Abstract
All-trans retinoic acid (ATRA) is a potent inducer of cell differentiation and growth arrest. Here, we investigated ATRA-induced regulatory cascades associated with growth arrest of the human hepatoma cell line HepG2. ATRA induced >2-fold changes in the expression of 402 genes including 55 linked to cell-cycle regulation, cell growth or apoptosis during 48 h treatment. Computational search predicted that 250 transcriptional regulatory factors (TRFs) could recognize the proximal upstream regions of any of the 55 genes. Expression of 61 TRF genes was significantly changed during ATRA incubation, providing many potential regulatory edges. We focused on six TRFs that could regulate many of the 55 genes and found a total of 160 potential edges in which the expression of each of the genes was changed later than the expression change of the corresponding regulator. RNAi knockdown of the selected TRFs caused perturbation of the respective potential targets. The genes showed an opposite regulation pattern by ATRA and specific siRNA treatments were selected as strong candidates for direct TRF targets. Finally, 36 transcriptional regulatory edges were validated by chromatin immunoprecipitation. These analyses enabled us to depict a part of the transcriptional regulatory cascades closely linked to ATRA-induced cell growth arrest.
INTRODUCTION
All-trans retinoic acid (ATRA), a derivative of retinol (vitamin A), can regulate important biological processes such as cell differentiation and proliferation (1,2). ATRA binds to retinoic acid receptors (RARs), which are heterodimerized with the retinoid receptors (RXRs) and induces a protein conformational change to recruit coactivators leading to the transcriptional activation of their target genes (3,4). In the absence of the ligand, RAR/RXR heterodimer binds to its specific DNA sequences, retinoic acid response elements (RAREs) composed typically of two direct repeats of a core motif, PuG(G/T)TCA, and represses their target genes through recruitment of the corepressors NCoR and SMRT (5,6). When ATRA binds to RAR, RAR/RXR can activate transcription of their target genes (3,4). Because each of the RAR genes has a recognition site for their own protein products in their regulatory region, once it is activated by ATRA, its expression is auto-activated (7) except for the RARG1 gene, one of the isoforms of RARG, which can repress the activation of RARE (8,9). Many genes have been reported as retinoic acid responsive genes (10). However, how the transcriptional cascades and networks relevant to ATRA-induced biological events function remains unclear.
Dynamic transcriptional regulation is a key event that leads to time-, tissue- and/or cell-specific eukaryotic gene expression in response to extracellular signals. Identification of the transcriptional regulatory edges consisting of transcriptional regulatory factors (TRFs) and their regulated genes is important for understanding the mechanisms of a given biological phenomenon. We have been working to establish a system to identify the regulatory edges by using overexpression or RNAi knockdown of the TRF genes triggering perturbation of their regulated genes and cross-linking-chromatin immunoprecipitation (X-ChIP) to confirm the interactions between TRFs and their target DNA elements (11,12). In the present study, we applied this experimental scheme in combination with time-course expression profiling to analyze the dynamic transcriptional regulatory cascades related to ATRA-induced biological events as a model system.
In human hepatoma-derived HepG2 cells, ATRA inhibits G1/S transition in the cell cycle and results in growth arrest (13). We investigated the transcriptional regulatory cascades involved in the growth arrest of HepG2 cells induced by ATRA. First, the time-dependent ATRA-induced perturbation of gene expression was examined to extract the potential ATRA-responsive genes, which were then filtered according to gene ontology for cell-cycle regulation, cell growth or apoptosis. Second, TRFs that could regulate the filtered ATRA-responsive genes were selected by examining the location of their potential binding sites in the proximal upstream regions of their potential targets and their time-course expression profiles. Third, highly reliable edges were detected by RNAi knockdown of the selected six TRF genes and by quantifying the perturbation levels of their potential targets. Finally, X-ChIP analysis validated a number of direct and nondirect regulatory edges, leading to depiction of the transcriptional regulatory cascades linked to growth arrest of HepG2 cells.
MATERIALS AND METHODS
Reagents
ATRA and DMSO were purchased from Sigma (St Louis, MO, USA).
Cell culture and ATRA treatment
HepG2 cells were obtained from RIKEN Bioresource Center (Tsukuba, Japan) and cultured in Minimum Essential Medium Eagle (Sigma) with 1 mM sodium pyruvate (Sigma) supplemented with 10% fetal bovine serum at 37°C in a 5% CO2 and 95% atmosphere. HepG2 cells were seeded in 15 cm dishes and cultured for 24 h so that they would be 50% confluent at the time of ATRA treatment. Cells were washed with PBS twice, and then exposed to fresh medium with 50 μM ATRA dissolved in DMSO or with only DMSO. These cells were incubated at 37°C in a CO2 incubator for 72 h. Total RNAs were prepared at 6, 12, 24, 36 and 48 h and used for expression analysis. To investigate the cell growth rates, we counted viable cells by trypan blue exclusion assays at each time point.
siRNA transfection and RNA extraction
The synthetic siRNAs targeting human CEBPA, DDIT3, EGR1, RARA, RARB and SREBF1 were purchased from Invitrogen (Supplementary Table 1). Transfection of HepG2 cells with siRNAs (at a final concentration of 20 nM) in Opti-MEM medium (Invitrogen, Carlsbad, CA, USA) by using siPORT NeoFX (Ambion, Woodward, TX, USA) was done 24 h before ATRA treatment according to the manufacturer's protocol. We used Negative Control #1 siRNA (Ambion) as the control. Total RNA was extracted 48 h after siRNA administration from cells treated with ATRA or only DMSO with the NucleoSpin RNA II total RNA isolation kit (Macherey-Nagel, Germany) containing RNase-free DNaseI according to the manufacturer's instructions. The yield and purity of RNA were spectrophotometrically determined.
Microarray analysis
First-strand cDNA was synthesized from 500 ng of total RNA, then second-strand DNA was synthesized with DNA polymerase and used to degrade the RNA with RNase H simultaneously. After purification of the cDNA, this became a template for in vitro transcription with T7 RNA polymerase. In this step, multiple copies of biotinylated cRNA were produced. The purified cRNA was ready for hybridization. All steps were done with the Illumina RNA Amplification Kit (Ambion) according to the manufacturer's instructions. The concentration of the cRNA solution was determined by spectrophotometric measurement, and the size distribution of cRNA was evaluated using an Agilent 2100 Bioanalyzer. Next, cRNA was hybridized using Sentrix Human-6 Expression BeadChips (Illumina, San Diego, CA, USA) for gene expression profiling according to the manufacturer's instructions. Hybridization was done four times for each cRNA sample, and the signal intensity of each data set was cut off at <0.01 P-values. The data were analyzed with BeadStudio (Illumina), and genes with the expression ratio of ATRA-treated samples compared with DMSO-treated samples that showed >2-fold increase (>2.0) or decrease (<0.5) in signal intensity ratio were considered to be ATRA-modulated genes and the remaining genes were excluded for further examination. We registered all Illumina microarray data to CIBEX (DDBJ, accession number CBX36).
Search for potential transcription factor-binding sites
The upstream regions (2.2 kb) extending from −2000 to +200 relative to the transcription start sites of the ATRA-modulated genes were extracted from the UCSC database. We searched the proximal promoter region of transcription factor-binding sites using the MATCH program (14) of the Transfac database (Biobase) and the TESS program (http://www.cbil.upenn.edu/tess/techreports/1997/CBIL-TR-1997-1001-v0.0.pdf and http://www.cbil.upenn.edu/tess?RQ=WELCOME for WEB searching). The cut-off to minimize false positive matches (minFP) was applied when searching the DNA sequences.
Quantitative real-time RT–PCR (qRT–PCR)
Total RNA was reverse-transcribed using Ex-script (Takara Bio, Shiga, Japan) with random 6mer primers. The resultant cDNAs were used as templates for PCR reaction with SYBR Premix Ex Taq Perfect Real Time (Takara Bio), 2.5 μl of 1:5 diluted cDNA and gene-specific primers (final concentrations of 200 nM) on a 10-μl scale. The primer sets used for real-time RT–PCR analysis are shown in Supplementary Tables 2 and 3 (for expression analysis of TRF genes and validation of the TRF-regulated gene pairs, respectively). PCR reaction was carried out on the ABI PRISM 7500 Fast Real-Time PCR System (Applied Biosystems) by denaturation at 95°C for 10 s, followed by running for 40 cycles at 95°C for 5 s and 62.5°C for 20 s. The expression ratio was calculated according to the 2−ΔΔCT method (12,15).
X-ChIP/quantitative real-time PCR (qPCR)
The procedures for X-ChIP were essentially as described elsewhere (11) with some modifications. The soluble chromatin was incubated with anti-CEBPA, DDIT3, EGR1, RARA, RARB, SREBF1 specific antibodies (sc-61, sc-7351, sc-189, sc-551, sc-552 and sc-17755, respectively; SantaCruz Biotechnology, Santa Cruz, CA, USA) for >12 h at 4°C. The chromatin–antibody mixture was incubated with Dynabeads Protein G (Dynal Biotech, Oslo, Norway) for 1 h at 4°C and the immunoprecipitates were captured using magnets. The recovered immunoprecipitates were washed once with IP wash buffer Low (2 mM EDTA, 20 mM Tris–HCl pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.1% SDS), once with IP wash buffer High (2 mM EDTA, 20 mM Tris–HCl pH 8.0, 500 mM NaCl, 1% Triton X-100, 0.1% SDS), once with IP wash buffer LiCl (1 mM EDTA, 10 mM Tris–HCl pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate) and twice with TE buffer (10 mM Tris–HCl pH 8.0, 1 mM EDTA). The reversal of formaldehyde-induced cross links was carried out by heating at 65°C for 3.5 h with 200 mM NaCl and RNaseI (Nippon Gene, Tokyo, Japan) treated at 65°C for 0.5 h. The reversal cross-linked complexes were treated with 0.05 mg/ml proteinase K (Nippon Gene) at 45°C for 1 h. Released DNA was extracted with phenol and phenol:chloroform:isoamyl alcohol (25:24:1), then ethanol-precipitated and dissolved in 100 μl of H2O. DNA obtained by ChIP with TRF-specific antibody, from the precipitates without any antibody and input DNA (total chromatin DNA) were used as templates for qRT–PCR. The PCR mixture contained 2 μl of DNA preparation, 200 nM of each of the specific primer sets, and SYBR Premix ExTaq (Takara) in a total volume of 10 μl. The specific primer sets were designed so as to amplify the DNA fragments surrounding the potential binding sequence of TRF detected as described in the analysis of consensus TRF binding sequences. These primer sequences are presented in Supplementary Table 4. The PCR reaction was carried out under the same conditions as those for expression analysis.
RESULTS
ATRA-induced growth arrest of HepG2 cells
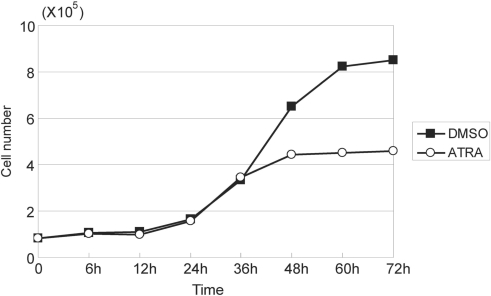
First, the concentration of ATRA to induce arrest of the growth of HepG2 cells was determined. HepG2 cells were cultured in the presence or absence of various concentrations of ATRA ranging from 10 to 50 µM, and the cell count was determined by trypan blue exclusion assay. HepG2 cells treated with 50 μM of ATRA reproducibly showed clear growth inhibition within 48 h after the start of treatment (Figure 1). On the other hand, lower concentrations of ATRA did not exhibit complete growth arrest of HepG2 cells under the conditions used.
Figure 1.
Cell number of HepG2 cells treated with 50 μM ATRA or 0.1% DMSO. ATRA induced growth arrest of HepG2 within 48 h.
Time-course analysis of ATRA-induced gene expression changes
To search for the genes affected by ATRA treatment, we comprehensively analyzed expression profiles at 6, 12, 24, 36 and 48 h after the start of incubation with the retinoid with Sentrix Human-6 Expression BeadChips covering about 47 300 different human transcripts. The time-course expression profiling data were grouped into three: two biological replicates, each of which consisted of two technical replicates, and the set of all the four replicates (two biological and two technical replicates). The data in each of these three groups were normalized independently. Only genes whose expression levels were significantly changed in two or all of the three groups were selected for further analysis. We identified 402 genes that were upregulated or downregulated in ATRA-treated cells >2-fold as compared with the levels of genes in DMSO-treated cells at any of the time points examined (Supplementary Table 5). In HepG2 cells, the growth arrest induced by ATRA was not accompanied by any change in the cyclin D1 expression level, whereas a reduction was observed in a previous study (16). Instead, expression of cyclin D2 was increased at an early stage of ATRA treatment and the mRNA level remained high up to 48 h.
RARA and RARB genes were upregulated by ATRA, but the expression signal of the latter was not detected with the RNA sample prepared from the DMSO-treated cells. On the other hand, no detectable level of hybridization signals for RARG gene expression was obtained, indicating that its expression was very low even after ATRA induction in HepG2 cells. We then tried to analyze their expression levels by qRT–PCR and found that the mRNA level of RARB was greatly increased within 6 h after ATRA administration, but the RARG gene expression remained at very low levels throughout the 48-h cell culture with ATRA.
ATRA significantly induced expression of 47 genes within 6 h. Because these immediate early genes may be the promising candidates for the direct targets of RARs, we examined whether they had a RARE in their upstream regions from −2000 to +200 bp relative to their transcriptional start sites. To search for potential RAR-binding sites with the MATCH program of TRANSFAC (Table 1), we used two kinds of matrix profiles (V$DR1_Q3 and V$DR4_Q2 with the accession numbers M00762 and M00965, respectively) defined on the basis of the sequence data including RAR–RXR heterodimer-binding sites. The genomic DNA sequences of two genes (MATND6 and Hs.508390) on the Sentrix Human-6 Expression BeadChips were not found in public DNA databases. This search detected 27 genes (60% of the 45 genes) that had a potential RAR recognition site in their proximal upstream regions (Table 1).
Table 1.
Immediate early ATRA-inducible genes having a RARE in their proximal upstream regions
| Gene | Refseq ID | Position | Matrix | Sequence |
|---|---|---|---|---|
| ADAMTS4 | NM_005099 | −403(+) | V$DR1_Q3 | gggcCAAAGggca |
| BHLHB2 | NM_003670 | −1934(+) | V$DR1_Q3 | agcgCAGAGgtca |
| DDIT3 | NM_004083 | −735(+) | V$DR4_Q2 | tgacctcaagTGATCcg |
| DNAJB9 | NM_012328 | −487(+) | V$DR1_Q3 | agccCAAAGgcca |
| −480(+) | V$DR1_Q3 | aggcCACAGgtcc | ||
| FSTL3 | NM_005860 | −731(+) | V$DR1_Q3 | aggaCAAAGgcca |
| +63(−) | V$DR1_Q3 | tggcCTCTGccct | ||
| HMGCS1 | NM_002130 | +154(−) | V$DR1_Q3 | tggaCTTTGcccg |
| HOZFP | NM_152995 | −1706(+) | V$DR4_Q2 | tgacctaaggTGATCca |
| −296(−) | V$DR4_Q2 | caGGTGAgtttagggca | ||
| IGFBP6* | NM_002178 | −667(+) | GGTCaNNNNNNGTTCA | |
| KCNJ10 | NM_002241 | −1709(+) | V$DR4_Q2 | tgcccgtgaaTGACCtc |
| KNG | NM_000893 | −1798(−) | V$DR4_Q2 | caGATCActagaggtca |
| LCAT | NM_000229 | −1827(+) | V$DR4_Q2 | tgccctgcccTGGCCcc |
| LOC389058 | NM_001003845 | −1694(+) | V$DR1_Q3 | gggaGATAGgtca |
| LOC144100 | NM_175058 | −527(+) | V$DR4_Q2 | tggactcgatTAACCtt |
| −217(+) | V$DR1_Q3 | gggaCAAAGgccg | ||
| LOC55971 | NM_018842 | −1580(+) | V$DR4_Q2 | tgacctcaagTGATCcg |
| −1488(−) | V$DR1_Q3 | agacCTCTGccct | ||
| NR0B2 | NM_021969 | −787(+) | V$DR4_Q2 | tgacctcaaaTGATCcc |
| OSTalpha | NM_152672 | −1757(+) | V$DR1_Q3 | cggcCACAGctca |
| OSTbeta | NM_178859 | −1191(−) | V$DR4_Q2 | tgGATCAcctgaggtca |
| PGRP-L | NM_052890 | −134(−) | V$DR1_Q3 | tgacCACTGacct |
| RBP1 | NM_002899 | −547(+) | V$DR4_Q2 | tgacctcaggTGATCcg |
| SDS-RS1 | NM_138432 | −1235(−) | V$DR1_Q3 | tgatCTCTGccca |
| −1119(−) | V$DR1_Q3 | agacCTTTGcact | ||
| −901(+) | V$DR1_Q3 | aggcCAAAGttcc | ||
| −380(+) | V$DR4_Q2 | agacctaagcTGACCcc | ||
| +158(+) | V$DR1_Q3 | tgggCAAAGtcca | ||
| SGK | NM_005627 | −1940(−) | V$DR4_Q2 | gaGCTCActttagggca |
| −1775(−) | V$DR4_Q2 | gaGGTCAtggcagggca | ||
| SLC16A5 | NM_004695 | −1214(+) | V$DR1_Q3 | gaggCAAAGgtca |
| −826(−) | V$DR4_Q2 | tgGATCAcctgaggtca | ||
| SLC22A3 | NM_021977 | −569(+) | V$DR4_Q2 | tgccctgcccTGCCCca |
| −176(−) | V$DR1_Q3 | cgacCTGTGgccc | ||
| SOS1 | NM_005633 | −1797(+) | V$DR4_Q2 | tgacctcaggTGATCca |
| TCEA2 | NM_003195 | −700(+) | V$DR4_Q2 | tgacctcaggTGATCca |
| TGM2* | NM_198951 | −969(+) | AGGTcANNNNNGGGTGA | |
| −1528(+) | GGtCANNNNNNGGTCA | |||
| +152(+) | GGtCANNNNNGGTCA | |||
| UPP1 | NM_003364 | −888(+) | V$DR4_Q2 | tgacctcaggTGATCca |
Forty-seven genes induced within 6 h after the start of ATRA treatment were detected by microarray analysis using Sentrix Human-6 Expression BeadChips. A search for the potential RAR binding sequences in their proximal upstream regions from −2000 to +200 bp relative to their transcriptional start sites was carried out with the MATCH program of TRANSFAC and the TESS program. Twenty-seven genes were found to have one or more RAREs. Two genomic sequences corresponding to the transcripts (MTND6 and Hs.508390) included in the Sentrix Human-6 Expression BeadChips were not found in public databases. The potential RAR-binding sites of the two genes with an asterisk were searched for using the TESS program because no RARE was detected in their proximal upstream regions by using TRANSFAC matrices.
To probe the cascades involved in the ATRA-induced cell growth arrest of HepG2, we selected 55 from 402 genes based upon their known relatedness to the processes of cell cycle, cell growth and/or apoptosis according to Gene Ontology (Table 2).
Table 2.
ATRA-induced expression changes of genes linked to cell growth, apoptosis and/or cell cycle
| Symbol | RefSeq ID | 6 h | 12 h | 24 h | 36 h | 48 h |
|---|---|---|---|---|---|---|
| ANXA3* | NM_005139 | 2.56 | 2.83 | 2.24 | ||
| AREG | NM_001657 | 3.88 | ||||
| BHD | NM_144606 | 2.50 | ||||
| BMP4 | NM_130851 | 0.44 | ||||
| C20orf97 | NM_021158 | 2.51 | 3.12 | 3.84 | ||
| CARD9 | NM_052814 | 2.63 | 2.48 | |||
| CCND2 | NM_001759 | 2.09 | ||||
| CNK | NM_004073 | 3.52 | ||||
| CTGF | NM_001901 | 0.46 | ||||
| DDIT3 | NM_004083 | 2.22 | 2.01 | 2.38 | 4.09 | 3.98 |
| DTNA | NM_001392 | 2.26 | ||||
| DUSP1 | NM_004417 | 2.69 | 2.65 | |||
| DUSP6 | NM_022652 | 0.42 | 0.37 | 0.38 | ||
| ELF5 | NM_001422 | 0.40 | ||||
| FABP3 | NM_004102 | 0.42 | ||||
| FGA | NM_021871 | 2.37 | 2.44 | |||
| FGB | NM_005141 | 2.55 | 3.44 | 2.36 | ||
| FGG | NM_000509 | 2.44 | 2.06 | 2.08 | ||
| FLJ30999 | NM_152461 | 2.94 | ||||
| FLJ31051 | NM_153687 | 0.43 | ||||
| FSCN1 | NM_003088 | 0.39 | ||||
| FSTL3 | NM_005860 | 4.99 | 3.80 | 4.30 | 7.88 | 9.95 |
| GPC6 | NM_005708 | 2.26 | 2.14 | 2.82 | ||
| HYPE | NM_007076 | 2.91 | 2.30 | |||
| IFITM1 | NM_003641 | 0.41 | 0.29 | |||
| IGF2 | NM_000612 | 2.39 | 2.09 | 4.50 | 5.11 | 9.80 |
| IGFBP1 | NM_000596 | 2.27 | ||||
| IGFBP6 | NM_002178 | 2.15 | ||||
| IGFBP7 | NM_001553 | 0.48 | 0.33 | |||
| IL17RB | NM_018725 | 0.43 | ||||
| INHBE | NM_031479 | 2.94 | 6.65 | 5.63 | 4.71 | |
| LIF | NM_002309 | 4.69 | ||||
| LOC117584 | NM_057178 | 0.47 | ||||
| MAPK13 | NM_002754 | 2.36 | ||||
| MDK | NM_002391 | 2.37 | ||||
| MGC8685 | NM_178012 | 3.24 | 3.78 | 4.38 | ||
| MLLT3 | NM_004529 | 0.45 | ||||
| MLLT7 | NM_005938 | 0.49 | ||||
| MXD4 | NM_006454 | 0.36 | ||||
| NEDD9 | NM_182966 | 0.40 | ||||
| OKL38 | NM_182981 | 2.40 | 2.23 | |||
| PDZK1 | NM_002614 | 0.47 | 0.45 | 0.31 | 0.23 | |
| PEG10 | NM_015068 | 0.36 | 0.29 | |||
| PLAB | NM_004864 | 2.72 | 2.45 | |||
| PLAU | NM_002658 | 0.43 | 0.38 | 0.34 | ||
| PPP1R15A | NM_014330 | 3.66 | 3.87 | |||
| RARA | NM_000964 | 2.21 | 2.65 | |||
| SESN2 | NM_031459 | 2.29 | 2.39 | 2.85 | ||
| SGK | NM_005627 | 0.33 | 0.23 | 0.18 | 0.26 | |
| TGM2* | NM_198951 | 2.24 | 2.25 | |||
| TNFRSF12A | NM_016639 | 2.70 | ||||
| TNFRSF19 | NM_018647 | 0.34 | ||||
| TNFSF10 | NM_003810 | 0.42 | 0.46 | |||
| TUBE1 | NM_016262 | 2.66 | 2.88 | |||
| WARS | NM_004184 | 2.40 |
The expression ratio represents the ratio of the expression level in the cells treated with ATRA to that in those incubated with only DMSO for the periods of time indicated. The four profiling data sets (two biological and two technical replicates) were categorized into three groups: each of the biological replicates and a sum of all the four replicates. Only data exhibited a P-value <0.01 were extracted and only genes whose expression levels were significantly changed in two or all of the three groups were selected. Results are the average of two or three groups of data analyzed. Genes with an asterisk were not assigned to any GO category, but are known to be related to cell growth or apoptosis (34,35).
Search of transcription factor genes involved in ATRA response
Next, we searched for the potential TRF-binding sites in the proximal upstream regions of the 55 selected ATRA-induced genes by using the MATCH program in TRANSFAC and detected them for a total of 250 different TRFs in the upstream regions of these genes. We then analyzed the time-dependent changes in the expression levels of these TRF genes. Among them, the expression levels of 109 TRF genes were high enough to be estimated by microarray analysis. However, the BeadChips could not be used to evaluate the expression levels of 141 genes because their expressions were too weak to be detected effectively by this technique. Their levels were examined using qRT–PCR, and 62 genes were found to be poorly expressed in HepG2 cells either before or after ATRA treatment. Therefore, the remaining 79 genes were examined for the time-dependent changes in their expressions with qRT–PCR. We found that ATRA treatment changed the expression levels of 61 out of the 188 TRF genes (109 and 79 genes based on microarray and qRT–PCR data, respectively) >1.5-fold as compared with the control (only incubation with DMSO) at any of the time points tested (Table 3). If a TRF regulates expression of some other genes, the time points when the expression levels of the regulated genes are changed are expected to be later than when the TRF gene expression is changed. Based on this assumption, we selected a total of 886 pairs of TRFs and their regulated genes from all combinations of 55 genes perturbed >2-fold by ATRA treatment and classified by GO as the genes related to the cell growth regulatory processes with 61 TRFs whose expression levels were changed >1.5-fold (based on the data of qRT–PCR analysis) before the 2-fold changes in the expression of the corresponding candidate regulated genes.
Table 3.
TRF genes whose expression levels were changed >1.5-fold by ATRA at one or more time points
| Symbol | RefSeq ID | 6 h | 12 h | 24 h | 36 h | 48 h | Detection |
|---|---|---|---|---|---|---|---|
| BHLHB2 | NM_003670 | 2.06 | 2.02 | 2.21 | 2.28 | 2.46 | Illumina |
| CEBPA | NM_004364 | 1.49 | 1.49 | 1.69 | 1.89 | 2.04 | Illumina |
| CEBPD | NM_005195 | 0.89 | 0.74 | 0.72 | 0.63 | 0.56 | Illumina |
| CEBPG | NM_001806 | 1.12 | 1.29 | 1.49 | 1.56 | 1.81 | Illumina |
| CUTL1 | NM_181500 | 1.36 | 1.07 | 1.01 | 2.20 | 2.09 | Illumina |
| DDIT3 | NM_004083 | 2.22 | 2.01 | 2.38 | 4.09 | 3.98 | Illumina |
| DEAF1 | NM_021008 | 0.75 (0.06) | 0.67 (0.13) | 0.75 (0.13) | 0.85 (0.20) | 0.91 (0.29) | qPCR |
| EGR1 | NM_001964 | 0.94 | 0.69 | 0.94 | 3.15 | 5.15 | Illumina |
| ELF4 | NM_001421 | 1.36 | 1.90 | 1.16 | 0.92 | 1.46 | Illumina |
| ETV6 | NM_001987 | 1.00 | 1.03 | 1.12 | 1.56 | 1.20 | Illumina |
| FOS | NM_005252 | 0.51 (0.22) | 0.27 (0.06) | 0.55 (0.18) | 0.46 (0.08) | 0.76 (0.21) | qPCR |
| FOSL2 | NM_005253 | 0.80 | 0.64 | 0.98 | 0.88 | 1.10 | Illumina |
| FOXF1 | NM_001451 | 0.91 (0.03) | 1 (0.05) | 0.88 (0.13) | 1.11 (0.14) | 2.19 (0.41) | qPCR |
| FOXF2 | NM_001452 | 1.14 (0.06) | 1.12 (0.03) | 1 (0.31) | 2.19 (0.62) | 4.32 (1.13) | qPCR |
| FOXJ2 | NM_018416 | 0.68 (0.05) | 0.79 (0.11) | 1.11 (0.12) | 1.56 (0.01) | 4.63 (0.33) | qPCR |
| GABPB2 | NM_016655 | 1.21 | 1.14 | 1.09 | 1.36 | 1.79 | Illumina |
| GFI1 | NM_005263 | 0.83 | 0.84 | 0.89 | 0.57 | 0.93 | Illumina |
| GFI1B | NM_004188 | 0.42 (0.08) | 0.39 (0.08) | 0.35 (0.06) | 0.4 (0.05) | 0.34 (0.02) | qPCR |
| GLI1 | NM_005269 | 1.86 (0.46) | 1.5 (0.43) | 0.72 (0.26) | 0.91 (0.06) | 1.02 (0.50) | qPCR |
| HIF1A | NM_181054 | 0.97 (0.21) | 0.82 (0.12) | 0.6 (0.11) | 1.03 (0.17) | 0.77 (0.2) | qPCR |
| HNF4A | NM_178850 | 0.62 (0.08) | 0.9 (0.1) | 0.95 (0.15) | 0.99 (0.06) | 1.02 (0.17) | qPCR |
| HNF4G | NM_004133 | 0.67 (0.1) | 0.72 (0.14) | 0.94 (0.21) | 1.24 (0.24) | 1.23 (0.32) | qPCR |
| HOXA5 | NM_019102 | 4.50 | 3.59 | 3.70 | 4.34 | 4.27 | Illumina |
| IRF1 | NM_002198 | 1.01 | 0.86 | 1.19 | 1.35 | 1.73 | Illumina |
| JUN | NM_002228 | 0.87 (0.23) | 0.82 (0.25) | 0.79 (0.20) | 1.41 (0.35) | 1.68 (0.37) | qPCR |
| JUNB | NM_002229 | 0.75 (0.13) | 0.63 (0.05) | 0.86 (0.23) | 1.09 (0.13) | 1.53 (0.35) | qPCR |
| MAD | NM_002357 | 1.5 (0.09) | 1.17 (0.13) | 1.25 (0.21) | 1.79 (0.08) | 1.87 (0.43) | qPCR |
| MAFF | NM_152878 | 1.1 (0.08) | 1.03 (0.49) | 1.11 (0.25) | 1.49 (0.16) | 2.6 (0.46) | qPCR |
| MAFK | NM_002360 | 1.31 (0.09) | 1.21 (0.18) | 1.06 (0.14) | 1.22 (0.20) | 2.2 (0.49) | qPCR |
| NFATC2 | NM_012340 | 0.36 (0.07) | 0.54 (0.03) | 0.81 (0.05) | 1.02 (0.09) | 1.12 (0.21) | qPCR |
| NR1H3 | NM_005693 | 0.98 (0.38) | 0.93 (0.18) | 1.07 (0.11) | 0.94 (0.38) | 1.98 (0.44) | qPCR |
| NR1H4 | NM_005123 | 0.90 | 1.20 | 1.49 | 1.58 | 1.47 | Illumina |
| NR1I2 | NM_022002 | 0.85 | 0.58 | 0.63 | 0.60 | 0.58 | Illumina |
| NR2F1 | NM_005654 | 0.86 | 0.53 | 0.67 | 0.71 | 0.63 | Illumina |
| NRF1 | NM_005011 | 0.63 (0.06) | 0.84 (0.17) | 1.4 (0.33) | 1.28 (0.33) | 1.51 (0.34) | qPCR |
| PAX6 | NM_000280 | 2.14 (0.23) | 1.58 (0.28) | 1.5 (0.05) | 1.47 (0.23) | 2.97 (0.40) | qPCR |
| PAX8 | NM_013992 | 1.04 | 0.77 | 1.08 | 0.76 | 0.67 | Illumina |
| PBX3 | NM_006195 | 0.55 | 1.08 | 1.06 | 0.90 | 1.13 | Illumina |
| PGR | NM_000926 | 1 (0.23) | 0.93 (0.26) | 0.98 (0.28) | 1.03 (0.27) | 1.99 (0.25) | qPCR |
| POU6F1 | XM_352901 | 1.6 (0.34) | 1.53 (0.24) | 1.12 (0.21) | 1.11 (0.05) | 0.67 (0.15) | qPCR |
| RARA | NM_000964 | 1.48 | 1.56 | 1.52 | 2.21 | 2.65 | Illumina |
| RARB | NM_016152 | 7.37 (2.74) | 7.11 (2.12) | 9.64 (3.05) | 10.62 (3.62) | 12.44 (3.96) | qPCR |
| RARG | NM_000966 | 0.57 (0.24) | 0.78 (0.16) | 0.56 (0.19) | 0.74 (0.09) | 0.75 (0.07) | qPCR |
| RFX5 | NM_000449 | 0.84 | 0.84 | 0.94 | 0.74 | 0.58 | Illumina |
| RFXANK | NM_134440 | 0.82 | 0.64 | 1.05 | 1.39 | 1.10 | Illumina |
| RXRA | NM_002957 | 0.66 | 0.73 | 0.88 | 0.81 | 0.67 | Illumina |
| SP2 | NM_003110 | 0.98 | 0.95 | 0.63 | 0.90 | 0.77 | Illumina |
| SREBF1 | NM_004176 | 2.26 | 2.31 | 2.31 | 2.21 | 2.20 | Illumina |
| SREBF2 | NM_004599 | 0.66 | 0.77 | 1.35 | 1.44 | 1.14 | Illumina |
| SRY | NM_003140 | 0.5 (0.08) | 0.69 (0.04) | 0.82 (0.05) | 0.59 (0.05) | 0.51 (0.04) | qPCR |
| STAT2 | NM_005419 | 0.99 | 1.08 | 0.97 | 0.72 | 0.64 | Illumina |
| STAT6 | NM_003153 | 1.19 | 0.91 | 1.04 | 0.65 | 0.81 | Illumina |
| TCF2 | NM_000458 | 0.80 | 1.07 | 0.86 | 0.86 | 0.68 | Illumina |
| TCF7 | NM_003202 | 1.40 | 1.17 | 1.33 | 1.45 | 1.95 | Illumina |
| TP53 | NM_000546 | 1.20 | 1.27 | 0.69 | 0.94 | 1.59 | Illumina |
| USF2 | NM_003367 | 1.34 | 1.09 | 1.25 | 1.50 | 1.51 | Illumina |
| VDR | NM_000376 | 1.53 (0.16) | 1.45 (0.13) | 0.92 (0.08) | 0.84 (0.03) | 0.77 (0.16) | qPCR |
| ZBTB33 | NM_006777 | 0.7 (0.01) | 0.56 (0.01) | 0.69 (0.00) | 0.66 (0.04) | 0.56 (0.09) | qPCR |
| ZIC2 | NM_007129 | 1.15 | 1.14 | 1.23 | 1.24 | 1.93 | Illumina |
| ZNF219 | NM_016423 | 0.84 | 0.73 | 0.65 | 0.91 | 0.77 | Illumina |
| ZNF238 | NM_006352 | 0.68 (0.38) | 0.72 (0.18) | 0.74 (0.13) | 1.08 (0.08) | 0.63 (0.15) | qPCR |
Expression levels of the genes not detected by microarray were analyzed using qRT–PCR. The expression ratio represents the ratio of expression levels in the cells treated with ATRA to that in those incubated with only DMSO for the periods of time indicated. Results are the average of two or three groups of data analyzed with Illumina microarrays and the average of four data sets (a combination of two sets of biological replicates) obtained by qRT–PCR analysis with SD values in parenthesis.
Validation of the regulatory relationships by expression perturbation upon knockdown of specific TRFs
To confirm the TRF-regulated gene pairs, we analyzed expression perturbation of the candidate-regulated genes by knockdown of the corresponding TRF genes. From a list of 61 TRFs, we chose six TRFs, RARA, RARB, CEBPA, DDIT3, EGR1 and SREBF1 having candidate regulated genes of 18, 18, 39, 26, 16 and 43, respectively (160 genes in total). All of these six TRFs are upregulated by ATRA treatment. Both RARA and RARB were chosen because the ATRA-induced regulatory cascades should originate from them. It was found that CEBPA and SREBF1 had many regulated gene candidates, 39 and 43 genes, respectively, and therefore were considered to play important roles in ATRA-induced regulatory cascades. DDIT3 (also known as CHOP-10) is a CEBP family member and known to be induced by stress leading to cell growth arrest and/or apoptosis (17). It has been reported that EGR1 induces cell growth arrest and regulation of EGR1 is involved in the MAPK pathway (18), a master regulatory process of the G1 to S-phase transition (19).
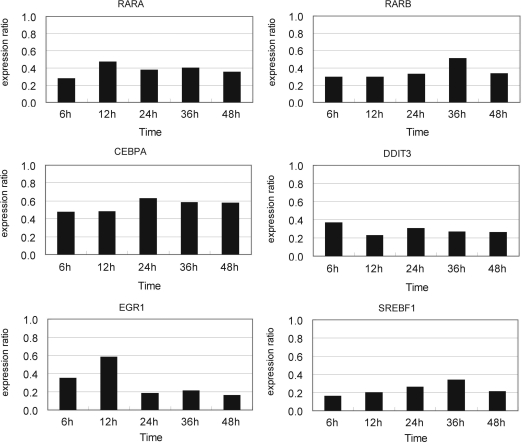
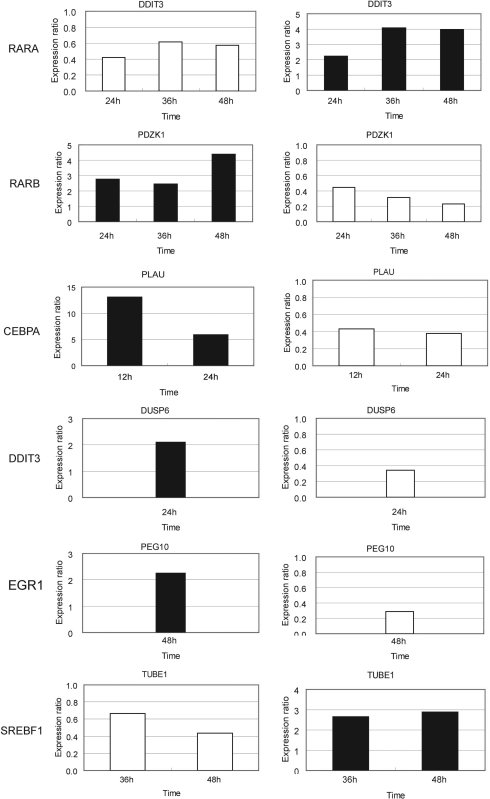
Knockdown of the six TRF genes was performed twice by addition of 20 nM (final concentration) of specific siRNAs 24 h before the start of ATRA treatment. Expression of all these six TRF genes was repressed throughout the 48 h ATRA treatment (Figure 2). We analyzed the expression levels of all of their regulated gene candidates at 6, 12, 24, 36 and 48 h after ATRA addition with qRT–PCR and selected 47 genes (9, 8, 13, 5, 2 and 10 genes for RARA, RARB, CEBPA, DDIT3, EGR1 and SREBF1, respectively) whose expression levels were changed >1.5-fold by siRNA targeting the respective TRF as compared with negative control siRNA. On the basis that all of these six TRF genes were upregulated by ATRA treatment (Table 3), we considered that genes regulated by each of these six TRFs would be reversely regulated by ATRA treatment and RNAi knockdown of the TRF gene. Knockdown of each of the six TRF genes led to detection of a total of 36 out of 160 genes (7/18, 6/18, 11/39, 3/26, 1/16, 8/43 for RARA, RARB, CEBPA, DDIT3, EGR1 and SREBF1, respectively) that clearly exhibited such a reverse regulatory pattern by ATRA treatment and knockdown of the respective TRF (Figure 3 for several examples; Supplementary Table 6). These genes and the corresponding TRFs are strong candidates for direct transcriptional regulatory edges in the regulatory cascades related to ATRA-induced growth arrest. In addition, 11 other genes were significantly perturbed by a specific siRNA in either of the two experiments and showed the reverse regulatory pattern. Therefore, these genes were also selected as potential candidates for direct regulatory node.
Figure 2.
Expression levels of the six selected TRF genes suppressed by respective siRNAs at several time points. HepG2 cells were transfected with each of the specific siRNAs (20 nM, final concentration) 24 h before ATRA addition, and the total RNA was extracted 6, 12, 24, 36 and 48 h after ATRA addition. The expression ratio was calculated according to the 2−ΔΔCT method as described in ‘Materials and methods’ section with GAPDH mRNA and negative control siRNA as the internal reference and the calibrator, respectively. The data are the averages of two biological replicates.
Figure 3.
Reverse perturbation patterns of gene expression induced by RNAi knockdown and ATRA treatment. Expression perturbations of the selected TRF-regulated genes by knockdown of the corresponding TRF genes are on the left. The expression ratio was calculated as in the legend to Figure 2. The reversed regulatory pattern was observed by ATRA treatment (right). The expression ratio was normalized by the expression level observed with DMSO treatment and the data are the averages as described in the caption of Table 3. Black and white bars indicate upregulation and downregulation, respectively. Data for the time points when the clear reverse regulatory patterns were confirmed are shown.
Confirmation of the deduced edges by X-ChIP/qPCR
Next, we confirmed whether these candidate edges would be really direct or indirect ones by examining the physical binding of the TRFs to their regulated genes with X-ChIP/qPCR technique. The cross-linked DNA samples were extracted from HepG2 cells at five time points after ATRA or DMSO treatment, and these samples were immunoprecipitated with antibodies specific for each of the six TRFs. DNA fragments were liberated from the proteins, purified and used as templates for qPCR amplification. Primer sets that were specific to the DNA regions of the candidate regulated genes containing a potential recognition sequence for the TRFs were used. We determined the Ct-values of the specific DNA regions by using DNA samples recovered with or without the specific antibody to calculate the difference ΔCt as an indicator of the specific DNA fragment enrichment. ΔCt-values >1.0 were evaluated as a positive binding of each TRF to its cognate DNA regions. Bindings of a total of 36 genes by any of the five TRFs were demonstrated (Table 4) and these genes could qualify as highly reliable direct regulatory edges. No significant chromatin binding was observed in X-ChIP/qPCR for SREBF1. This is due to the low specificity and/or affinity of the SREBF1 antibody used because it has failed to recover the positive control DNA region derived from the PPARG gene (data not shown).
Table 4.
X-ChIP/qPCR analysis of the five TRFs and their regulated gene candidates
| TF | Time | Regulated gene | ΔCt | TF | Time | Regulated gene | ΔCt |
|---|---|---|---|---|---|---|---|
| CEBPA | 36 h | DDIT3 | 3.58 | RARA | 24 h | DDIT3 | 1.47 |
| CEBPA | 12 h | DUSP6 | 2.61 | RARA | 36 h | DDIT3 | 1.13 |
| CEBPA | 36 h | DUSP6 | 2.48 | RARA | 48 h | DDIT3 | 1.23 |
| CEBPA | 36 h | FGA | 2.98 | RARA | 06 h | FSTL3 | 5.59 |
| CEBPA | 36 h | FGB | 2.07 | RARA | 12 h | FSTL3 | 3.42 |
| CEBPA | 48 h | IFITM1 | 1.77 | RARA | 36 h | FSTL3 | 5.07 |
| CEBPA | 36 h | INHBE | 3.71 | RARA | 48 h | FSTL3 | 4.75 |
| CEBPA | 48 h | MLLT3 | 2.78 | RARA | 48 h | MAPK13 | 2.37 |
| CEBPA | 36 h | MLLT7 | 2.07 | RARA | 48 h | MDK | 5.11 |
| CEBPA | 12 h | PDZK1 | 2.45 | RARA | 48 h | NEDD9 | 4.23 |
| CEBPA | 48 h | PEG10 | 3.27 | RARA | 12 h | PDZK1 | 3.06 |
| CEBPA | 48 h | PLAB | 2.33 | RARA | 24 h | PDZK1 | 3.14 |
| CEBPA | 12 h | PLAU | 1.79 | RARA | 36 h | PDZK1 | 3.38 |
| CEBPA | 24 h | PLAU | 1.29 | RARA | 48 h | PDZK1 | 2.67 |
| CEBPA | 36 h | SESN2 | 3.03 | RARA | 48 h | PEG10 | 3.91 |
| CEBPA | 36 h | TUBE1 | 1.28 | RARA | 06 h | RARA | 1.88 |
| RARA | 12 h | RARA | 2.46 | ||||
| DDIT3 | 06 h | DDIT3 | 1.70 | RARA | 24 h | RARA | 3.45 |
| DDIT3 | 36 h | DDIT3 | 1.81 | RARA | 36 h | RARA | 3.08 |
| DDIT3 | 48 h | DDIT3 | 3.00 | RARA | 48 h | RARA | 2.91 |
| DDIT3 | 24 h | DUSP6 | 1.26 | RARA | 06 h | RARB | 2.32 |
| RARA | 12 h | RARB | 3.00 | ||||
| RARB | 48 h | FABP3 | 1.84 | RARA | 24 h | RARB | 4.54 |
| RARB | 48 h | MAPK13 | 2.41 | RARA | 36 h | RARB | 4.23 |
| RARB | 36 h | PDZK1 | 2.63 | RARA | 48 h | RARB | 3.84 |
| RARB | 48 h | PDZK1 | 2.36 | RARA | 48 h | SESN2 | 2.67 |
| RARB | 36 h | PEG10 | 1.91 | ||||
| RARB | 48 h | PEG10 | 1.14 | EGR1 | 48 h | LIF | 3.68 |
| RARB | 06 h | RARB | 1.74 | EGR1 | 48 h | PEG10 | 6.56 |
| RARB | 12 h | RARB | 3.19 | EGR1 | 48 h | TGM2 | 3.50 |
| RARB | 24 h | RARB | 2.21 | EGR1 | 48 h | TNFRSF12A | 5.75 |
| RARB | 36 h | RARB | 2.62 | ||||
| RARB | 48 h | RARB | 3.20 | ||||
| RARB | 48 h | SREBF1 | 2.63 |
The extent of enrichment of DNA regions was calculated by using the following equation: ΔCt = Ct (sample recovered with antibody) − Ct (sample recovered without antibody). The data are the averages from two independent experiments.
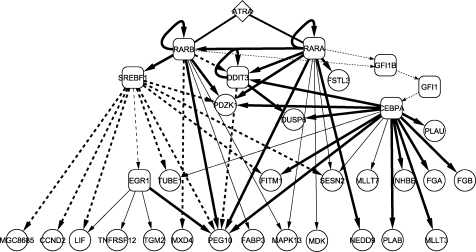
Integration of all the data obtained from time-course expression analysis with ATRA treatment, searching for the potential TRF recognition sequences, perturbation upon RNAi knockdown of TRFs and X-ChIP/qPCR enables us to depict the transcriptional regulatory cascades as shown in Figure 4. To estimate statistical significance, we measured P-values with four replicates for each gene described in ‘Time-course analysis of ATRA-induced gene expression changes’ by a t-test. This analysis extracted 5593 and 2540 genes at all time points (6, 12, 24, 36 and 48 h) with the filters of P < 0.05 and 0.01, respectively. Selection based on GO classification left 425 and 192 genes with the filters of P < 0.05 and 0.01, respectively. Finally, 39 and 29 genes in 55 genes, which were extracted by our original analysis with the criterion of >2-fold perturbation and GO selection, were found in the 425 and 192 genes, respectively. We found 23 (85%) out of the 27 genes finally selected to depict the transcriptional cascades (Figure 4; RARB was excluded because its expression level was not assessed by microarray but by qRT–PCR, EGR1 and SREBF1 were excluded because GO did not suggest any relatedness of these TRFs to cell growth regulation) in the 39 genes with a P-value <0.05. This implies that four out of the 27 genes (15%) are left out of the most probable regulated genes that appear in the cascades. If we would apply P < 0.01 to a threshold, 10 genes (37%) would be omitted.
Figure 4.
Transcriptional cascades involving six ATRA-stimulated TRFs. Bold lines indicate the edges validated by both RNAi knockdown of TRFs and X-ChIP/qPCR. The edges shown as thin lines were revealed by TRF binding but the perturbation was not highly reproducible. The edges indicated by broken lines were not detected by X-ChIP/qPCR but by perturbation experiments. The broken lines from SREBF1 were drawn on the basis of RNAi knockdown data. Perturbation of EGR1 expression was less reproducible. The dotted lines emitting from or directed to GFI1 and GFI1B were speculated from our experiments and the literature information (see Discussion section for details). These cascades were drawn by Cytoscape 2.4.1(http://www.cytoscape.org/).
The thresholds of P = 0.05 and 0.01 correspond to the false discovery rates (FDR) or q-values of 0.058–0.153 and 0.030–0.086, which have been estimated by using QVALUE software (20). Q-values were calculated based on P-values estimated with the data sets (four replicates of ATRA/DMSO- and DMSO-treated HepG2 cells) at the time points of 6, 12, 24, 36 and 48 h after the start of ATRA/DMSO and DMSO treatment. There are 163 uncharacterized genes (192 – 29 genes) with a P-value <0.01. Although we have not yet searched for potential recognition sites for six selected TRFs in the neighboring DNA region of each of the 163 genes, they might contain a few genes to be regulated by these TRFs.
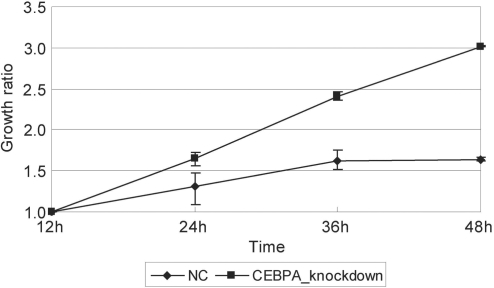
Knockdown of CEBPA gene induced growth arrest inhibition
If transcriptional regulation by a TRF is critical for the control of cell growth arrest, perturbation of its expression may lead to some phenotypic change. Therefore, we checked the growth of the cells treated with siRNA specific to each of the TRFs as the typical phenotypic change and found that the ATRA-induced growth arrest of HepG2 cells was suppressed when the CEBPA gene was repressed by a specific siRNA (Figure 5). In sharp contrast, knockdown of any other of the five TRF genes did not cause any detectable phenotypic changes. In the present study, we identified several CEBPA-regulated genes (Figure 4). Among them, we noted that ATRA markedly induced downregulation of PLAU, urokinase plasminogen activator gene, between 12 and 36 h after ATRA addition. In addition, PLAU was upregulated (>13-fold on average) by CEBPA knockdown (Figure 3). Moreover, we found that PLAU had multiple binding sites for CEBPA in its proximal upstream region and demonstrated the CEBPA binding to the PLAU DNA region in vivo. These results indicate that ATRA-induced upregulation of CEBPA repressed PLAU expression during the growth arrest of HepG2 cells.
Figure 5.
Knockdown of CEBPA inhibits ATRA-induced growth arrest of HepG2 cells. CEBPA-specific siRNA was administrated 24 h before the addition of ATRA and cell number was counted at the time points indicated in triplicate and the ratio to that at 12 h after ATRA treatment. HepG2 cells transfected with negative control siRNA (NC) showed growth arrest by ATRA, but CEBPA-suppressed cells did not.
DISCUSSION
The present study revealed a part of the ATRA-induced transcriptional cascades related to the growth arrest of HepG2 cells. For investigation of the cascades, we used a combination of time-course comprehensive expression profiling, searching for TRF recognizable sequences, time-course expression profiling of TRF genes and chromatin-binding assay. To probe the TRF-regulated genes, we noted the difference in the times for the changes in the mRNA levels of TRFs and their regulated genes. After a TRF gene is transcribed, the mRNA should be processed, exported from the nucleus, translated on cytoplasmic ribosomes and the resultant proteins return into the nucleus to promote transcription of its target genes. Therefore, there should be a time lag between the induction of the TRF gene and their regulated genes. For example, expression of EGR1 was markedly changed 36 h after ATRA treatment and the known EGR1-regulated gene TP53 changed its expression level by >1.5-fold at 48 h (Table 3). This supports the idea that the TRF-regulated genes can be selected by noting the time lag between the abrupt change in the expression of the TRF gene in question and its regulated gene candidates.
The present study provides new insights into the mechanisms of cell growth arrest by ATRA. ATRA exhibits crosstalks with various signaling pathways such as PI3K/Akt (21), p38 MAPK pathway (22) and TGFβ/Smad pathway (23). Cellular retinol-binding protein 1 (RBP1) inhibits in an ATRA-dependent manner the assembly of p85 and p110 subunits that form activated PI3 kinase (PI3K) leading to the activation of Akt (protein kinase B) (24). We found that RBP1 gene was induced immediately after ATRA addition (Supplementary Table 5) as RARA and RARB were and had a RARE in its proximal upstream region (Table 1), suggesting that it may be directly regulated by these RARs.
The complex of PLAU and its receptor interacts with integrin leading to signal transduction via the MAPK pathway that stimulates cell growth (25). It has been reported that ATRA treatment increased the levels of expression of PLAU and its receptor in human epidermal keratinocyte (26). In sharp contrast, we found that PLAU expression was repressed by ATRA treatment in HepG2 cells. We also demonstrated that PLAU was negatively regulated by CEBPA in HepG2 cells, where CEBPA gene expression is stimulated by ATRA leading to effective suppression of PLAU. On the other hand, the EGR1 gene is functionally implicated in cell proliferation and differentiation processes (27,28) and plasminogen/plasmin regulates EGR1 expression via the MEK/ERK pathway (29). Consistent with this, we found EGR1 induction at mid-late stage of ATRA-induced HepG2 cell growth arrest after the significant decrease of PLAU expression had started at an early stage. These results strongly suggest a multitiered regulatory network comprised of CEBPA, PLAU, plasminogen/plasmin, EGR1, integrin and MAPK pathway.
The p38 MAP kinase pathway is activated in an ATRA-dependent manner in acute promyelocytic leukemia and breast carcinoma cell lines (22). However, its direct regulator has not yet been identified. We found that p38δ (MAPK13) expression was significantly increased in HepG2 cells by ATRA treatment (Table 2). Moreover, our RNAi knockdown and X-ChIP/qPCR results offer evidence of the direct regulation of p38δ MAPK by RARA and RARB. Interestingly, ATRA also markedly stimulates INHBE encoding activin βE and this gene was identified to be regulated by CEBPA. Specific inhibitors of the p38 kinase completely abolish the activin-mediated cell growth inhibition of human breast cancer T47D cells (30). Therefore, we propose a cascade of ATRA stimulation of RARA, RARB and CEBPA genes, stimulation of p38 by RARs and activin-mediated activation of the MAPK leading to cell growth arrest.
CEBPA has been known as a target gene of GFI1 in promyelocytic KG-1 cells, monocytic cells and Jurkat T cells (31). The transcriptional activity of GFI1 was regulated by GFI1B (32). GFI1B and GFI1 are zinc finger transcriptional repressors, which in turn repress their own expression (32,33). Because GFI1B has a RARE in its regulatory region and its expression level was repressed for 48 h after ATRA treatment (Table 3), it may be directly regulated by RARs. In our analysis, the GFI1 gene expression was repressed at 36 h and then CEBPA gene expression was significantly upregulated. These findings with literature information suggest a possible ATRA-GFI1B-GFI1-CEBPA transcriptional regulatory cascade.
We confirmed the growth arrest of HeLa cells by ATRA treatment as observed with HepG2 cells and found that several genes showed quite similar time-lapse expression patterns as found in HepG2 cells but several genes did not (our unpublished data). This finding suggests that some of the gene regulatory events occurred in HepG2 cells may be common (or partially common) to another cell types with different genetic background. The marked differences in the expressions of some genes between HepG2 and HeLa cells may be indicators of some different mechanism of cell growth arrest in these two cell types. Extensive investigation with HeLa cells by the same approach as applied to HepG2 cells could afford insights into the regulatory mechanisms relevant to cell growth arrest.
Our experimental approach focusing on the six TRFs successfully revealed the transcriptional regulatory cascades related to the growth arrest of HepG2 cells. Although these cascades are only a portion of the entire process, this approach could be applied to further elucidate the transcriptional regulatory events involved in the cell growth arrest. The present study also showed that RNAi knockdown of TRFs is a powerful tool for investigating dynamically changing transcriptional regulatory networks.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Ms Judy Noguchi for editing our manuscript. This work was supported by the Research Grant for the RIKEN Genome Exploration Research Project from the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government to Y.H. and a grant to the Genome Network Project from the Ministry of Education, Culture, Sports, Science and Technology, Japan. Funding to pay the Open Access publication charges for this article was provided by the grant to the Genome Network Project.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gudas LJ. Retinoids and vertebrate development. J. Biol. Chem. 1994;269:15399–15402. [PubMed] [Google Scholar]
- 2.Blomhoff R, Green MH, Berg T, Norum KR. Transport and storage of vitamin A. Science. 1990;250:399–404. doi: 10.1126/science.2218545. [DOI] [PubMed] [Google Scholar]
- 3.Bastien J, Rochette-Egly C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene. 2004;328:1–16. doi: 10.1016/j.gene.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Lefebvre P, Martin PJ, Flajollet S, Dedieu S, Billaut X, Lefebvre B. Transcriptional activities of retinoic acid receptors. Vitam. Horm. 2005;70:199–264. doi: 10.1016/S0083-6729(05)70007-8. [DOI] [PubMed] [Google Scholar]
- 5.Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 6.Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiol. Rev. 2001;81:1269–1304. doi: 10.1152/physrev.2001.81.3.1269. [DOI] [PubMed] [Google Scholar]
- 7.Takeyama K, Kojima R, Ohashi R, Sato T, Mano H, Masushige S, Kato S. Retinoic acid differentially up-regulates the gene expression of retinoic acid receptor alpha and gamma isoforms in embryo and adult rats. Biochem. Biophys Res. Commun. 1996;222:395–400. doi: 10.1006/bbrc.1996.0755. [DOI] [PubMed] [Google Scholar]
- 8.Leroy P, Nakshatri H, Chambon P. Mouse retinoic acid receptor alpha 2 isoform is transcribed from a promoter that contains a retinoic acid response element. Proc. Natl Acad. Sci. USA. 1991;88:10138–10142. doi: 10.1073/pnas.88.22.10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Husmann M, Lehmann J, Hoffmann B, Hermann T, Tzukerman M, Pfahl M. Antagonism between retinoic acid receptors. Mol. Cell Biol. 1991;11:4097–4103. doi: 10.1128/mcb.11.8.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balmer JE, Blomhoff R. Gene expression regulation by retinoic acid. J. Lipid Res. 2002;43:1773–1808. doi: 10.1194/jlr.r100015-jlr200. [DOI] [PubMed] [Google Scholar]
- 11.Tomaru Y, Kondo S, Suzuki M, Hayashizaki Y. A comprehensive search for HNF-3alpha-regulated genes in mouse hepatoma cells by 60K cDNA microarray and chromatin immunoprecipitation/PCR analysis. Biochem. Biophys. Res. Commun. 2003;310:667–674. doi: 10.1016/j.bbrc.2003.08.148. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka T, Tomaru Y, Nomura Y, Miura H, Suzuki M, Hayashizaki Y. Comprehensive search for HNF-1beta-regulated genes in mouse hepatoma cells perturbed by transcription regulatory factor-targeted RNAi. Nucleic Acids Res. 2004;32:2740–2750. doi: 10.1093/nar/gkh597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung HY, Park SH, Yoo YD, Kim JS, Kim YH. CDK2/4 regulate retinoic acid-induced G1 arrest in hepatocellular carcinoma cells. Hepatol. Res. 2005;31:143–152. doi: 10.1016/j.hepres.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Kel AE, Gossling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Suzui M, Masuda M, Lim JT, Albanese C, Pestell RG, Weinstein IB. Growth inhibition of human hepatoma cells by acyclic retinoid is associated with induction of p21(CIP1) and inhibition of expression of cyclin D1. Cancer Res. 2002;62:3997–4006. [PubMed] [Google Scholar]
- 17.Fontanier-Razzaq NC, Hay SM, Rees WD. Upregulation of CHOP-10 (gadd153) expression in the mouse blastocyst as a response to stress. Mol. Reprod. Dev. 1999;54:326–332. doi: 10.1002/(SICI)1098-2795(199912)54:4<326::AID-MRD2>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 18.Lim CP, Jain N, Cao X. Stress-induced immediate-early gene, egr-1, involves activation of p38/JNK1. Oncogene. 1998;16:2915–2926. doi: 10.1038/sj.onc.1201834. [DOI] [PubMed] [Google Scholar]
- 19.Meloche S, Pouyssegur J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene. 2007;26:3227–3239. doi: 10.1038/sj.onc.1210414. [DOI] [PubMed] [Google Scholar]
- 20.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gianni M, Kopf E, Bastien J, Oulad-Abdelghani M, Garattini E, Chambon P, Rochette-Egly C. Down-regulation of the phosphatidylinositol 3-kinase/Akt pathway is involved in retinoic acid-induced phosphorylation, degradation, and transcriptional activity of retinoic acid receptor gamma 2. J. Biol. Chem. 2002;277:24859–24862. doi: 10.1074/jbc.C200230200. [DOI] [PubMed] [Google Scholar]
- 22.Alsayed Y, Uddin S, Mahmud N, Lekmine F, Kalvakolanu DV, Minucci S, Bokoch G, Platanias LC. Activation of Rac1 and the p38 mitogen-activated protein kinase pathway in response to all-trans-retinoic acid. J. Biol. Chem. 2001;276:4012–4019. doi: 10.1074/jbc.M007431200. [DOI] [PubMed] [Google Scholar]
- 23.Pendaries V, Verrecchia F, Michel S, Mauviel A. Retinoic acid receptors interfere with the TGF-beta/Smad signaling pathway in a ligand-specific manner. Oncogene. 2003;22:8212–8220. doi: 10.1038/sj.onc.1206913. [DOI] [PubMed] [Google Scholar]
- 24.Farias EF, Marzan C, Mira-y-Lopez R. Cellular retinol-binding protein-I inhibits PI3K/Akt signaling through a retinoic acid receptor-dependent mechanism that regulates p85-p110 heterodimerization. Oncogene. 2005;24:1598–1606. doi: 10.1038/sj.onc.1208347. [DOI] [PubMed] [Google Scholar]
- 25.Laufs S, Schumacher J, Allgayer H. Urokinase-receptor (u-PAR): an essential player in multiple games of cancer: a review on its role in tumor progression, invasion, metastasis, proliferation/dormancy, clinical outcome and minimal residual disease. Cell Cycle. 2006;5:1760–1771. doi: 10.4161/cc.5.16.2994. [DOI] [PubMed] [Google Scholar]
- 26.Braungart E, Magdolen V, Degitz K. Retinoic acid upregulates the plasminogen activator system in human epidermal keratinocytes. J. Invest. Dermatol. 2001;116:778–784. doi: 10.1046/j.1523-1747.2001.01310.x. [DOI] [PubMed] [Google Scholar]
- 27.Hill CS, Treisman R. Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell. 1995;80:199–211. doi: 10.1016/0092-8674(95)90403-4. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen HQ, Hoffman-Liebermann B, Liebermann DA. The zinc finger transcription factor Egr-1 is essential for and restricts differentiation along the macrophage lineage. Cell. 1993;72:197–209. doi: 10.1016/0092-8674(93)90660-i. [DOI] [PubMed] [Google Scholar]
- 29.De Sousa LP, Brasil BS, Silva BM, Freitas MH, Nogueira SV, Ferreira PC, Kroon EG, Bonjardim CA. Plasminogen/plasmin regulates c-fos and egr-1 expression via the MEK/ERK pathway. Biochem. Biophys. Res. Commun. 2005;329:237–245. doi: 10.1016/j.bbrc.2005.01.123. [DOI] [PubMed] [Google Scholar]
- 30.Cocolakis E, Lemay S, Ali S, Lebrun JJ. The p38 MAPK pathway is required for cell growth inhibition of human breast cancer cells in response to activin. J. Biol. Chem. 2001;276:18430–18436. doi: 10.1074/jbc.M010768200. [DOI] [PubMed] [Google Scholar]
- 31.Duan Z, Horwitz M. Gfi-1 takes center stage in hematopoietic stem cells. Trends Mol. Med. 2005;11:49–52. doi: 10.1016/j.molmed.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 32.Vassen L, Fiolka K, Mahlmann S, Moroy T. Direct transcriptional repression of the genes encoding the zinc-finger proteins Gfi1b and Gfi1 by Gfi1b. Nucleic Acids Res. 2005;33:987–998. doi: 10.1093/nar/gki243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doan LL, Porter SD, Duan Z, Flubacher MM, Montoya D, Tsichlis PN, Horwitz M, Gilks CB, Grimes HL. Targeted transcriptional repression of Gfi1 by GFI1 and GFI1B in lymphoid cells. Nucleic Acids Res. 2004;32:2508–2519. doi: 10.1093/nar/gkh570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harashima M, Niimi S, Koyanagi H, Hyuga M, Noma S, Seki T, Ariga T, Kawanishi T, Hayakawa T. Change in annexin A3 expression by regulatory factors of hepatocyte growth in primary cultured rat hepatocytes. Biol. Pharm. Bull. 2006;29:1339–1343. doi: 10.1248/bpb.29.1339. [DOI] [PubMed] [Google Scholar]
- 35.Nanda N, Iismaa SE, Owens WA, Husain A, Mackay F, Graham RM. Targeted inactivation of Gh/tissue transglutaminase II. J. Biol. Chem. 2001;276:20673–20678. doi: 10.1074/jbc.M010846200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.