unstructured (Supplementary Figure 1), and thus could be classified as an intrinsically unstructured protein (IUP), a hallmark of cellular and viral RNA chaperones (28,40). In order to examine the nucleic acid binding and potential chaperoning properties of Tat, full-length Tat (1–86) and Tat peptides corresponding to the N-terminal domain I (aa 1–20), to the cysteine-rich domain (aa 21–43), to part of the core and RNA-binding domains III/IV (aa 44–61) and to the glutamine-rich region together with the C-terminus (aa 48–86) were synthesized and purified to homogeneity in conditions preventing oxidation in vitro (see Materials and methods section).
Figure 1.
Domain organization of HIV-1 Tat protein, and peptides used in this study.
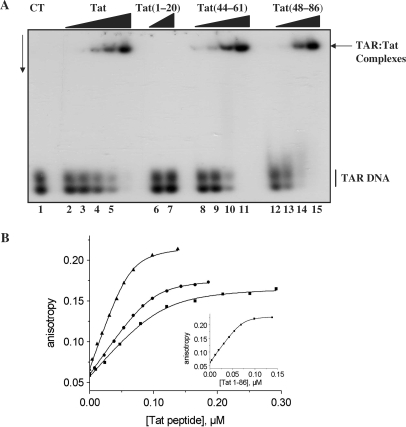
Tat and the peptides were examined for their ability to bind nucleic acids in vitro, using TAR DNA at a concentration of 3 × 10−9 M and stringent conditions (see Materials and methods section). As shown in Figure 2A, Tat protein and the basic peptides 44–61 and 48–86, but not peptide 1–20, were found to tightly bind TAR DNA at concentrations of 2–4 × 10−8 M, leading to the formation of stable nucleoprotein complexes. Similar data were obtained with the HIV-1 5′ leader RNA containing the 5′ TAR sequence (data not shown). The cysteine-rich peptide 21–43 was found to aggregate, precluding a clear examination of its DNA-binding property by gel retardation assay (data not shown). The binding of the Tat peptides to TAR(−) was further investigated by fluorescence anisotropy using TAR(−) labelled with 5(and6)-carboxyfluorescein (Fl). The binding stoichiometry was inferred from the intersection of the initial slope of the titration with the fluorescence anisotropy plateau in the absence of salt to maximize the affinity of the peptides for cTAR (Figure 2B inset). Initial experiments with various concentrations of TAR(−)-3′-Fl showed that below 20 nM, TAR aggregation caused by the Tat peptides was avoided.
Figure 2.
Binding of Tat to the viral 5′TAR DNA. (A) Analysis by gel electrophoresis. TAR DNA/Tat nucleoprotein complexes were analysed by 1% agarose gel electrophoresis in 0.5× TBE pH 8.3. The gel was dried and autoradiographed. Lane 1: control 32P-labelled TAR DNA at 3 × 10−9 M; lanes 2–5: with Tat(1–86) at 0.3, 0.6, 1.2, 2.4 and 4 × 10−8 M (corresponding to protein to nt molar ratios of respectively 1:57, 1:28.5, 1:14.2, 1:7.1 and 1:4.2); lanes 6 and 7: Tat(1–20) at 1–2 × 10−7 M; lanes 8–11: Tat(44–61) and 12–15: Tat(48–86), at concentrations of 1 to 8 × 10−8 M (corresponding to peptide to nt molar ratios of respectively 1:18.5, 1:9, 1:4.5 and 1:2.2). TAR DNA migrates as a double band due to its extensive secondary structure. (B). Binding curves of Tat derivatives to TAR(−) monitored by steady-state fluorescence anisotropy. The concentration of TAR(−)-3′-Fl was 10 nM. Anisotropy titrations were performed by adding increasing concentrations of Tat(1–86) (triangles), Tat(48–86) (circles), Tat(44–61) (squares) in 25 mM Tris (pH 7.5), 30 mM NaCl, 0.2 mM MgCl2. Excitation wavelength was 480 nm. Solid lines correspond to the fit of the experimental points with Equation (1) and the parameters of Table 1. Inset: determination of the binding stoichiometry in the absence of NaCl. The intercept of the initial slope with the plateau gives the number of Tat(1–86) binding sites on TAR(−).
For Tat(1–86), the number of binding sites was found to be about seven (Table 1), corresponding to an occluded binding site of about ≈8 nt per protein. The large increase in the fluorescence anisotropy (from 0.06 to 0.22) could be readily explained by the 5-fold mass increase of TAR when it was bound to seven Tat protein molecules. A slightly larger number of binding sites (≈10) was observed for both Tat(48–86) and Tat(44–61) peptides. Since the molar mass of Tat(48–86) is about twice that of Tat(44–61), this explains the higher anisotropy plateau observed with the former peptide. No DNA binding was observed for Tat(1–20). The apparent binding constant, Kapp, was determined for the peptides (Figure 2B), by assuming that the binding sites on TAR DNA are identical and independent. This approach was shown to be reasonable to assess the affinities of other viral proteins and their mutants to TAR DNA (41,42). By fitting the binding curves with Equation (1), a Kapp value of 2.3 ± 0.8 × 108 M−1 was obtained for Tat(1–86). Similar values were obtained with both Tat(48–86) and Tat(44–61) peptides. Due to its aggregating properties, only a rough estimation of the Kapp value of Tat(21–43) could be obtained, indicating an about two orders of magnitude decrease with respect to Tat(48–86) and Tat(44–61). Thus, the binding of Tat to TAR DNA is mainly mediated by its nucleic acid-binding (48–61) domain.
Table 1.
Binding parameters of Tat peptides to TAR(−)a
| Peptide | nb | bc | Kappb, M−1 |
|---|---|---|---|
| Tat(1–86) | 7.0 ± 0.7 | 7.8 | 2.3 ± 0.8 × 108 |
| Tat(48–86) | 10 ± 1 | 5.5 | 1.9 ± 0.7 × 108 |
| Tat(44–61) | 9.9 ± 0.5 | 5.5 | 0.9 ± 0.2 × 108 |
| Tat(21–43) | −d | – | 1 × 106 |
aThe buffer was 25 mM Tris (pH 7.5), 30 mM NaCl, 0.2 mM MgCl2, except for Tat(21–43) where the buffer was 50 mM Hepes (pH 6.5), 30 mM NaCl, 0.2 mM MgCl2.
bThe number of binding sites, n, and the apparent binding constant, Kapp, were determined as described in Figure 2.
cThe occluded binding size was calculated by dividing the number of TAR(−) nucleotides by n.
dIt was not possible to accurately determine the number of binding sites for Tat(21–43) because of partial peptide aggregation.
The basic and intrinsically unstructured nature of Tat, together with its ability to strongly bind nucleic acids suggested to us that Tat could have nucleic acid-chaperoning properties, similar to the NC protein of HIV-1, that is a key viral factor involved in proviral DNA synthesis (5,6,29,43). To investigate how Tat may assist nucleic acid folding, we used standard nucleic acid chaperone assays, namely DNA annealing and strand exchange (44), ribozyme-directed cleavage of an RNA substrate (45) and RNA trans-splicing (32,33,46).
Tat stimulates DNA annealing and strand exchange
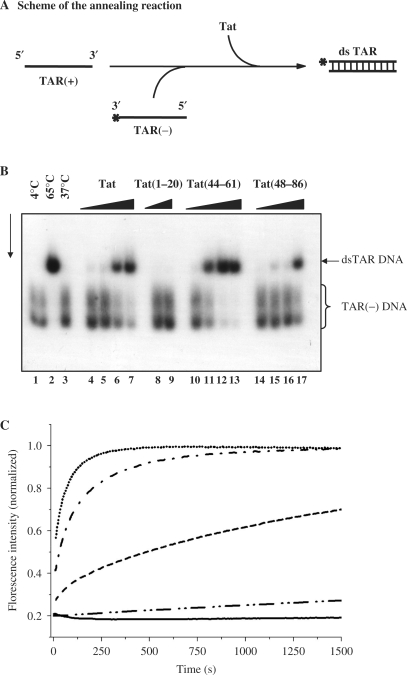
The DNA annealing assays used the TAR(+) and TAR(−) sequences of 56 nt in length, which fold into stable stem–loop structures. Hybridization of TAR(+) to TAR(−) to form a double-stranded DNA was examined by PAGE in native conditions (Figure 3A). Formation of TAR(+):TAR(−) occurred upon heating at 65°C (Figure 3B, lane 2) or upon addition of the HIV-1 NC protein (data not shown; see also ref. 29). Tat(1–86) promoted hybridization of TAR(+) to TAR(−) (Figure 3B, lanes 4–7) at protein concentrations of 2–4 × 10−8 M while DNA concentration was 0.6 × 10−8 M (see Materials and methods section). These results are similar to those found for NCp7 (see Figure 3 legend and ref. 29). Tat peptides (44–61) and (48–86) but not peptide 1–20 promoted Tar DNA annealing at concentrations of 2–4 × 10−8 M (lanes 10–13 and 14–17). It is interesting to note that Tat(44–61) was about two times more active than the Tat peptide (48–86) (compare lanes 12 and 16 and see figure 3 legend). The annealing activity of peptide (21–43) was found to be about 10 times less than that of peptide (44–61) (70% annealing at peptide to nt molar ratios of 1:60 and 1:4 for 44–61 and 21–43, respectively) (lane 12 and data not shown).
Figure 3.
HIV-1 Tat has DNA annealing activity. (A) Schematic representation of the DNA annealing assay (see explanation in text). (B) TAR(+) and 32P-labelled TAR(−) DNA at 6 × 10−9 M were incubated together in the presence of increasing concentrations of Tat or a Tat derived peptide (see Materials and methods section). Upon annealing at 65°C for 30 min a double-stranded TAR DNA was formed (lane 2) but not at 37°C (lane 3). Lanes 4–7: Tat at increasing concentrations of 0.5, 1, 2 and 4 × 10−8 M (corresponding to protein to nt molar ratios of respectively 1:68, 1:34, 1:17 and 1:8.5), for 5 min at 37°C; lanes 8 and 9: no dsTAR DNA formation occurred with Tat(1–20) even at 1–2 × 10−7 M; lanes 10–13: annealing occured with Tat peptide (44–61) at peptide to nt molar ratios of 1:120, 1:60, 1:30 and 1:15. Ds TAR DNA formation also occurred with Tat(48–86) but only at a concentration of 4 × 10−8 M (corresponding to a peptide to nt molar ratio of 1:8.5) (lane 17). TAR DNA migrates as a double band due to its extensive secondary structure. (C) Annealing of TAR(−) to TAR(+) promoted by Tat. The kinetic traces were recorded with 10 nM TMR-5′-TAR(−)-3′-Fl and 300 nM TAR(+) with Tat(44–61) (dots) and Tat(1–20) (solid line) at a molar ratio of three peptides per ODN. Kinetic traces are also shown at one peptide per ODN for the complete Tat(1–86) (dash dot) and Tat(44–61) (dash). The kinetic trace with Tat(21–43) was performed at a ratio of 13 peptides per ODN (dash dot dot). The kinetic trace with Tat(48–86) at a ratio of three peptides per ODN is fully superimposable with that of Tat(44–61) at the same ratio and is thus not represented. Excitation and emission wavelength were 480 nm and 520 nm, respectively. The lines correspond to the fit of the kinetic traces with Equation (2) and the parameters of Table 2.
To quantify the Tat-promoted TAR(+)/TAR(−) annealing, we monitored their real-time annealing kinetics in pseudo-first-order conditions by mixing 10 nM TMR-5′-TAR(−)-3′-Fl with a 30-fold excess of non-labelled TAR(+). Formation of the TAR(+)/TAR(−) extended duplex (ED) strongly increases the interchromophore distance, leading to a full recovery of Fl emission (47). In the absence of Tat, TAR(+)/TAR(−) DNA annealing was previously shown to involve two distinct kinetic components characterized by two slow second-order rate constants (48). Due to the high concentration of TAR(+) needed for the pseudo-first-order conditions, distorted kinetic traces owing to aggregation were observed when Tat(1–86) was added at a molar ratio of protein to TAR ≥ 2 or when Tat(48–86) or Tat(44–61) was added at a ratio of protein to TAR ≥ 5. As a consequence, the annealing reactions were monitored at lower ratios where no aggregation occured. In the presence of Tat(48–86) or Tat(44–61) at a molar ratio of three peptide molecules per TAR, the same fluorescence plateau was reached as with the Tat protein, indicating that the reaction went to completion generating the ED (Figure 3C). Tat and the two peptides dramatically increased the annealing kinetics, since the reaction was complete in <10 min, instead of more than one day in the absence of Tat peptide. The observed kinetic rate constants (kobs1 and kobs2 values) with the two peptides were respectively one and two orders of magnitude larger than the corresponding values in the absence of peptide (Table 2). The similar annealing activities of the two peptides suggested that these activities are mainly supported by the nucleic acid-binding (aa 48–61) domain. In agreement with this, monitoring of the Tat(21–43) annealing activity by fluorescence intensity showed that it was about 10 times less active than Tat(44–61) (Table 2). Comparison of Tat(44–61) with Tat(1–86) at a molar ratio of one peptide per TAR further revealed that the full-length Tat was substantially more active than the peptide (Table 2). This indicates that residues flanking the basic (48–61) domain contribute to the Tat-promoted TAR(+)/TAR(−) annealing and that a substantial promotion of TAR(+)/TAR(−) annealing can be achieved at a molar ratio as low as one Tat per TAR sequence. In sharp contrast, no promotion of TAR(+)/TAR(−) DNA annealing was observed with Tat(1–20) (Figure 3C and Table 2).
Table 2.
Kinetic parameters of TAR(+)/TAR(−) annealing with Tat peptidesa
| Peptide | Peptide to oligonucleotide ratio | kobs1, s−1 | kobs2, s−1 |
|---|---|---|---|
| – | – | 2.7 × 10−3b | 2.5 × 10−5b |
| Tat(44–61) | 3 | 3.5 ± 0.2 × 10−2 | 9.7 ± 1 × 10−3 |
| 1 | 1.01 ± 0.04 × 10−2 | 4.8 ± 0.3 × 10−4 | |
| Tat(48–86) | 3 | 5.0 ± 0.8 × 10−2 | 9.8 ± 0.8 × 10−3 |
| Tat(1–86) | 1 | 2.6 ± 0.8 × 10−2 | 4.2 ± 0.5 × 10−3 |
| Tat(21–43) | 13 | – | 1.4 ± 0.3 × 10−4 |
aExperimental conditions were as described in Table 1. The kobs values were obtained by fitting the kinetic traces in Figure 2 by Equation 2.
bFrom Godet et al. (48).
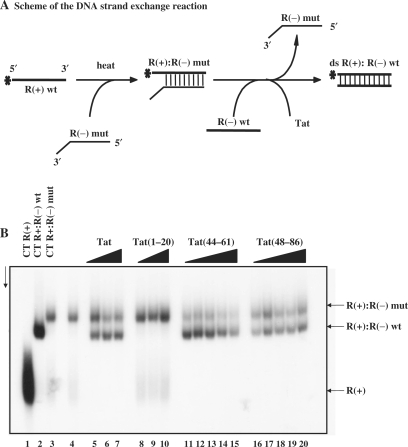
The next assays aimed at investigating the ability of Tat to promote formation of the most stable double stranded nucleic acid. These assays used three DNA molecules of 96 nt in length representing the R sequence of the HIV-1 LTR (Figure 4A). First, R(+) was annealed to a mutant R(−) containing seven mutations at its 3′ end (Materials and methods section) by heating at 62°C for 30 min (Figure 4B, lane 3 to be compared with lane 2 corresponding to the R(+):R(−) DNA). Next, R(−) DNA and Tat were added to the imperfect double-stranded DNA and incubated for 5 min at 37°C (Figure 4A). The final DNA products were analysed by PAGE in native conditions. As previously reported, HIV-1 NCp7 directed DNA strand exchange by replacing the mutated R(−) by the wild type R(−) in the double stranded DNA (data not shown and refs 5,6,43). The basic peptide NC(12–53) was able to bind DNA but unable to promote the DNA strand exchange under these in vitro conditions (data not shown). Tat was efficient at directing the DNA strand exchange (Figure 4B, lanes 5–7) at concentrations of 1–2 × 10−7 M. Tat peptides (44–61) (lanes 11–15) and (48–86) (lanes 16–20) also promoted strand exchange at concentrations of 1–2 × 10−7 M. Again, Tat(44–61) was clearly more active than Tat(48–86) (compare lanes 12 and 17) while Tat(1–20) was inactive even at 10−6 M (lanes 8–10).
Figure 4.
Tat has DNA strand exchange activity. (A) Schematic representation of the DNA strand exchange assay using the HIV-1 R sequence of 96 nt in length (see explanation in the text). (B) 10−8 M R(+)DNA (lane 1) was annealed to an equal amount of mutant R(−) DNA with three mismatched regions at its 3′ end (see Materials and methods section) by heating at 62°C for 30 min (lane 3). The complete double-stranded R(+):R(–) DNA is shown in lane 2. R(–) DNA and Tat were added to the R(+):R(–) mutant and reactions were conducted for 5 min at 37°C. After Tat removal, the final DNA products were analysed by PAGE in native conditions (see Materials and methods section) Lanes 5–7: Tat concentrations were 0.5, 1 and 2 × 10−7 M, corresponding to protein to nt molar ratios of respectively 1:18, 1:9 and 1:4.5. Lanes 8–10: Tat(1–20) at concentrations of 0.2, 0.5 and 1 × 10−6 M. Lanes 11–15: Tat(44–61) at concentrations of 0.25, 0.5, 1, 2 and 4 × 10−7 M, corresponding to protein to nt molar ratios of respectively 1:36, 1:18, 1:9, 1:4.5 and 1:2.2. Lanes 16–20: Tat(48–86) at concentrations of 0.25, 0.5, 1, 2 and 4 × 10−7 M, corresponding to protein to nt molar ratios of respectively 1:36, 1:18, 1:9, 1:4.5 and 1:2.2. Note that Tat(44–61) was as active as Tat(1–86) and more active than Tat(48–86) (compare lanes 6,12 and 17).
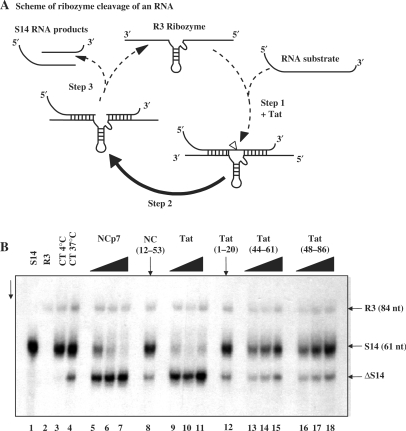
Tat enhances hammerhead ribozyme cleavage of an RNA substrate
Ribozyme-directed cleavage of an RNA substrate allows examination of both the RNA annealing and unwinding activities of nucleic acid chaperones (31). Chaperones are thought to enhance the rate of ribozyme cleavage by activating the annealing of the substrate RNA to the ribozyme (Figure 5A, step 1) and the unwinding and release of the RNA products (Figure 5A, step 3), thus allowing fast recycling of the ribozyme. The ribozyme cleavage assay examines whether Tat can accelerate ribozyme cleavage of an RNA substrate in a manner similar to the viral NCp7 chaperone (see Figure 5A) (31,49). We selected the R3 hammerhead ribozyme and two RNA substrates, namely S14 with a 14-nt substrate-ribozyme duplex length (7 nt on either side of the cleavage site) and S20 with 10 nt on either side of the cleavage site. The above RNA substrate model system was chosen due to its likely biological relevance as evidenced by the similarity of data obtained in vitro and in cultured cells (49). Labelled RNA S14 (Figure 5B, lane 1), the ribozyme R3 (lane 2) and Tat or a Tat peptide were mixed and incubated. Next, RNAs were deproteinized, recovered and analysed by PAGE under denaturing conditions. In the absence of a chaperone, ribozyme-directed cleavage of the RNA substrate occurred only slowly at 37°C (Figure 5B, lanes 3 at 4°C and 4 at 37°C; labelled RNA substrate is S14 and cleaved product ΔS14). In agreement with previous reports (49), HIV-1 NCp7 caused ribozyme cleavage of RNA S14 to reach completion at a concentration as low as 2 × 10−8 M (lanes 5–7). On the other hand, mutant NC(12–53) was inactive even at a concentration of 2 × 10−7 M (lane 8). Interestingly, Tat strongly enhanced ribozyme-directed cleavage of RNA S14 at a concentration of 1–2 × 10−8 M (lanes 9–11). The basic Tat peptides (44–61) and (48–86) only slightly enhanced ribozyme-directed cleavage of S14 RNA (lanes 15 and 18), and levels were only a fifth of that with Tat protein at peptide concentration of 1 × 10−7 M (see figure 5 legend). The Tat peptides (1–20) and (21–43) were inactive even at 1 × 10−7 M (lane 12 and data not shown). As expected, RNA template S20 was not cleaved and there was no enhancement of ribozyme cleavage by Tat under these conditions (data not shown; 49).
Figure 5.
Enhancement of hammerhead ribozyme cleavage by the HIV-1 Tat protein. (A) Schematic representation of the hammerhead ribozyme RNA cleavage reaction (see explanation in text). The S14 RNA cleavage site is indicated by the arrow. (B) R3 ribozyme and S14 substrate RNA were incubated with or without a chaperone, and after protein removal by phenol, RNAs were analysed by PAGE in denaturing conditions (see Materials and methods section). Lanes 1 and 2: S14 RNA and R3 alone, respectively. Lanes 3 and 4: S14 RNA cleavage by R3 in the absence of protein at 4°C and 37°C. Note that cleavage by R3 alone was limited (ΔS14). Lanes 5–7: HIV-1 NCp7 at concentrations of 1, 2 and 4 × 10−8 M, corresponding to protein to nt molar ratios of 1:60, 1:30 and 1:15. Lane 8: mutant NC(12–53) at a concentration of 2 × 10−7 M (peptide to nt ratio of 1:3). Lanes 9–11: Tat(1–86) at concentrations of 1, 2 and 4 × 10−8 M, corresponding to peptide to nt ratios of 1:60, 1:30 and 1:15. Lanes 12: Tat(1–20) at 1 × 10−7 M (ratio of 1:6). Lanes 13–15: Tat(44–61) at concentrations of 0.5, 1 and 2 × 10−7 M, corresponding to peptide to nt ratios of 1:12, 1:6 and 1:3. Lanes 16–18: Tat(48–86) at concentrations of 0.5, 1 and 2 × 10−7 M (peptide to nt ratios of 1:12, 1:6 and 1:3). Note that Tat(1–86) was as active as NCp7 (see lanes 6 and 10), but that the Tat basic petides (44–61) and (48–86) were only poorly active in this ribozyme assay. Indeed levels of cleavage in the presence of the Tat peptides at 2 × 10−7 M amounted to 15–20% of that with Tat protein at a concentration of 1 × 10−8 M. RNA template S20 was not cleaved and there was no enhancement of ribozyme cleavage by HIV-1 NCp7 and Tat under these conditions (data not shown and see previously published data on NCp7; refs. 31,50).
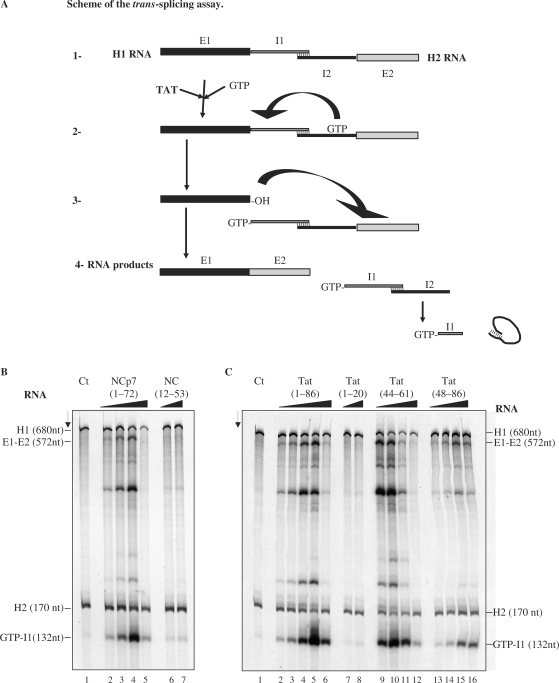
Tat enhances RNA trans-splicing
To evaluate the role of the Tat chaperoning activity in splicing (21) we used the previously established trans-splicing assay (33,50), where the pre-mRNA of the thymidylate synthase (td) gene containing a group I intron was split into two halves. The first RNA transcript, H1, corresponds to the 5′ exon sequence of 549 nt and 131 nt of the intron while the second RNA transcript, H2, corresponds to the 3′ part of the intron (147 nt) and part of exon 2 (23 nt) (Figure 6A). The two RNAs were 35S-UMP labelled during transcription and were incubated together. Reactions were started by adding 32P-GTP, so that the resulting spliced RNA was doubly labelled, internally and at the 5′ end with 32P-GTP (Figure 6A). In the absence of protein, the reaction was carried out at 55°C to allow a productive interaction between H1 and H2 RNAs. Trans-splicing was indeed found to take place at 55°C but only to a limited extend (data not shown and refs 33,50) but not at 37°C according to the accumulation of 32P-GTP-I1 final product (lane 1 in Figure 6B and C).
Figure 6.
Enhancement of trans-splicing by HIV-1 Tat. (A) Schematic representation of the trans-splicing assay: The two RNA constructs H1 (encoding exon 1 and the 5′ part of the intron) and H2 (encoding the 3′ part of the intron and exon 2) have to fold into a splicing competent structure (1). The splicing reaction was started by addition of the 32P-labeled GTP. The nucleic acid chaperone was eventually added at step 2. The final RNA products are represented in (4). The splicing rates are based on the levels of GTP-I1 (see B). (B and C). Assays where the H1 and H2 RNAs were incubated with or without a nucleic acid chaperone. At the end of the reaction RNAs were phenol treated to remove the protein and analysed by PAGE in denaturing conditions (see Materials and methods section). The RNA substrates (H1, H2), the ligated exons (E1-E2) and the product (guanosine-5′-intron G-I1) are indicated. Panel B. Lane 1: splicing reaction at 37°C with RNA alone at a concentration of 4 × 10−8 M. Lanes 2–5: HIV-1 NCp7 at concentrations of 2.5, 5, 10 and 20 × 10−7 M, corresponding to protein to nt ratios of 1:128, 1:64, 1:32 and 1:16. Lanes 6 and 7: NC(12–53) at concentrations of 1 and 2 × 10−6 M, corresponding to peptide to nt ratios of 1:32 and 1:16. Panel C. Lane 1: splicing reaction at 37°C with RNA alone at a concentration of 4 × 10−8 M. Lanes 2–6: Tat(1–86) at concentrations of 1.25, 2.5, 5, 10 and 20 × 10−7 M, corresponding to protein to nt molar ratios of 1:256, 1:128, 1:64, 1:32 and 1:16. Lanes 7 and 8: Tat(1–20) at concentrations of 1 and 2 × 10−6 M. Lanes 9–12: Tat(44–61) at concentrations of 2, 4, 8 and 16 × 10−7 M, corresponding to peptide to nt molar ratios of 1:170, 1:85, 1:42.5 and 1:21.2. Lanes 13–16: Tat(48–86) at concentrations of 2, 4, 8 and 16 × 10−7 M, corresponding to molar ratios of 1:170, 1:85, 1:42.5 and 1:21.2. Note that HIV-1 NCp7 and Tat strongly activated trans-splicing at protein to nt molar ratios of 1:32 (compare lanes 1 and 4 in B, and lanes 1 and 5 in C). Tat peptide (44–61) was also very active (lane 10 in C). Peptides NC(12–53) and Tat(1–20) were very poorly active.
HIV-1 NCp7 strongly stimulated trans-splicing (lanes 2–5), with an optimal enhancement at a concentration of 8 × 10−7 M (panel B, lane 4). Mutant NC(12–53) was found to be poorly active even at 2 × 10−6 M (panel B, lane 7).
Tat protein was found to be a strong activator of trans-splicing (Figure 6C, lanes 2–6), with an optimal activation at concentrations of 4–8 × 10−7 M (lanes 4 and 5). Interestingly the Tat peptide (44–61) also strongly activated trans-splicing (Figure 6C, lanes 9–12) at concentrations of 2–4 × 10−7 M (lanes 9 and 10) while the Tat peptide (48–86) was less efficient in activating trans-splicing at 8 × 10−7 M (lanes 15 and 16). Tat peptide (1–20) was totally inactive in the trans-splicing assay even at a concentration of 2 × 10−6 M (compare lanes 7 and 8 with lane 1 in Figure 6C).
DISCUSSION
The Tat protein plays a pivotal role in HIV-1 replication by strongly transactivating proviral DNA transcription, which necessitates the binding of Tat to the 5′ trans-activation response element (TAR) of the nascent viral RNA (9). This in turn causes the recruitment of the positive elongation factor P-TEFB allowing the rapid synthesis of the full-length HIV-1 mRNA by RNAP II and the cyclic reuse of the viral promoter (16–19,51–53). The interplay of Tat with the viral RNA extends beyond transcription activation since Tat also influences viral mRNA capping and splicing (20,21). In agreement with the view that Tat is multi-functional, it was recently found that Tat can suppress the cellular RNA interference machinery by interacting with DICER, TRBP, siRNA and the mRNA (22,23).
A computer analysis of Tat predicts that this small basic protein is essentially unstructured and thus belongs to the growing family of intrinsically unstructured proteins (IUP) (Supplementary Figure 1, ref. 25). A disordered nature of a protein is thought to render possible interactions with multiple molecular partners (28,54,55), which may account for different functions as in the case of retroviral NC proteins (56). Intrinsic disorder may also be important for the nucleic acid chaperone activity of Tat, contributing folding energy to nucleic acid rearrangement by way of an entropy exchange mechanism (40). Interestingly, the majority of proteins involved in transcription regulation was found to contain long disordered regions, indicating that protein chain flexibility is pivotal for this function (57,58). To better understand the multiple roles of Tat, we have investigated in vitro its interplay with nucleic acids, notably with DNA, RNA and DNA–RNA hybrid molecules.
Results reported here show that Tat has potent DNA and RNA chaperoning activities as evidenced by its ability to strongly enhance DNA–DNA annealing and exchange (Figures 3 and 4), ribozyme-directed cleavage of an RNA substrate (Figure 5) and RNA trans-splicing (Figure 6). In agreement with these findings, Tat can activate an RNA–DNA exchange within an RNA–DNA hybrid representing the HIV-1 promoter sequences (Supplementary Figure 2). Interestingly, Tat has a window of chaperoning activity corresponding to about one protein per 20 to 40 nt (Figures 2–6), also found for HIV-1 NC protein with similar values of window of activity (see Figure 5 for example and ref. 46). Previously, Guo et al. (59) reported that Tat could replace the HIV-1 NC protein in the initiation of reverse transcription by promoting primer tRNALys,3 placement onto the primer binding site and strand transfer in vitro. However, the relevance of these findings is still a matter of speculation, because only very small amounts of Tat are found in virions (60). Else Tat could well activate the intra-cellular viral DNA synthesis, which occurs in the case of HIV-1 with mutations in the NC zinc finger motifs (61).
Taken together, these results indicate that Tat is a nucleic acid chaperone with potent nucleic acid annealing and strand-displacement activities, that may act at the levels of proviral DNA transcription, viral RNA splicing and suppression of silencing (Figures 3–6) (8,9,21–23).
In an attempt to map the Tat sequences responsible for the chaperoning activities, we used peptides representing the N-terminal domain, peptide 1–20, the cysteine-rich domain, peptide 21–43, part of the core and nucleic acid-binding domains, peptide (44–61), and the nucleic acid-binding region together with the C-terminal fragment (48–86). We also used smaller Tat peptides, namely (44–57) and (48–61) (see Supplementary Data). Interestingly, the Tat peptide (44–61) was found to be almost as active as the Tat protein in three chaperoning assays (Figures 2, 3, 5 and 6). However, the use of a large panel of assays to dissect the chaperoning activities allowed us to find that the Tat peptides (44–61), (44–57), (48–61) and (48–86) were poorly active in the ribozyme-directed cleavage of an RNA substrate (Figure 5B, Supplementary Figure 3 and Table 3). This assay monitors three reactions that are RNA annealing and cleavage, and the release of the end products to allow a cyclic reuse of the R3 ribozyme (Figure 5A and Materials and methods section). Yet, we do not know what is the limiting step for the Tat peptides (44–61) and (48–86) in the ribozyme-directed cleavage reaction (Figure 5A). RNA chaperoning is a complex process, that involves molecular crowding achieved via the formation of nucleoprotein complexes (Figures 2 and 3; refs 46,62) where annealing of complementary strands and eventually their destabilization can take place (Figure 5). The degree of RNA occupancy by Tat peptides (44–61) and (48–86) is thought to be higher than that by Tat protein because the peptides bind 5 nt while it is 8 nt for the complete Tat protein (Figure 2). This could preclude the release of the cleaved RNA substrate and thus the cyclic reuse of the ribozyme. Strand displacement and release could be influenced by the cysteine-rich domain (Figure 1) as suggested by the work of Guo et al. (59) on DNA strand displacement, but this remains to be demonstrated.
Table 3.
Summary of the nucleic acid chaperoning activities of the Tat peptides
| Tat peptide | Assay | |||
|---|---|---|---|---|
| DNA annealing | DNA strand exchange | RNA ribozyme cleavage | RNA trans-splicing | |
| Tat(1–86) | + + + + | + + + + | + + + + | + + + + |
| Tat(1–20) | − | − | − | − |
| Tat(44–61) | + + + + | + + + + | + | + + + + |
| Tat(48–86) | + + | + + | + | + + |
| Tat(21–43) | + | ND | − | ND |
−/+/++/+ + +/+ + + + The crosses indicate increasing DNA/RNA chaperoning activities, from low (+) to high (++++) as compared with the Tat protein (++++). ND is not done.
Our results also show that the 18-residue-long peptide (Tat 44–61) is the smallest known peptide with most, but not all, DNA/RNA chaperoning properties (Table 3). The use of such a small peptide should facilitate high-throughput screening aimed at identifying small molecules capable of inhibiting Tat and thus HIV-1 proviral DNA transcription and viral RNA splicing. This is presently in progress.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This study was supported by ANRS, CNRS, INSERM (France), TRIoH (EU, 6th PCRDT) and Sidaction. Funding to pay the Open Access publication charges for this article was provided by ANRS (France).
Conflict of interest statement. None declared.
REFERENCES
- 1.Barre-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vezinet-Brun F, Rouzioux C, et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 2.Cullen BR. Regulation of HIV-1 gene expression. FASEB J. 1991;5:2361–2368. doi: 10.1096/fasebj.5.10.1712325. [DOI] [PubMed] [Google Scholar]
- 3.Strebel K. Virus-host interactions: role of HIV proteins Vif, Tat, and Rev. AIDS. 2003;17(Suppl. 4):S25–S34. doi: 10.1097/00002030-200317004-00003. [DOI] [PubMed] [Google Scholar]
- 4.Argyris EG, Pomerantz RJ. HIV-1 Vif versus APOBEC3G: newly appreciated warriors in the ancient battle between virus and host. Trends Microbiol. 2004;12:145–148. doi: 10.1016/j.tim.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 5.Darlix JL, Cristofari G, Rau M, Pechoux C, Berthoux L, Roques B. Nucleocapsid protein of human immunodeficiency virus as a model protein with chaperoning functions and as a target for antiviral drugs. Adv. Pharmacol. 2000;48:345–372. doi: 10.1016/s1054-3589(00)48011-7. [DOI] [PubMed] [Google Scholar]
- 6.Darlix JL, Lapadat-Tapolsky M, de Rocquigny H, Roques BP. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J. Mol. Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 7.Ciuffi A, Bushman FD. Retroviral DNA integration: HIV and the role of LEDGF/p75. Trends Genet. 2006;22:388–395. doi: 10.1016/j.tig.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 8.Jeang KT, Xiao H, Rich EA. Multifaceted activities of the HIV-1 transactivator of transcription, Tat. J. Biol. Chem. 1999;274:28837–28840. doi: 10.1074/jbc.274.41.28837. [DOI] [PubMed] [Google Scholar]
- 9.Gatignol A. Transcription of HIV: Tat and cellular chromatin. Adv. Pharmacol. 2007;55:137–159. doi: 10.1016/S1054-3589(07)55004-0. [DOI] [PubMed] [Google Scholar]
- 10.Xiao H, Lis JT, Jeang KT. Promoter activity of Tat at steps subsequent to TATA-binding protein recruitment. Mol. Cell. Biol. 1997;17:6898–6905. doi: 10.1128/mcb.17.12.6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brady J, Kashanchi F. Tat gets the “green” light on transcription initiation. Retrovirology. 2005;2:69. doi: 10.1186/1742-4690-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veschambre P, Roisin A, Jalinot P. Biochemical and functional interaction of the human immunodeficiency virus type 1 Tat transactivator with the general transcription factor TFIIB. J. Gen. Virol. 1997;78(Pt 9):2235–2245. doi: 10.1099/0022-1317-78-9-2235. [DOI] [PubMed] [Google Scholar]
- 13.Kashanchi F, Piras G, Radonovich MF, Duvall JF, Fattaey A, Chiang CM, Roeder RG, Brady JN. Direct interaction of human TFIID with the HIV-1 transactivator tat. Nature. 1994;367:295–299. doi: 10.1038/367295a0. [DOI] [PubMed] [Google Scholar]
- 14.Marzio G, Tyagi M, Gutierrez MI, Giacca M. HIV-1 tat transactivator recruits p300 and CREB-binding protein histone acetyltransferases to the viral promoter. Proc. Natl Acad. Sci. USA. 1998;95:13519–13524. doi: 10.1073/pnas.95.23.13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parada CA, Roeder RG. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature. 1996;384:375–378. doi: 10.1038/384375a0. [DOI] [PubMed] [Google Scholar]
- 16.Okamoto H, Sheline CT, Corden JL, Jones KA, Peterlin BM. Trans-activation by human immunodeficiency virus Tat protein requires the C-terminal domain of RNA polymerase II. Proc. Natl Acad. Sci. USA. 1996;93:11575–11579. doi: 10.1073/pnas.93.21.11575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chun RF, Jeang KT. Requirements for RNA polymerase II carboxyl-terminal domain for activated transcription of human retroviruses human T-cell lymphotropic virus I and HIV-1. J. Biol. Chem. 1996;271:27888–27894. doi: 10.1074/jbc.271.44.27888. [DOI] [PubMed] [Google Scholar]
- 18.O’Brien T, Hardin S, Greenleaf A, Lis JT. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- 19.Dahmus ME. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 20.Chiu YL, Ho CK, Saha N, Schwer B, Shuman S, Rana TM. Tat stimulates cotranscriptional capping of HIV mRNA. Mol. Cell. 2002;10:585–597. doi: 10.1016/s1097-2765(02)00630-5. [DOI] [PubMed] [Google Scholar]
- 21.Berro R, Kehn K, de la Fuente C, Pumfery A, Adair R, Wade J, Colberg-Poley AM, Hiscott J, Kashanchi F. Acetylated Tat regulates human immunodeficiency virus type 1 splicing through its interaction with the splicing regulator p32. J. Virol. 2006;80:3189–3204. doi: 10.1128/JVI.80.7.3189-3204.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennasser Y, Le SY, Benkirane M, Jeang KT. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22:607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 23.Bennasser Y, Jeang KT. HIV-1 Tat interaction with Dicer: requirement for RNA. Retrovirology. 2006;3:95. doi: 10.1186/1742-4690-3-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Viglianti GA, Mullins JI. Functional comparison of transactivation by simian immunodeficiency virus from rhesus macaques and human immunodeficiency virus type 1. J. Virol. 1988;62:4523–4532. doi: 10.1128/jvi.62.12.4523-4532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shojania S, O’Neil JD. HIV-1 Tat is a natively unfolded protein: the solution conformation and dynamics of reduced HIV-1 Tat-(1-72) by NMR spectroscopy. J. Biol. Chem. 2006;281:8347–8356. doi: 10.1074/jbc.M510748200. [DOI] [PubMed] [Google Scholar]
- 26.Ivanyi-Nagy R, Lavergne JP, Gabus C, Ficheux D, Darlix JL. RNA chaperoning and intrinsic disorder in the core proteins of Flaviviridae. Nucleic Acids Res. 2008;36:712–725. doi: 10.1093/nar/gkm1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gabus C, Mazroui R, Tremblay S, Khandjian EW, Darlix JL. The fragile X mental retardation protein has nucleic acid chaperone properties. Nucleic Acids Res. 2004;32:2129–2137. doi: 10.1093/nar/gkh535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivanyi-Nagy R, Davidovic L, Khandjian EW, Darlix JL. Disordered RNA chaperone proteins: from functions to disease. Cell Mol. Life Sci. 2005;62:1409–1417. doi: 10.1007/s00018-005-5100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Rocquigny H, Gabus C, Vincent A, Fournie-Zaluski MC, Roques B, Darlix JL. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc. Natl Acad. Sci. USA. 1992;89:6472–6476. doi: 10.1073/pnas.89.14.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gabus C, Ficheux D, Rau M, Keith G, Sandmeyer S, Darlix JL. The yeast Ty3 retrotransposon contains a 5′-3′ bipartite primer-binding site and encodes nucleocapsid protein NCp9 functionally homologous to HIV-1 NCp7. EMBO J. 1998;17:4873–4880. doi: 10.1093/emboj/17.16.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsuchihashi Z, Khosla M, Herschlag D. Protein enhancement of hammerhead ribozyme catalysis. Science. 1993;262:99–102. doi: 10.1126/science.7692597. [DOI] [PubMed] [Google Scholar]
- 32.Rajkowitsch L, Semrad K, Mayer O, Schroeder R. Assays for the RNA chaperone activity of proteins. Biochem. Soc. Trans. 2005;33:450–456. doi: 10.1042/BST0330450. [DOI] [PubMed] [Google Scholar]
- 33.Coetzee T, Herschlag D, Belfort M. Escherichia coli proteins, including ribosomal protein S12, facilitate in vitro splicing of phage T4 introns by acting as RNA chaperones. Genes Dev. 1994;8:1575–1588. doi: 10.1101/gad.8.13.1575. [DOI] [PubMed] [Google Scholar]
- 34.Chaloin O, Peter JC, Briand JP, Masquida B, Desgranges C, Muller S, Hoebeke J. The N-terminus of HIV-1 Tat protein is essential for Tat-TAR RNA interaction. Cell Mol. Life Sci. 2005;62:355–361. doi: 10.1007/s00018-004-4477-1. [DOI] [PubMed] [Google Scholar]
- 35.de Rocquigny H, Petitjean P, Tanchou V, Decimo D, Drouot L, Delaunay T, Darlix JL, Roques BP. The zinc fingers of HIV nucleocapsid protein NCp7 direct interactions with the viral regulatory protein Vpr. J. Biol. Chem. 1997;272:30753–30759. doi: 10.1074/jbc.272.49.30753. [DOI] [PubMed] [Google Scholar]
- 36.Kuipers BJ, Gruppen H. Prediction of molar extinction coefficients of proteins and peptides using UV absorption of the constituent amino acids at 214 nm to enable quantitative reverse phase high-performance liquid chromatography-mass spectrometry analysis. J. Agric. Food Chem. 2007;55:5445–5451. doi: 10.1021/jf070337l. [DOI] [PubMed] [Google Scholar]
- 37.Berkhout B, Jeang KT. Trans activation of human immunodeficiency virus type 1 is sequence specific for both the single-stranded bulge and loop of the trans-acting-responsive hairpin: a quantitative analysis. J. Virol. 1989;63:5501–5504. doi: 10.1128/jvi.63.12.5501-5504.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dingwall C, Ernberg I, Gait MJ, Green SM, Heaphy S, Karn J, Lowe AD, Singh M, Skinner MA. HIV-1 tat protein stimulates transcription by binding to a U-rich bulge in the stem of the TAR RNA structure. EMBO J. 1990;9:4145–4153. doi: 10.1002/j.1460-2075.1990.tb07637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuppuswamy M, Subramanian T, Srinivasan A, Chinnadurai G. Multiple functional domains of Tat, the trans-activator of HIV-1, defined by mutational analysis. Nucleic Acids Res. 1989;17:3551–3561. doi: 10.1093/nar/17.9.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18:1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- 41.Beltz H, Clauss C, Piemont E, Ficheux D, Gorelick RJ, Roques B, Gabus C, Darlix JL, de Rocquigny H, Mely Y. Structural determinants of HIV-1 nucleocapsid protein for cTAR DNA binding and destabilization, and correlation with inhibition of self-primed DNA synthesis. J. Mol. Biol. 2005;348:1113–1126. doi: 10.1016/j.jmb.2005.02.042. [DOI] [PubMed] [Google Scholar]
- 42.Egele C, Piemont E, Didier P, Ficheux D, Roques B, Darlix JL, Rocquigny HD, Mely Y. The single-finger nucleocapsid protein of moloney murine leukemia virus binds and destabilizes the TAR sequences of HIV-1 but does not promote efficiently their annealing. Biochemistry. 2007;46:14650–14662. doi: 10.1021/bi7012239. [DOI] [PubMed] [Google Scholar]
- 43.Darlix JL, Vincent A, Gabus C, de Rocquigny H, Roques B. Trans-activation of the 5′ to 3′ viral DNA strand transfer by nucleocapsid protein during reverse transcription of HIV1 RNA. C R Acad. Sci. III. 1993;316:763–771. [PubMed] [Google Scholar]
- 44.Tsuchihashi Z, Brown PO. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 1994;68:5863–5870. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herschlag D, Khosla M, Tsuchihashi Z, Karpel RL. An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J. 1994;13:2913–2924. doi: 10.1002/j.1460-2075.1994.tb06586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cristofari G, Darlix JL. The ubiquitous nature of RNA chaperone proteins. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:223–268. doi: 10.1016/s0079-6603(02)72071-0. [DOI] [PubMed] [Google Scholar]
- 47.Bernacchi S, Piemont E, Potier N, Van Dorsselaer A, Mély Y. Excitonic heterodimer formation in an HIV-1 oligonucleotide labeled with a donor-acceptor pair used for florescence resonance energy transfer. Biophys. J. 2003;84:643–654. doi: 10.1016/S0006-3495(03)74884-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Godet J, de Rocquigny H, Raja C, Glasser N, Ficheux D, Darlix JL, Mely Y. During the early phase of HIV-1 DNA synthesis, nucleocapsid protein directs hybridization of the TAR complementary sequences via the ends of their double-stranded stem. J. Mol. Biol. 2006;356:1180–1192. doi: 10.1016/j.jmb.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 49.Bertrand EL, Rossi JJ. Facilitation of hammerhead ribozyme catalysis by the nucleocapsid protein of HIV-1 and the heterogeneous nuclear ribonucleoprotein A1. EMBO J. 1994;13:2904–2912. doi: 10.1002/j.1460-2075.1994.tb06585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Semrad K, Green R, Schroeder R. RNA chaperone activity of large ribosomal subunit proteins from Escherichia coli. RNA. 2004;10:1855–1860. doi: 10.1261/rna.7121704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laspia MF, Rice AP, Mathews MB. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell. 1989;59:283–292. doi: 10.1016/0092-8674(89)90290-0. [DOI] [PubMed] [Google Scholar]
- 52.Kao SY, Calman AF, Luciw PA, Peterlin BM. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- 53.Feinberg MB, Baltimore D, Frankel AD. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc. Natl Acad. Sci. USA. 1991;88:4045–4049. doi: 10.1073/pnas.88.9.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 55.Dosztanyi Z, Chen J, Dunker AK, Simon I, Tompa P. Disorder and sequence repeats in hub proteins and their implications for network evolution. J. Proteome Res. 2006;5:2985–2995. doi: 10.1021/pr060171o. [DOI] [PubMed] [Google Scholar]
- 56.Darlix JL, Garrido JL, Morellet N, Mely Y, de Rocquigny H. Properties, functions, and drug targeting of the multifunctional nucleocapsid protein of the human immunodeficiency virus. Adv. Pharmacol. 2007;55:299–346. doi: 10.1016/S1054-3589(07)55009-X. [DOI] [PubMed] [Google Scholar]
- 57.Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry. 2006;45:6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Minezaki Y, Homma K, Kinjo AR, Nishikawa K. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcriptional regulation. J. Mol. Biol. 2006;359:1137–1149. doi: 10.1016/j.jmb.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 59.Guo X, Kameoka M, Wei X, Roques B, Gotte M, Liang C, Wainberg MA. Suppression of an intrinsic strand transfer activity of HIV-1 Tat protein by its second-exon sequences. Virology. 2003;307:154–163. doi: 10.1016/s0042-6822(02)00068-5. [DOI] [PubMed] [Google Scholar]
- 60.Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess J.W., Jr, Sowder R.C., II, Barsov E, Hood BL, Fisher RJ, et al. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J. Virol. 2006;80:9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Houzet L, Morichaud Z, Didierlaurent L, Muriaux D, Darlix JL, Mougel M. Nucleocapsid mutations turn HIV-1 into a DNA-containing virus. Nucleic Acids Res. 2008;36:2311–2319. doi: 10.1093/nar/gkn069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rajkowitsch L, Schroeder R. Dissecting RNA chaperone activity. RNA. 2007;13:2053–2060. doi: 10.1261/rna.671807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.