Abstract
Long single-stranded DNAs and RNAs possess considerable secondary structure under conditions that support stable hybrid formation with oligonucleotides. Consequently, different oligomeric probes can hybridize to the same target with efficiencies that vary by several orders of magnitude. The ability to enzymatically generate structure-free single-stranded copies of any nucleic acid without impairing Watson–Crick base pairing to short probes would eliminate this problem and significantly improve the performance of many oligonucleotide-based applications. Synthetic nucleic acids that exhibit these properties are defined as pseudo-complementary. Previously, we described a pseudo-complementary A-T couple consisting of 2-aminoadenine (nA) and 2-thiothymine (sT) bases. The nA-sT couple is a mismatch even though nA-T and A-sT are stable base pairs. Here we show that 7-alkyl-7-deazaguanine and N4-alkylcytosine (where alkyl = methyl or ethyl) can be used in conjunction with nA and sT to render DNA largely structure-free and pseudo-complementary. The deoxynucleoside triphosphates (dNTPs) of these bases are incorporated into DNA by selected mesophilic and thermophilic DNA polymerases and the resulting primer extension products hybridize with good specificity and stability to oligonucleotide probes composed of the standard bases. Further optimization and characterization of the synthesis and properties of pseudo-complementary DNA should lead to an ideal target for use with oligonucleotide probes that are <25 nt in length.
INTRODUCTION
Long single-stranded DNA and RNA contain secondary and tertiary structures whose sequences are sequestered from oligonucleotide probes (1–5). Accessibility to these sequences drops off with decreasing probe length and becomes a significant limitation when using probes <20 nt in length. Secondary structure in nucleic acid targets impairs the performance of oligonucleotide probes by reducing hybridization efficiency and increasing probe-to-probe variability (6–9). In some applications, the impact of secondary structure can be diminished by testing multiple probes to different regions of the same target and selecting one that hybridizes efficiently. In other applications, however, such as SNP detection or re-sequencing this is not an option since the target sequences are fixed. Despite numerous attempts, no effective solution has been found. For example, in most microarray protocols the target nucleic acid is cleaved into 100–200 nt long fragments partly to reduce secondary structure. In some instances long probes (e.g. 60-mer) provide an effective alternative by allowing the use of more stringent hybridization conditions and improving strand invasion of any remaining secondary structure. Long probes, however, tolerate mismatches and are inappropriate for certain applications.
Conversion of naturally occurring DNA or RNA into single-stranded pseudo-complementary targets could improve the performance of short probes by eliminating interference from secondary structure. Pseudo-complementary nucleic acid is composed of base analogs that pair poorly or not at all to each other but are able to Watson–Crick pair to the standard bases of a complementary probe. Consequently, such nucleic acid is single-stranded and ‘structure-free’ in the Watson–Crick sense. Because each analog can pair to one of the standard bases, the corresponding dNTPs of these analogs should be suitable substrates for DNA polymerases thus providing a way for converting a regular DNA into a pseudo-complementary DNA. This strategy should not be confused with the more simplistic approach of synthesizing probes with duplex stabilizing modifications or preparing targets with duplex destabilizing modifications. While these efforts can improve the efficiency of probe-target hybridization, they do not provide a general solution to the challenge of competing structure in the target.
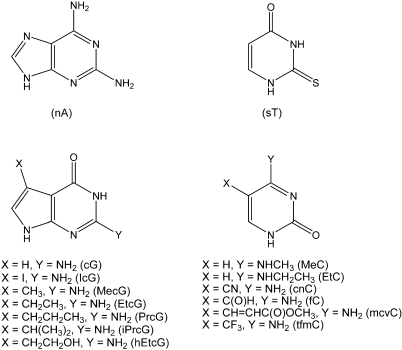
In earlier work we showed that 2-aminoadenine (nA) and 2-thiothymine (sT) meet the criteria for a pseudo-complementary A-T couple in DNA and that 2-aminoadenine and 2-thiouracil (sU) function similarly in RNA (10,11). Steric clash between the 2-amino group of nA and the bulky 2-thio group of sT or sU prevents stable Watson–Crick pairing. A pseudo-complementary G-C couple has not yet been reported, underscoring the difficulty of rationally designing such a couple. In this study, we have screened a limited number of dGTP and dCTP analogs (Figure 1) for combinations that are incorporated together with dnATP and dsTTP into DNA to give a pseudo-complementary product. Our study has shown that 7-deazaguanine (cG), 7-iodo-7-deazaguanine (IcG), 7-methyl-7-deazaguanine (MecG) or 7-ethyl-7-deazaguanine (EtcG) can function in conjunction with N4-methylcytosine (MeC) or N4-ethylcytosine (EtC) to reduce or eliminate secondary structure in DNA that is also substituted with nA and sT. Very stable DNA hairpins substituted with these analogs readily hybridized to short probes whereas the unmodified hairpins were inaccessible. The significant improvement in hybridization efficiency achieved when oligonucleotide probes are presented with pseudo-complementary DNA targets could improve the performance of oligonucleotide microarrays and jumpstart the development of new technologies for sequencing and genotyping DNA (12–14).
Figure 1.
Analogs of adenine, thymine, cytosine and guanine used in this study.
MATERIALS AND METHODS
dNTPs of nA, sT, cG and MeC were purchased from TriLink Biotechnologies. dNTPs of 7-alkyl- and 7-iodo-7-deazaguanine were prepared as described by McDougall et al. (15). The synthesis of all other dNTPs is included as Supplementary Data. Locked nucleic acid (LNA) oligonucleotides were prepared by Sigma-Proligo. Oligonucleotide primers were purchased from IDT and purified by electrophoresis through a denaturing 12% polyacrylamide gel. ϕ29 N62D DNA polymerase was expressed and purified by the Salas laboratory. 9°N DNA polymerase was obtained from New England Biolabs.
Primer extension reactions
Radiolabeled primer was hybridized to 2 mol equivalents of template in kinase buffer and then used directly for DNA synthesis. Reactions ranged in volume from 5 μl to 75 μl and contained 0.05 μg/μl ϕ29 N62D DNA polymerase, 0.6 pmoles/μl primer-template and 200 μM each dNTP in 40 mM Tris–HCl, pH 7.5, 50 mM KCl, 10 mM MgCl2 and 4 mM (NH4)2SO4. Synthesis was terminated after 30 min at 30°C by adding EDTA to a final concentration of 12 mM. The extent of synthesis was determined by electrophoresing 1 μl aliquots (in 4 μl of loading buffer) through a 12% polyacrylamide–7 M urea gel. After visualization of the dried gel by phosphorimaging, bands were quantified by ImageQuant. The yields of hairpins substituted with nA, sT, cG/MecG/EtcG/IcG and mcvC/cnC/fC/MeC/EtC were 60–90% of that obtained when using standard dNTPs. For subsequent hybridization studies single-stranded primer extension products, which were shorter than their respective templates, were gel purified under denaturing conditions. Reactions were extracted with phenol, ethanol precipitated in the presence of glycogen and the pellets dissolved at high temperature in 7 M urea with indicator dyes. Following electrophoresis in a 12% denaturing polyacrylamide gel radioactive bands were detected in the wet gel by autoradiography. Excised bands were crushed and extracted overnight in HEPES, pH 7.5. The yield of extracted sample was improved by incubating 10 min at 94°C and then at –80°C. DNA was ethanol precipitated from the clarified extract in the presence of glycogen carrier. Dried pellets were dissolved in HEPES, pH 7.5, by incubation for 10 min at 80°C and stored at –20°C until use. Primer extension products with a hairpin configuration were not gel purified unless stated otherwise. Instead, these reactions were diluted ∼15-fold with water and heated 2 min at 95°C in the presence of an oligonucleotide that was complementary to the 3′ overhang of the template. Upon quick cooling in an ice bath the template and product strands collapsed into separate hairpins. By including the oligonucleotide at 0.4 μM concentration re-annealing of template and product hairpins through their complementary single-stranded tails was blocked.
Solution hybridization experiments
Protocols for determining the apparent melting temperatures (Tm) of Watson–Crick hybrids and hairpins by gel mobility shift assay have been previously described by us (11,16). The gel assay allowed us to characterize the hybridization of oligonucleotide probes to radiolabeled primer extension products with respect to accessibility, hybrid stability and specificity. All of the solution hybridization experiments were conducted in 5 mM MgCl2, 25 mM NaCl, 20 mM HEPES, pH 7.5. Melting of radiolabeled HP25 hairpins was determined using a 25-mer probe (5 μM) that was complementary to the 3′-arm of the hairpin. We used a standard DNA probe (5′-ACCTGACTCCTGAGGAGAAGTGTGC-3′) when the hairpins were substituted with two or more base analogs (unless otherwise specified) and a chimeric LNA–DNA probe (5′-ACCtGACtCCtGaGGaGAAGtCtGC-3′; where ‘a’ and ‘t’ are LNA residues) when the hairpin was unmodified or mono-substituted. The chimeric probe hybridized more effectively than the standard probe to stable hairpins and yielded melting curves for all of the mono-substituted hairpins. The Tm of intermolecular hybrids were determined using trace amounts of radiolabeled single-strand, 1 μM probe (8-mer LNA or 12-mer DNA) and 10 μM competitor oligonucleotide. The competitor, which was composed of regular bases, was identical in length and type of backbone to the probe and complementary to it. Replicate determinations of apparent Tm usually fell within a 3°C window. Hybridization of tiled probes (5 μM) to the HP25 and HP18 hairpins were monitored by gel mobility shift assay as previously described (11).
RESULTS
Screening strategy for G/C analogs
We restricted our initial screening of bases to those shown in Figure 1. These base analogs contained relatively conservative modifications chosen to attenuate the strength of Watson–Crick base pairing. All, however, retained the 2-carbonyl group of pyrimidine or the 3-imino group of purine since their presence is required for the efficient recognition of bases by polymerases (17,18). As long as base pairing was preserved, we assumed that dNTPs of these bases would be substrates for DNA polymerases. Modified DNAs prepared by primer extension provided an opportunity to characterize the base-pairing properties of the analogs by melting analysis and other methods. By carrying out this study we hoped to find analogs of G and C that supported DNA synthesis but interacted weakly or not at all with each other.
Particular attention was paid to several analogs of cG. This base is readily incorporated into DNA as a dNTP where it reduces the strength of G-C pairing and eliminates Hoogsteen type G-G pairing (19–22). Consistent with these properties, DNA substituted with cG exhibits fewer banding artifacts in sequencing gels (23). Given our earlier success in using cG to reduce secondary structure in DNA and RNA that also contained nA and sT/sU (16,24), we evaluated analogs of cG substituted at the C7 position with alkyl or halo groups. Indirect evidence suggested that these modifications could modulate the pairing strength of cG (15).
Several analogs of C were also evaluated, with MeC and EtC being the favored candidates. These alkylated bases are readily incorporated into DNA and retain the specificity of C (25–28). They pair more weakly to G than does C because the N4-alkyl group must rotate from its preferred cis orientation (relative to the hydrogen bonding edge of the base) to the trans orientation (29). Furthermore, the appended alkyl group has a positive inductive effect that attenuates hydrogen bonding between O6 of G and N4 of MeC or EtC. The other C analogs, none of which have been rigorously characterized for incorporation and pairing properties, were expected to lower hybrid stability. Cyano, formyl, trifluoromethyl and 2-(methoxycarbonyl)ethenyl groups were conjugated to C5 of cytosine with the expectation that their electron withdrawing character would reduce the strength of hydrogen bonding between N1 of G and N3 of C.
Since a primary criterion for further study was enzymatic incorporation of the analogs into DNA, we evaluated each candidate base as a dNTP. Templates with a stable hairpin structure provided a rigorous test for whether dNTPs with attenuated base-pairing properties could support strand displacement synthesis. Those dNTPs that participated in primer extension reactions were used to prepare short radiolabeled single-stranded and hairpin DNAs (Figure 2) that were substituted in turn with each of the analogs. Melting studies of the single-stranded and hairpin products were determined by forming hybrids with complementary oligonucleotide probes at different temperatures followed by gel mobility shift analysis. The apparent Tm generated by this protocol permitted us to rank the analogs according to the thermostability of the corresponding mono-substituted hybrids. In a similar manner the relative stabilities of different G-C couples were inferred from apparent Tm of hairpins substituted with analogs of G and C. Fully substituted hairpins, which also contained nA and sT, were prepared and functioned as models for evaluating the pseudo-complementary properties of base analogs in sets of four. A hairpin was deemed to be pseudo-complementary if it failed to give a melting curve and readily hybridized to short oligonucleotide probes. Only a small quantity of each radiolabeled primer extension product was required for these solution hybridization studies, thus avoiding the expense, time and challenge of chemical synthesis. Our systematic studies have revealed several G/C analogs that function synergistically with nA and sT to destabilize secondary structure in DNA and increase its accessibility to oligonucleotide probes.
Figure 2.
Products of primer extension used in this study. Primer extension reactions were catalyzed by ϕ29 N62D DNA polymerase using different combinations of regular and modified dNTPs to prepare short single-stranded and hairpin DNAs. The 5′-radiolabeled products were used in solution hybridization experiments with unmodified probes to characterize the pairing properties of the modified DNAs. The hairpin (HP25 and HP18) and half-hairpin (SS37) products were prepared using one to four modified dNTPs. Note that SS37 recapitulates the 3′ arm of HP25. The three shorter primer extension products, which were used as complementary targets for 12-mer DNA or 8-mer LNA probes, were prepared with three regular and one modified dNTPs (dCTP analogs for the target in hybrid 1 and dGTP analogs for the target in hybrids 2 and 3) and gel purified prior to use. Primers were end-labeled with 32P and contained standard bases.
Enzymatic incorporation of dNTP analogs
Each analog was substituted for dGTP or dCTP in primer extension reactions catalyzed by the N62D variant of ϕ29 DNA polymerase (DNAP), an enzyme which has potent strand displacement activity and can readily synthesize DNA using both single-stranded and double-stranded templates. The catalytic center of this enzyme resides within a ‘tunnel’ that accommodates the template and nascent product strands (30) and conceivably reduces fraying by stabilizing the nascent hybrid. The nontemplate strand of a dsDNA template is excluded from the active site thus preventing strand displacement. We have found that ϕ29 DNAP can utilize very weakly pairing dNTPs to synthesize both single-stranded and hairpin products (data not shown). Incorporation of such dNTPs could be further enhanced by using the N62D mutant of ϕ29 DNAP (31). This mutation severely depresses the proofreading function of wild-type enzyme without impairing its ability to carry out strand displacement synthesis. By nearly eliminating 3′ to 5′ exonuclease activity, potential hydrolysis of base analogs from the 3′ end of the nascent DNA was minimized.
Most of the dNTP analogs supported synthesis of A/T- or nA/sT-containing single-strand (SS37) and hairpin (HP25) primer extension products at levels at least 70% efficient as the corresponding dGTP or dCTP. The two exceptions were dNTPs of hEtcG and tfmC. These dNTPs may interact poorly with the active site of ϕ29 DNAP. Tabular summaries of the primer extension yields are in Supplementary Data.
Relative pairing strengths of G and C analogs
The radiolabeled primer extension products generated above were used in solution hybridization experiments to determine base-pairing properties of the G/C analogs. In this regard, mono-substituted HP25 hairpins were particularly useful since they provided a convenient readout of how the analogs affected duplex stability. Apparent Tm were determined using a gel mobility shift assay (11). In this assay, a small amount of radiolabeled hairpin was mixed with a large molar excess of 25-mer probe that was complementary to one arm of the hairpin stem. Aliquots were incubated at increasing temperature, quenched in an ice bath and then analyzed by electrophoresis through a nondenaturing polyacrylamide gel at low temperature to resolve free hairpin from hairpin hybridized to probe. A plot of percent hybrid versus temperature was fit to a sigmoidal curve from which an apparent Tm could be calculated. It should be pointed out that Tm determined by this method do not have thermodynamic significance and that primer extension did not generate sufficient product to monitor melting spectrophotometrically. The physical meaning of the apparent Tm values is a measure of the ability of the probe to invade the hairpin. To improve the efficiency of hybrid formation, the 25-mer probe that we used in the analysis of mono-substituted hairpins contained a DNA backbone interspersed with six LNA monomers bearing A/T bases. By increasing hybrid stability, these modifications facilitated strand invasion of the probe into the hairpin leading to an underestimation of the true Tm. For instance, mfold predicted a Tm of 86°C for the unmodified 25-bp hairpin, but in the gel shift assay this hairpin had a Tm of only 76°C. Nonetheless, the values reported here are useful as indicators of the relative base-pairing strength between each base analog and its regular base complement.
Table 1 lists the base analogs in order of decreasing Tm for the corresponding mono-substituted 25-bp hairpins. Also listed is the average drop in Tm per incorporated base analog taking into account that the hairpin stem contained 14 G-C base pairs. In previous work, EtC paired less strongly to G than MeC, but in this model system the two bases had identical pairing strengths. However, when the same mono-substituted hairpins were melted in the presence of a DNA probe, the expected relationship was observed with MeC being more stabilizing than EtC (Tm of 66°C and 64°C, respectively). This was also the case in a short Watson–Crick duplex. The EtC-G base pair has been reported to be equal in stability to an A-T base pair (26). A comparison of Tm for HP25 when C was substituted by EtC (64–66°C; experimentally determined) or when G-C pairs were replaced by T-A pairs [62°C; predicted by mfold (32)] supports this observation. The C5 substituted analogs of C were less destabilizing than MeC and EtC. All of the G analogs were successfully incorporated into HP25 and all were destabilizing. The pairing strength of cG was modulated by the presence of substituents at the C7 position. Methyl, ethyl, propyl and hydroxylethyl groups strengthened pairing strength of cG whereas isopropyl and iodo groups reduced it. The pairing strength of MecG relative to cG is consistent with an earlier study of this base analog by Seela (33).
Table 1.
Apparent melting temperatures of the HP25 hairpin substituted with analogs of G or C
| Substitution | Tm (°C) | ΔTm/analog |
|---|---|---|
| None | 76 | – |
| G Analogs | ||
| MecG | 74 | −0.14 |
| hEtcG | 72 | −0.29 |
| EtcG | 71 | −0.36 |
| PrcG | 70 | −0.43 |
| cG | 68 | −0.57 |
| IcG | 67 | −0.64 |
| iPrcG | 66 | −0.71 |
| C Analogs | ||
| tfmC | 75 | −0.07 |
| mcvC | 73 | −0.21 |
| cnC | 72 | −0.29 |
| fC | 70 | −0.43 |
| MeC | 66 | −0.71 |
| EtC | 66 | −0.71 |
Additional melting studies were conducted on short Watson–Crick hybrids in which one strand was substituted with different analogs of G or C. Ranking of the analogs according to hybrid stability (see Supplementary Data) was similar to that reported above using the HP25 hairpin.
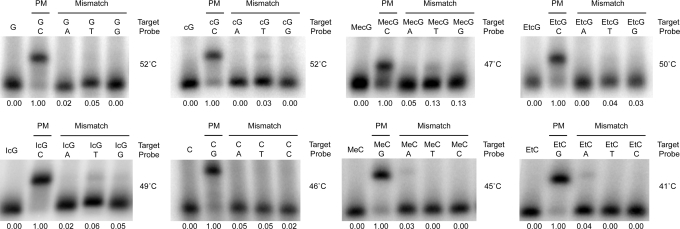
Specificity of pairing by analogs of G and C
Specificities of selected base analogs were determined by measuring hybrid formation when the analog of interest was paired to each of the four regular bases in a short duplex under stringent conditions (i.e. 8°C below the Tm of the corresponding perfect match hybrid). Short hybrids were formed between single-stranded primer extension products that contained a given G or C analog and 12-mer DNA probes (hybrid 1 for the C-substituted targets) or 8-mer LNA probes (hybrid 3 for the G substituted targets). One perfect-match and three single mismatch probes were hybridized to each target. A mismatch in the hybrid involved a single G or C analog near the middle of the targeted sequence. Reactions were quenched by adding an unlabeled competitor oligonucleotide that was composed entirely of regular bases and was identical in backbone and complementary in sequence to the perfect-match probe. After cooling in an ice bath, aliquots were analyzed by gel mobility shift assay. Phosphorimages of the gels are compiled in Figure 3. In each image one perfect-match and three mismatch hybrids were electrophoresed alongside a no-probe control. The relative amount of hybrid detected in each lane is indicated below the gel. Both MeC and EtC exhibited good discrimination of mismatches, confirming the results of an earlier study (26). Several cG bases were also evaluated. While cG, EtcG and IcG exhibited good discrimination, MecG did not fully discriminate against T and G in a LNA-DNA hybrid. Better specificity, however, was observed for this analog in DNA hybrids (data not shown).
Figure 3.
Specificity of base pairing by analogs of G and C. Gel-purified and end-labeled single-stranded primer extension products that were substituted with different analogs of G or C were hybridized under stringent conditions to oligonucleotide probes and then analyzed by gel mobility shift assay. One probe was a perfect-match to the target sequence and three other probes contained a mismatched base to a single G or C analog near the middle of the same targeted sequence. Hybrid 1 was used for the analysis of C analogs and hybrid 3 was used for the analysis of G analogs (see Figure 2). Competitor oligonucleotides used to quench the hybridization reactions were identical in backbone and complementary in sequence to the perfect-match probe. The first lane in each gel contains the single-stranded target. Other lanes contain target plus probe. The temperature of hybridization is shown to the right of each gel and is 8°C below the apparent Tm of the perfect-match hybrid. The relative amount of hybrid formed by each probe is indicated below the gel.
Thermostability of hairpins substituted with multiple base analogs
Based on incorporation, stability and specificity considerations, we further investigated the cG, IcG, MecG and EtcG analogs of G and the MeC and EtC analogs of C. All of these analogs were readily incorporated into DNA, formed relatively stable pairs with C or G and exhibited good specificity. Given that each of these analogs alone decreased the strength of G-C pairing, we investigated whether using them in combination might lead to a synergistic decrease in pairing strength. Different combinations of analogs were enzymatically incorporated (along with A and T) into the same 25-bp DNA hairpin used previously (HP25). Tm of the modified hairpins, which were efficiently synthesized using ϕ29 N62D DNAP, were determined by gel mobility shift assay using 25-mer probes that had either a DNA or a chimeric LNA-DNA backbone. Table 2 lists the experimentally determined Tm in order of decreasing hairpin stability. The extent to which each pair of G-C analogs reduced hairpin stability is reflected by its PC ratio, defined as the decrease in Tm attributable to substitution by that pair of analogs divided by the sum of Tm decreases attributable to mono-substitution by the same analogs of G and C. When this ratio is >1.0 there is a synergistic effect.
Table 2.
Apparent melting temperatures of HP25 hairpin substituted with different G-C couplesa
| LNA–DNA probe | DNA probe | ||||
|---|---|---|---|---|---|
| Couple | Tm (°C) | bPC | Couple | Tm (°C) | bPC |
| G-C | 76 | – | |||
| IcG-MeC | 67 | 0.47 | cG-MeC | 65 | 0.61 |
| cG-MeC | 66 | 0.56 | cG-EtC | 57 | 1.05 |
| cG-EtC | 64 | 0.67 | IcG-MeC | 52 | 1.26 |
| EtcG-MeC | 62 | 0.93 | IcG-EtC | 50 | 1.37 |
| IcG-EtC | 56 | 1.05 | EtcG-MeC | 50 | 1.73 |
| EtcG-EtC | 53 | 1.53 | MecG-MeC | 55 | 1.75 |
| MecG-MeC | 56 | 1.67 | EtcG-EtC | 42 | 2.27 |
| MecG-EtC | 55 | 1.75 | MecG-EtC | 46 | 2.50 |
aTwo different 25-mer probes were used to determine the apparent melting temperatures of HP25 hairpins substituted with the indicated G-C couples. bThe PC ratio is a measure of the extent to which a G-C couple is pseudo-complementary. PC = (ΔTm of HP25 with indicated G-C couple)/(ΔTm of HP25 with G analog only + ΔTm of HP25 with C analog only).
A comparison of the PC values in Table 2 shows that regardless of which probe was used, couples between MecG/EtcG and MeC/EtC were most likely to act synergistically in reducing hairpin stability. With respect to Tm values, MecG-EtC and EtcG-EtC couples were more destabilizing than MecG-MeC and EtcG-MeC, thus reflecting the dominant role of N4-alkylcytosine relative to 7-alkyl-7-deazaguanine in reducing hybrid stability. Couples involving cG/IcG and MeC/EtC had the lowest PC values and were the least destabilizing. Every di-substituted hairpin had a higher Tm when measured with the chimeric LNA–DNA probe than with the DNA probe, suggesting that the chimeric probe was less effective than the DNA probe in strand invading these hairpins.
To test whether the G/C analogs could eliminate secondary structure in DNA when used in combination with nA and sT, we enzymatically prepared different tri- and tetra-substituted HP25 hairpins (with yields 70% of the standard hairpin) and determined their apparent Tm using the unmodified 25-mer DNA probe (Table 3). The results allowed us to rank the analogs according to the average Tm of each set of hairpins that contain the same modified base. These rankings, which we assume to reflect the relative strength of base pairing, were EtC < MeC << C << fC ≈ cnC < mcvC and EtcG < MecG ≈ cG < IcG < G. Relative to A/T-containing hairpins that were mono-substituted with the same analogs (Table 3), IcG and cG moved up in stability while MecG and EtcG moved down. Notably, the C5 substituted analogs of C, which were weakly destabilizing in the presence of A and T, became strongly stabilizing in the presence of nA and sT. From among the destabilizing analogs the most promising pseudo-complementary candidates were MeC, EtC, MecG and EtcG. Hairpins that contained any combination of these bases readily hybridized to the 25-mer probe at temperatures as low as 10°C. By this criterion, nA/sT-containing hairpins that were also substituted with MeC and MecG/EtcG or with EtC and cG/MecG/EtcG/IcG could be inferred to be ‘structure-free’ and pseudo-complementary. Such a conclusion, however, is probably unwarranted for the more stable G-C couples since the 25-mer probe used in these experiments might hybridize to the hairpin through a strand invasion mechanism at low temperature.
Table 3.
Apparent melting temperatures of HP25 hairpin substituted with different G-C couples in the presence of nA and sTa
| Couple | G | IcG | cG | MecG | EtcG |
|---|---|---|---|---|---|
| mcvC | 42 | 52 | 39 | 38 | 41 |
| cnC | – | 38 | 41 | 41 | 32 |
| fC | 44 | 40 | 38 | 37 | 36 |
| C | 40 | 36 | 26 | 27 | 18 |
| MeC | 28 | 29 | 12 | <10 | <10 |
| EtC | 25 | <10 | <10 | <10 | <10 |
aTemperature in °C.
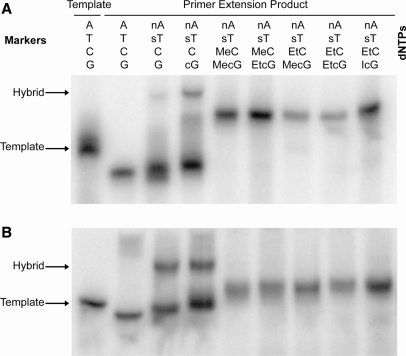
Electrophoretic mobility of hairpins substituted with multiple base analogs
Further confirmation of the base-pairing properties of fully modified DNA hairpins was obtained using a gel mobility shift assay. Electrophoretic mobility in a nondenaturing gel is dependent upon the global conformation of a hairpin (34). Taking into consideration that a hairpin electrophoreses much faster than a single strand of the same length, loss of base pairing in the stem region of a hairpin should be accompanied by a decrease in electrophoretic mobility. We observed this phenomenon when hairpins substituted with nA, sT, MecG/EtcG/IcG and MeC/EtC were electrophoresed in 12% polyacrylamide gels run under two conditions. Relative to the markers, the fully modified hairpins migrated more slowly when electrophoresis was carried out in Tris–borate buffer at room temperature than when electrophoresis was conducted in a cold room with buffer that contained 5 mM MgCl2 (Figure 4). We attribute the drop in mobility to a change in hairpin conformation from stem–loop to single strand. Even at the lower temperature the fully modified hairpins trailed the unmodified control, suggesting that their stem was imperfect and more ‘open’. As pointed out earlier, the 25-mer probe used in the hairpin melting experiments might readily strand invade such duplexes thus leading to an underestimation of the true Tm. Modified hairpins substituted with nA + sT or nA + sT + cG gave two bands, with the faster moving species corresponding to the intact hairpin and the slower moving species representing a duplex between the hairpin and the template used for its synthesis. In the more stringent gel, a subpopulation of these hairpins had a more open conformation and ran behind the main band.
Figure 4.
Gel mobility shift analysis of modified HP25 hairpins under nondenaturing conditions. Modified hairpins were prepared by primer extension and purified by denaturing PAGE. The hairpins were then electrophoresed through 12% nondenaturing polyacrylamide gels in (A) Tris–borate buffer at room temperature or in (B) Tris–borate buffer with 5 mM MgCl2 at 4°C. Radiolabeled template was run in the same gels as a marker. Primer extension products that contained nA, sT and C formed a hybrid with the template that survived gel purification; it functions as a second marker in the gels. The slow-moving band in the second lane of gel (B) has not been identified. Using the two markers as a ruler with the template assigned a value of 0 and the hybrid assigned a value of 1.0, the fully modified hairpins are found at positions 0.70 in gel (A) and 0.43 in gel (B).
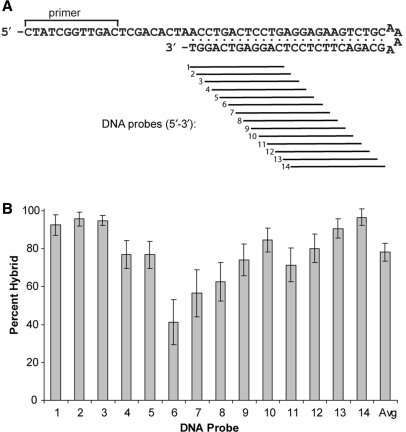
Hybridization of tiled oligonucleotide probes to hairpins substituted with multiple base analogs
As a practical demonstration of pseudo-complementarity, gel-purified HP25 hairpins substituted with different G/C analogs in the presence of nA and sT were hybridized to short 8-mer LNA or 12-mer DNA probes tiled across the 3′ half of the hairpin stem (see Figure 5A). Unless otherwise indicated, hybridizations were conducted for 10 min at 30°C in the presence of 5 mM MgCl2 and 25 mM NaCl. Hybrids were detected by gel mobility shift analysis after quenching the reactions in an ice bath. Quantification showed that hairpins substituted with any of the cG/MecG/EtcG-MeC/EtC couples were accessible to both types of probes. The average signal was 77% for the DNA probes and 85% for the LNA probes. In Figure 5B, the hybridization signal obtained for each 12-mer DNA probe is averaged across all the modified hairpins. The probe-to-probe variation in signal implies that there are sequence-dependent differences in accessibility and/or hybrid stability. Probes 6–8 formed the least amount of hybrid but no obvious signature was found in the target sequences. The tiled 8-mer LNA probes performed better than the DNA probes, consistently hybridizing to a greater fraction of hairpin target (see Supplementary Data). Nonetheless, the LNA probes also had difficulty accessing the same sequences addressed by DNA probes 6–8 (data not shown). Overall, the tiling experiments confirmed that the fully substituted HP25 hairpins had significant pseudo-complementary character. By contrast, the same hairpin composed of regular bases did not hybridize to any of the probes.
Figure 5.
Hybridization of tiled 12-mer DNA probes to HP25 hairpins. The probes, which were tiled across the 3′-arm of HP25 (A), were separately hybridized to six modified versions of the hairpin, which contained cG-MeC, cG-EtC, MecG-MeC, MecG-EtC, EtcG-MeC or EtcG-EtC couples in the presence of the nA-sT couple. The average hybridization efficiency of all the hairpins to each probe is plotted (B). Hybridizations were carried out at 30°C with 5 μM probe in 5 mM MgCl2, 25 mM NaCl, 20 mM HEPES pH 7.5.
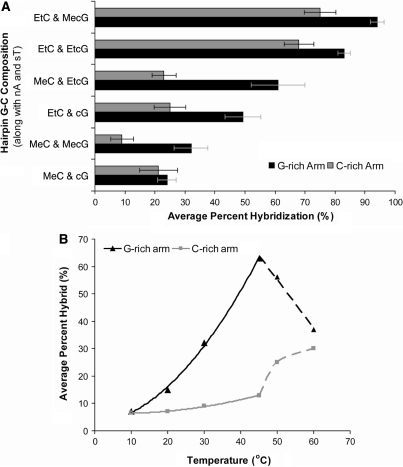
The experiments with HP25 left unanswered whether the same base analogs could eliminate secondary structure in a hairpin that possessed a much higher G/C content. To address this question we carried out tiling experiments with an 18-bp hairpin (HP18) that was substituted with different combinations of cG/MecG/EtcG and MeC/EtC in the presence of nA and sT. The HP18 hairpin had a G/C content of 78% and an estimated Tm of 96°C. Since one arm of the hairpin stem was G rich and other was C rich, each arm was addressed by a separate set of tiled 12-mer DNA probes. As before, hybridizations were conducted at 30°C for 10 min in 25 mM NaCl, 5 mM MgCl2 and 20 mM HEPES pH 7.5 and analyzed by gel mobility shift assay. The average hybridization efficiencies of the differently substituted hairpins to both probe sets varied significantly (Figure 6A). Hairpins substituted with EtC were much more accessible to the probes than those substituted with MeC. Among the G analogs, MecG and EtcG were the most effective. The extent to which different G-C couples promoted hybridization of 12-mer probes to the HP18 hairpin was roughly proportional to their PC values in Table 2. Hence EtcG-EtC and MecG-EtC couples were the best while the cG-MeC couple was the worst. While these relationships held for both arms of the hairpin, the C-rich arm hybridized less efficiently to probes than did the G-rich arm, a difference that held for all of the G-C couples evaluated.
Figure 6.
Hybridization of tiled 12-mer DNA probes to HP18 hairpins. (A) Average hybridization of probe sets to the G-rich and C-rich arms of HP18 hairpins substituted with nA, sT and the indicated analogs of G and C at 30°C with 5 μM probe in 5 mM MgCl2, 25 mM NaCl, 20 mM HEPES pH 7.5. (B) Temperature dependence for hybridization of tiled probe sets to the G-rich and C-rich arms of HP18 hairpin substituted with nA, sT, MecG and MeC. At temperatures above 45°C the hairpin is disrupted and the two probes sets acquire equivalent efficiencies of hybridization.
If residual secondary structure in the hairpin target is a limiting factor in hybridization, then the performance of less effective G-C couples should improve with increasing temperature, as long as the corresponding probe-target hybrids have sufficient stability at elevated temperature. We tested this prediction by repeating the tiling experiments at different temperatures for HP18 hairpin that was substituted with nA-sT and MecG-MeC couples. The results in Figure 6B show that probes to the G-rich arm of the hairpin hybridized more effectively as the temperature was increased from 10°C to 45°C. Reduction of secondary structure in the target is most likely responsible for the enhanced hybridization. In contrast, hybridization of probes to the C-rich arm of the hairpin remained constant over the same temperature range. Hybrids formed by these probes were probably too weak to support strand invasion of the structured target. At higher temperatures, up to 60°C, the extent of hybridization converged for the two arms of the hairpin. Under these conditions, we postulate that no base pairing occurred and that probe-target hybrid formed only when the reactions were quenched in an ice bath.
Hybridization of probes to the C-rich arm of the hairpin was increased 8-fold by replacing MeC with EtC in a target substituted with nA, sT and MecG (Figure 6A). Given that probe-target hybrids, which contain G-EtC are less stable than those which contain G-MeC, the improved hybridization of probes to the C-rich arm of HP18 when it is substituted with EtC instead of MeC can probably be attributed to the greater PC value of MecG-EtC (2.50) relative to MecG-MeC (1.75). In this instance, hybridization was more dependent on reduction of secondary structure in the target than on stability of the resulting probe-target hybrids. The enhanced hybridization with MecG/EtC versus MecG/MeC substitution argues against the G-rich probes having self-structure, which would inhibit intermolecular pairing to the C-rich arm of the hairpin.
Incorporation of dCTP and dGTP analogs by thermostable DNA polymerases
Regular use of pseudo-complementary dNTPs in diagnostics and genomics will require their efficient incorporation into long DNAs. We surveyed seven thermostable DNA polymerases for their ability to synthesize a 600-mer primer extension product using different sets of dNTPs. A synthetic 600-bp gene with extensive secondary structure and multiple regions of high G/C or A/T content was selected as a template. Radiolabeled primer was hybridized to the antisense strand of this gene by incubating the two at 95°C for 2 min prior to elongation at 65–72°C. Full-length product was resolved in a 10 M urea-6% polyacrylamide gel at 60°C and quantified by phosphorimaging. Five of the polymerases were able to synthesize full-length product substituted with nA, sT, MecG and MeC or EtC. 9°N was the best, retaining 67 or 49% of wild-type activity when asked to incorporate MeC or EtC along with nA, sT and MecG. Results for the other polymerases can be found in the Supplementary Data. The limited survey described here suggests that naturally occurring or engineered polymerases could be found that synthesize pseudo-complementary DNA nearly as efficiently as regular DNA.
DISCUSSION
Several sets of base analogs that confer pseudo-complementary properties to DNA have been described. Each set was incorporated into short primer extension products that exhibited reduced secondary structure and improved accessibility to oligonucleotide probes. Use of nA and sT for this purpose was previously reported by us (10,16). However, until now a satisfactory G-C pair was unknown. In this study, we screened a limited number of G/C analogs for enzymatic incorporation into DNA and for stability and specificity in base pairing. The most promising analogs, ranked according to their pairing strength opposite C or G in HP25 hairpins substituted with nA and sT, were EtcG < MecG ≈ cG < IcG < G and EtC < MeC < C (Table 2). All possible combinations of the G/C analogs were enzymatically incorporated as dNTPs into the same 25-bp hairpin so that the properties of the new base pairs could be indirectly analyzed by melting point determinations. Many of the modified hairpins were further analyzed by gel mobility shift and tiling experiments. The G-C couples differed significantly in their ability to alter hybrid stability and promote pairing to unmodified probes. Careful consideration of all the results suggests an ordering of couples from least to most stable of EtcG-EtC ≈ MecG-EtC < EtcG-MeC ≈ MecG-MeC < cG-EtC ≈ IcG-EtC <cG-MeC ≈ IcG-MeC < G-C. In tiling experiments, the ability of these couples to impart pseudo-complementary properties to DNA when used in conjunction with nA and sT followed the same approximate ranking with the least stable couples being the most effective. Thus, EtcG-EtC and MecG-EtC rendered a highly G/C-rich hairpin accessible to short probes. The utility of other G/C couples was restricted to secondary structure that had a greater A/T content.
The methyl and ethyl analogs of cG and C interacted weakly with one another even though each formed a stable base pair with C or G, respectively. This, of course, was central to their success in promoting hybridization of unmodified probes to G/C-rich secondary structure. A plausible explanation for the synergistic loss of pairing strength between analogs is the significant increase in hydrophobicity of the major groove that is associated with the introduction of methyl or ethyl groups into that groove. The replacement of N7 of G with C7 of cG eliminates an important cation binding site in the major groove, alters the organization of water and salts in the major groove and decreases base stacking (35). Since cation binding to N7 of G is estimated to account for 20–30% of the stability of a G-C base pair (36,37), substitution of cG for G reduces hybrid stability. Some stability is regained if the C7 position of cG is substituted with a methyl or ethyl group. The electron donating character of these groups may partially overcome reduced cation binding by making O6 more electron rich. Alternatively, the hydrophobic alkyl groups might have a stabilizing effect on base stacking. Our results show, however, that duplex stability is greatly decreased when both C7 of cG and N4 of C are alkylated. With both positions located within the major groove, increased hydrophobicity associated with alkylation could lower cation occupancy and reduce base-pairing strength. Unfavorable steric interactions associated with the presence of two alkyl groups and possible disruption of the duplex hydration pattern could be another contributing factor. Finally, alkylation could also alter the electronic charge distribution of the modified nucleobase couples in ways that lower their pairing strength.
Successful hybridization of short oligonucleotides to ‘structure-free’ hairpins confirms that the different combinations of base analogs that we have used to generate pseudo-complementary DNA retain the ability to Watson–Crick pair to the standard bases even though the G/C analogs pair less strongly than the corresponding regular bases. Melting point determinations of model Watson–Crick hybrids have shown that pseudo-complementary DNA targets substituted with nA, sT, MecG/EtcG and MeC/EtC form less stable hybrids with standard probes than unmodified DNAs (data not shown). In this context, it may prove beneficial to utilize probes that are specifically modified to form more stable hybrids with the structure-free targets. One such modification is the LNA backbone. Our observation that 8-mer LNA probes hybridize more effectively than 12-mer DNA probes to modified HP25 hairpins is best explained in terms of hybridization strength of the respective probes. The mcvC, cnC and fC analogs of C may also have merit in this regard since they form unusually stable base pairs with MecG and EtcG (Table 3).
Although cG paired more strongly to MeC and EtC than did MecG or EtcG, the cG containing couples conferred pseudo-complementary properties to the HP25 hairpin when present with nA and sT. Given that nA-sT functions as a mismatch, only a moderate reduction in the pairing strength of G-C may be sufficient to eliminate duplex regions that are not too G/C rich. One common attribute shared by all the cG bases is an inability to self-pair. Normally, destabilization of Watson–Crick pairing would be expected to promote Hoogsteen pairing between adjacent G's. By eliminating this possibility, all of the cG bases promoted accessibility of the modified DNA to complementary probes.
Attempts to footprint the base pairing state of fully modified pseudo-complementary hairpins using reagents such as dimethyl sulfate and potassium permanganate were unsuccessful. These reagents gave uninterpretable results, which we attribute to the altered structure and hydration state of the DNA. Nuclease probes were not tested out of concern that fully modified DNAs might exhibit base specific modulation of enzymatic activity thus confounding analysis of the results.
In screening potential analogs of G and C we prepared dNTPs of interesting candidates and used them to enzymatically synthesize short single-stranded and hairpin oligonucleotides. Hybridization properties of the oligonucleotides were then determined using gel mobility shift assays. The small amounts of radiolabeled targets required for the assays were easily generated by primer extension reactions. Had we used phosphoramidites of the same base analogs to chemically synthesize larger quantities of oligonucleotides, the task would have required more time and incurred greater expense. In all likelihood, chemical synthesis of fully substituted versions of the longer oligonucleotides would have been difficult or impossible.
Further evaluation of the alkylated G-C couples is planned. The PC index should be validated by thermodynamic stability measurements using short chemically synthesized oligonucleotides and the hybridization properties of fully substituted pseudo-complementary DNAs need to be characterized with respect to the stability, specificity and cooperativity of hybrid formation using unmodified oligonucleotide probes with DNA or LNA backbones. Some of these studies could be carried out by hybridizing pseudo-complementary DNA to oligonucleotide microarrays. Practical use of the technology will require compilation of the nearest-neighbor free energy parameters for hybrids formed between regular probes and modified targets as well as development of straightforward and reliable protocols for faithfully generating pseudo-complementary DNA. Techniques that rely on the efficient hybridization of short oligonucleotide probes to DNA would be the primary beneficiaries of the work described here. Direct detection of SNPs by hybridization should become more reliable since shorter probes with better discrimination could be used. The availability of targets that are entirely accessible to oligonucleotide probes should improve the resolution of microarray-based genome scans of SNP status and DNA copy number. Emerging technologies such as sequencing by ligation, resequencing of genes on a microarray and the development of universal microarrays could benefit from the availability of pseudo-complementary DNA targets.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health (NIGMS Grant 74564 to H.G.) and Agilent Technologies. Dr Carl Fuller (GE Healthcare) kindly provided us with dNTPs of 7-alkyl-7-deazaguanines and 7-iodo-7-deazaguanine. We thank Dr Jeffrey Sampson (Agilent Technologies) for his constant encouragement and Caryn Evila (Idaho State University) for her early participation in the project. At TriLink Biotechnologies we are grateful to Dr Richard Hogrefe, Dr Gerald Zon, Dr Natasha Paul and Dr David Combs for support and helpful discussions and Dr Inna Koukhareva and Stephanie Perry for synthesis of dNTPs. Funding to pay the Open Access publication charges for this article was provided by NIH GM74564.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gamper HB, Cimino GD, Hearst JE. Solution hybridization of crosslinkable DNA oligonucleotides to bacteriophage M13 DNA. Effect of secondary structure on hybridization kinetics and equilibria. J. Mol. Biol. 1987;197:349–362. doi: 10.1016/0022-2836(87)90128-8. [DOI] [PubMed] [Google Scholar]
- 2.Ratushna VG, Weller JW, Gibas CJ. Secondary structure in the target as a confounding factor in synthetic oligomer microarray design. BMC Genomics. 2005;6:31. doi: 10.1186/1471-2164-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kierzek E, Kierzek R, Turner DH, Catrina IE. Facilitating RNA structure prediction with microarrays. Biochemistry. 2006;45:581–593. doi: 10.1021/bi051409+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hacia JG, Woski SA, Fidanza J, Edgemon K, Hunt N, McGall G, Fodor SP, Collins FS. Enhanced high density oligonucleotide array-based sequence analysis using modified nucleoside triphosphates. Nucleic Acids Res. 1998;26:4975–4982. doi: 10.1093/nar/26.21.4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen HK, Southern EM. Minimising the secondary structure of DNA targets by incorporation of a modified deoxynucleoside: implications for nucleic acid analysis by hybridisation. Nucleic Acids Res. 2000;28:3904–3909. doi: 10.1093/nar/28.20.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kushon SA, Jordan JP, Seifert JL, Nielsen H, Nielsen PE, Armitage BA. Effect of secondary structure on the thermodynamics and kinetics of PNA hybridization to DNA hairpins. J. Am. Chem. Soc. 2001;123:10805–10813. doi: 10.1021/ja016310e. [DOI] [PubMed] [Google Scholar]
- 7.Armitage BA. The impact of nucleic acid secondary structure on PNA hybridization. Drug Discov. Today. 2003;8:222–228. doi: 10.1016/s1359-6446(03)02611-4. [DOI] [PubMed] [Google Scholar]
- 8.Ramdas L, Cogdell DE, Jia JY, Taylor EE, Dunmire VR, Hu L, Hamilton SR, Zhang W. Improving signal intensities for genes with low-expression on oligonucleotide microarrays. BMC Genomics. 2004 doi: 10.1186/1471-2164-5-35. 5, Article 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chou CC, Chen CH, Lee TT, Peck K. Optimization of probe length and the number of probes per gene for optimal microarray analysis of gene expression. Nucleic Acids Res. 2004;32:e99. doi: 10.1093/nar/gnh099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kutyavin IV, Rhinehart RL, Lukhtanov EA, Gorn VV, Meyer R.B, Jr., Gamper H.B., Jr. Oligonucleotides containing 2-aminoadenine and 2-thiothymine act as selectively binding complementary agents. Biochemistry. 1996;35:11170–11176. doi: 10.1021/bi960626v. [DOI] [PubMed] [Google Scholar]
- 11.Gamper HB, Jr, Gewirtz A, Edwards J, Hou YM. Modified bases in RNA reduce secondary structure and enhance hybridization. Biochemistry. 2004;43:10224–10236. doi: 10.1021/bi049196w. [DOI] [PubMed] [Google Scholar]
- 12.Service RF. Gene sequencing. The race for the $1000 genome. Science. 2006;311:1544–1546. doi: 10.1126/science.311.5767.1544. [DOI] [PubMed] [Google Scholar]
- 13.Shendure J, Mitra RD, Varma C, Church GM. Advanced sequencing technologies: methods and goals. Nat. Rev. Genet. 2004;5:335–344. doi: 10.1038/nrg1325. [DOI] [PubMed] [Google Scholar]
- 14.Rhee M, Burns MA. Nanopore sequencing technology: research trends and applications. Trends Biotechnol. 2006;24:580–586. doi: 10.1016/j.tibtech.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 15.McDougall MG, Sun L, Livshin I, Hosta LP, McArdle BF, Samols SB, Fuller CW, Kumar S. Analogs of guanine nucleoside triphosphates for sequencing applications. Nucleosides Nucleotides Nucleic Acids. 2001;20:501–506. doi: 10.1081/NCN-100002325. [DOI] [PubMed] [Google Scholar]
- 16.Gamper HB, Jr, Arar K, Gewirtz A, Hou YM. Unrestricted hybridization of oligonucleotides to structure-free DNA. Biochemistry. 2006;45:6978–6986. doi: 10.1021/bi0600392. [DOI] [PubMed] [Google Scholar]
- 17.Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- 18.Steitz T. In: Biological Organization: Macromolecular Interactions at High Resolution. Burnett R, Vogel H, editors. New York: Academic Press; 1987. pp. 45–55. [Google Scholar]
- 19.Seela F, Roling A. 7-Deazapurine containing DNA: efficiency of c7GdTP, c7AdTP and c7IdTP incorporation during PCR-amplification and protection from endodeoxyribonuclease hydrolysis. Nucleic Acids Res. 1992;20:55–61. doi: 10.1093/nar/20.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seela F, Driller H. Solid-phase synthesis of the self-complementary hexamer d(c7GpCpc7GpCpc7GpC) via the O-3′-phosphoramidite of 7-deaza-2′-deoxyguanosine. Nucleic Acids Res. 1985;13:911–926. doi: 10.1093/nar/13.3.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seela F, Driller H. Alternating d(G-C)3 and d(C-G)3 hexanucleotides containing 7-deaza-2′-deoxyguanosine or 8-aza-7-deaza-2′-deoxyguanosine in place of dG. Nucleic Acids Res. 1989;17:901–910. doi: 10.1093/nar/17.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seela F, Tran-Thi QH, Franzen D. Poly(7-deazaguanylic acid), the homopolynucleotide of the parent nucleoside of queuosine. Biochemistry. 1982;21:4338–4343. doi: 10.1021/bi00261a024. [DOI] [PubMed] [Google Scholar]
- 23.Mizusawa S, Nishimura S, Seela F. Improvement of the dideoxy chain termination method of DNA sequencing by use of deoxy-7-deazaguanosine triphosphate in place of dGTP. Nucleic Acids Res. 1986;14:1319–1324. doi: 10.1093/nar/14.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gamper HB, Jr, Arar K, Gewirtz A, Hou YM. Unrestricted accessibility of short oligonucleotides to RNA. RNA. 2005;11:1441–1447. doi: 10.1261/rna.2670705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li S, Haces A, Stupar L, Gebeyehu G, Pless RC. Elimination of band compression in sequencing gels by the use of N4- methyl-2′-deoxycytidine 5′-triphosphate. Nucleic Acids Res. 1993;21:2709–2714. doi: 10.1093/nar/21.11.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen HK, Auffray P, Asseline U, Dupret D, Thuong NT. Modification of DNA duplexes to smooth their thermal stability independently of their base content for DNA sequencing by hybridization. Nucleic Acids Res. 1997;25:3059–3065. doi: 10.1093/nar/25.15.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen HK, Bonfils E, Auffray P, Costaglioli P, Schmitt P, Asseline U, Durand M, Maurizot JC, Dupret D, Thuong NT. The stability of duplexes involving AT and/or G4EtC base pairs is not dependent on their AT/G4EtC ratio content. Implication for DNA sequencing by hybridization. Nucleic Acids Res. 1998;26:4249–4258. doi: 10.1093/nar/26.18.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen HK, Fournier O, Asseline U, Dupret D, Thuong NT. Smoothing of the thermal stability of DNA duplexes by using modified nucleosides and chaotropic agents. Nucleic Acids Res. 1999;27:1492–1498. doi: 10.1093/nar/27.6.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fazakerley GV, Kraszewski A, Teoule R, Guschlbauer W. NMR and CD studies on an oligonucleotide containing N4-methylcytosine. Nucleic Acids Res. 1987;15:2191–2201. doi: 10.1093/nar/15.5.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamtekar S, Berman AJ, Wang J, Lazaro JM, de Vega M, Blanco L, Salas M, Steitz TA. Insights into strand displacement and processivity from the crystal structure of the protein-primed DNA polymerase of bacteriophage phi29. Mol. Cell. 2004;16:609–618. doi: 10.1016/j.molcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 31.de Vega M, Lazaro JM, Salas M, Blanco L. Primer-terminus stabilization at the 3′-5′ exonuclease active site of phi29 DNA polymerase. Involvement of two amino acid residues highly conserved in proofreading DNA polymerases. EMBO J. 1996;15:1182–1192. [PMC free article] [PubMed] [Google Scholar]
- 32.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seela F, Chen Y. Methylated DNA: The influence of 7-deaza-7-methylguanine on the structure and stability of oligonucleotides. Helvet. Chim. Acta. 1997;80:1073–1086. [Google Scholar]
- 34.Stellwagen E, Abdulla A, Dong Q, Stellwagen NC. Electrophoretic mobility is a reporter of hairpin structure in single-stranded DNA oligomers. Biochemistry. 2007;46:10931–10941. doi: 10.1021/bi701058f. [DOI] [PubMed] [Google Scholar]
- 35.Ganguly M, Wang F, Kaushik M, Stone MP, Marky LA, Gold B. A study of 7-deaza-2′-deoxyguanosine 2′-deoxycytidine base pairing in DNA. Nucleic Acids Res. 2007;35:6181–6195. doi: 10.1093/nar/gkm670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burda J, Sponer J, Leszczynski J, Hobza P. Nonempirical ab initio calculations of structures, energies, and nonadditivity of the interaction. J. Phys. Chem. B. 1997;101:9670–9677. [Google Scholar]
- 37.Sponer J, Leszczynski J, Hobza P. Electronic properties, hydrogen bonding, stacking, and cation binding of DNA and RNA bases. Biopolymers. 2002 doi: 10.1002/1097-0282(2001)61:1<3::AID-BIP10048>3.0.CO;2-4. 61, 3–31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.