Abstract
Previous work showed that, in the presence of DNA-dependent protein kinase (DNA-PK), Artemis slowly trims 3′-phosphoglycolate-terminated blunt ends. To examine the trimming reaction in more detail, long internally labeled DNA substrates were treated with Artemis. In the absence of DNA-PK, Artemis catalyzed extensive 5′→3′ exonucleolytic resection of double-stranded DNA. This resection required a 5′-phosphate, but did not require ATP, and was accompanied by endonucleolytic cleavage of the resulting 3′ overhang. In the presence of DNA-PK, Artemis-mediated trimming was more limited, was ATP-dependent and did not require a 5′-phosphate. For a blunt end with either a 3′-phosphoglycolate or 3′-hydroxyl terminus, endonucleolytic trimming of 2–4 nucleotides from the 3′-terminal strand was accompanied by trimming of 6 nt from the 5′-terminal strand. The results suggest that autophosphorylated DNA-PK suppresses the exonuclease activity of Artemis toward blunt-ended DNA, and promotes slow and limited endonucleolytic trimming of the 5′-terminal strand, resulting in short 3′ overhangs that are trimmed endonucleolytically. Thus, Artemis and DNA-PK can convert terminally blocked DNA ends of diverse geometry and chemical structure to a form suitable for polymerase-mediated patching and ligation, with minimal loss of terminal sequence. Such processing could account for the very small deletions often found at DNA double-strand break repair sites.
INTRODUCTION
Artemis is a multifunctional nuclease that is essential for hairpin opening in V(D)J recombination (1,2). Artemis was first identified as the causal defect in radiation-sensitive SCID (RS-SCID) and Athabascan SCID (SCIDA), defined as specific subsets of individuals with B– T– NK+ severe combined immune deficiency (1,3). Artemis also appears to be required for repair of a minor fraction of radiation-induced DNA double-strand breaks (DSBs) that in normal cells are rejoined very slowly (4,5). Such repair presumably accounts for the radiosensitivity of Artemis-deficient human and mouse cells (1,3,6,7). It has been suggested that some DSBs require Artemis for repair because they bear chemically modified termini that cannot otherwise be converted to the 5′-phosphate and 3′-hydroxyl ends required by gap-filling polymerases and DNA ligase (5). Consistent with this proposal, Artemis-deficient human and mouse cells are also sensitive to agents such as bleomycin and neocarzinostatin (6,8), that induce DSBs with well-defined chemically modified termini, and neocarzinostatin treatment promotes stable association of Artemis with chromatin in cells (9).
Studies with oligomeric substrates have shown that Artemis has constitutive 5′→3′ exonuclease activity toward single-stranded DNA, and that in the presence of catalytically active DNA-dependent protein kinase (DNA-PK), it acquires an endonucleolytic activity that removes 5′ overhangs and shortens 3′ overhangs, typically to 4–5 bases (2,8,10). We previously showed that with plasmid-length DNA substrates, Artemis exhibits additional DNA-PK-dependent activities that are not apparent with oligomeric substrates (8). For example, on these longer substrates, 3′ overhangs as short as 4–5 bases were efficiently and rapidly trimmed, with two terminal bases being removed in a reaction that was almost completely dependent on both Ku and DNA-PKcs. Unexpectedly, even a 3′-PG-terminated blunt end was slowly processed by Artemis/DNA-PK, with several bases being removed from the 3′-terminus. Inasmuch as about half the DSBs induced by bleomycin have 3′-PG-terminated blunt ends (11), processing of these lesions by Artemis could explain the bleomycin sensitivity of Artemis-deficient mouse and human cells (6,8). We therefore examined the action of Artemis/DNA-PK on blunt ends in greater detail. The results reveal a complex, conservative and stringently regulated process that slowly trims a few nucleotides from each strand, leaving a short 3′ overhang.
MATERIALS AND METHODS
Proteins
Recombinant histidine-tagged Artemis and untagged Ku70/80 were overproduced in baculovirus-infected insect cells and purified to apparent homogeneity by affinity and MonoQ chromatography, as described previously (4,8,12,13). DNA-PKcs was purified from HeLa cells, also to apparent homogeneity, as judged by denaturing gel electrophoresis (8,13). All other enzymes were from New England Biolabs (Beverly, MA, USA) and reactions were performed in buffers supplied by the vendor.
Substrates
An 11-mer bearing a 3′-PG terminus was prepared by treating a partial duplex with bleomycin (14). The modified oligomer was purified by gel electrophoresis and HPLC, and its structure verified by mass spectrometry (14). Internally labeled blunt-ended plasmid substrates were prepared by ligating this 5′-32P-labeled 11-mer, or an analogous 3′-hydroxyl 11-mer, into an 11-base 5′ overhang of plasmid pRZ56 and were purified by agarose gel electrophoresis as described (15,16). A substrate of identical sequence but with 3′-end label was generated by cutting circular pRZ56 with MluI, and filling in the 4-base 5′ overhang with dGTP and [α-32P]dCTP (3000 Ci/mmole, Perkin-Elmer), using exonuclease-deficient Klenow fragment. Alternatively, the MluI-cut plasmid was dephosphorylated and then 5′-end-labeled with polynucleotide kinase and [γ-32P]ATP (6000 Ci/mmole, Perkin-Elmer), and then similarly filled in with unlabeled nucleotides.
A substrate with internal label 25 bases from the 5′-terminus was prepared by ligating a 3′-end-labeled oligomeric duplex to linearized pUC19. The 26-mer CGCGTTCCTCGAGAATGTGGTGGTTG was annealed to its partial complement CTAGACAACCACCACATTCTCGAGG and a single labeled TMP residue was added to the 3′ end, by treatment with exonuclease-deficient Klenow fragment in the presence of [α-32P]dTTP. The resulting 3′-end-labeled duplex was then ligated to XbaI-cut pUC19. Following ethanol precipitation, monomer-length pUC19 was gel-purified and electroeluted. The recessed 3′ end was then filled in using exonuclease-deficient Klenow fragment and unlabeled dNTPs. An analogous labeled plasmid substrate with a 3′ overhang (Figure 1) was similarly prepared.
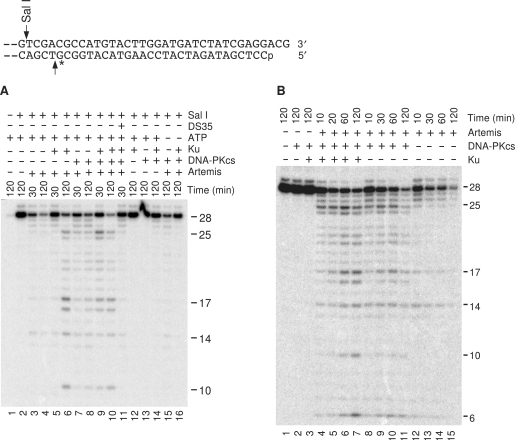
Figure 1.
Artemis-mediated 5′ resection of duplex DNA. (A) The substrate shown, bearing a 3-base 3′ overhang and internally labeled 28 bases from the 5′ end, was treated with 90 nM Artemis, 13 nM Ku and/or 35 nM DNA-PKcs for the times indicated. In some samples, various proteins or ATP were omitted from the reaction, or unlabeled 0.5 μM blunt 35-bp duplex DNA was added, as indicated. (B) Time course of 5′ resection. Reaction conditions are the same as in (A).
Nuclease assays
Reactions (usually 10 μl) were performed in 25 mM NaCl/25 mM Tris–HCl pH 8/10 mM MgCl2/0.25 mM ATP/50 μg/ml BSA. The labeled DNA substrate (10 ng) and Ku were added, and the sample was vortexed and briefly centrifuged. DNA-PKcs was then added, followed immediately by Artemis and the sample was mixed by pipetting and incubated for the indicated times at 37°C. Reactions were stopped by addition of 2 vol. of 0.45 M sodium acetate/10 mM EDTA/100 μg/ml tRNA, and immediately extracted with phenol and then with chloroform and ethanol-precipitated. Samples were dissolved in 25 or 50 μl of 10 mM Tris–HCl pH 8/0.1 mM EDTA and in some cases half of each sample was treated with 35 U/ml T4 DNA polymerase (T4 pol) in the presence of dNTPs (250 μM each) for 15 min at 12°C. The reaction was stopped by addition of EDTA to 20 mM, the sample heated to 75°C for 20 min to inactivate T4 pol, and the DNA again precipitated. All samples were then treated with TaqI (200 U/ml for 6 h at 65°C), precipitated and analyzed on 24% polyacrylamide sequencing gels, or treated with XbaI or SalI (200 U/ml for 4 h at 37°C) and analyzed on 20% gels. Phosphorimages of the gels were analyzed quantitatively using ImageQuant software (GE Healthcare, Piscataway NJ).
Reactions with end-labeled substrates were performed similarly except that samples were diluted with an equal volume of formamide, denatured for 1 min at 90°C and loaded onto 36% polyacrylamide nondenaturing gels without further treatment. For antibody inactivation, preimmune serum or antiserum raised against Artemis protein produced in Escherichia coli (4) was diluted in Artemis reaction buffer and incubated at 22°C for 30 min with 450 nM Artemis. The Artemis-antibody mixture was then added to nuclease reactions as above.
RESULTS
DNA-PK modulates 5′ resection of duplex DNA by Artemis
Previously, we showed that Artemis/DNA-PK can trim plasmid substrates bearing blocked 3′-termini on very short 3′ overhangs and even blunt ends, although much more slowly than longer 3′ overhangs (8). Inasmuch as Artemis has intrinsic 5′→3′ exonucleolytic activity (2), these results could be explained by Artemis-mediated 5′→3′ resection, followed by trimming of the resulting 3′ overhang.
To determine whether the 5′→3′ exonuclease of Artemis can act on long duplex DNA, a plasmid substrate was constructed with a 3-base 3′ overhang, internally labeled 28 bases from the 5′-terminus. This substrate was treated with Artemis, and then cut with SalI to release the terminal 28-base fragment (or any degradation products thereof). In such experiments, treatment with Artemis alone eliminated most of the 28-base fragment within 30 min, suggesting extensive degradation of the 5′-terminal strand (Figure 1A, lanes 3 and 4). Addition of Ku or DNA-PKcs did not block the loss of full-length 28-base fragment, but did increase the amount of shorter fragments, which are presumably intermediates in degradation of the strand (lanes 5–8). These results suggest that Artemis-mediated 5′ trimming proceeded more slowly in the presence of either Ku or DNA-PKcs. When both Ku and DNA-PKcs were present, a 25-base fragment, corresponding to removal of three bases from the 5′ end (presumably leaving a 6-base 3′ overhang) was particularly prominent (Figure 1A, lane 9; Figure 1B, lanes 4 and 5). Moreover, there was a time-dependent progression from longer to shorter fragments, consistent with progressive, possibly exonucleolytic digestion of the strand (Figure 1B, lanes 4–7). When Artemis, Ku and DNA-PKcs were present but ATP was absent, no digestion products were detected, suggesting that digestion required phosphorylation of either Artemis or DNA-PK, or both (Figure 1A, lane 16). However, in the presence of Artemis and DNA-PK but not Ku or ATP, some digestion was still detected, probably because DNA-PK was not as effectively recruited to DNA ends (lane 15).
Given the cellular abundance of DNA-PK, its high affinity for DNA termini, and the limiting amounts of Artemis in vivo, we presume the more relevant Artemis substrate is DNA-PK/DNA rather than naked DNA termini. For this reason, and to promote Artemis nuclease activity at all or most DNA ends, a molar excess of DNA-PK to DNA termini, and excess Artemis to DNA-PK was used in all reactions unless otherwise noted. Based on previous titrations with each protein (8), the specific concentrations used in this and subsequent experiments were chosen to saturate DNA ends with DNA-PK and ensure optimal nuclease activity, without using more of any protein than necessary.
Trimming of 3′-blocked blunt ends by Artemis and DNA-PK is accompanied by limited resection of the complementary strand
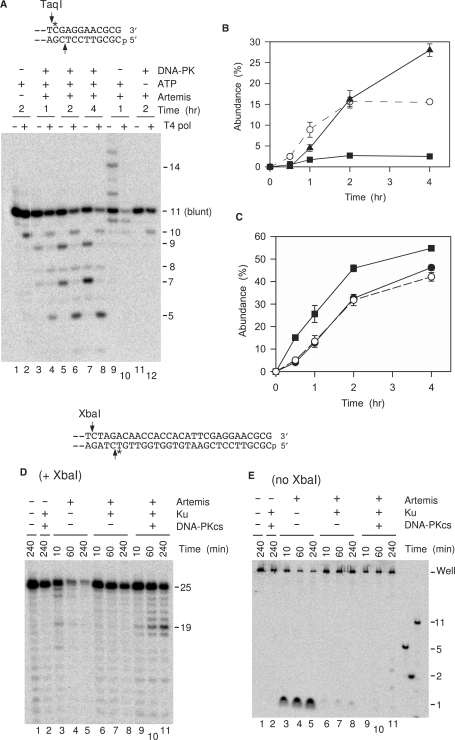
Our previous studies showed that treatment of a 3′-PG-terminated blunt end with Artemis/DNA-PK resulted in slow removal of 2–4 nt from the 3′ end (8). To determine whether this 3′ trimming was accompanied by 5′ trimming, an internally labeled blunt-ended 3′-PG substrate was treated with Artemis and/or DNA-PK, deproteinized, and then treated with T4 pol plus dNTPs (Figure 2). Under these conditions, 3′-termini that were trimmed by Artemis will be either filled in by the polymerase activity of T4 pol, or resected by its 3′→5′ exonuclease activity, in either case producing a blunt end and thus indicating whether the complementary strand had also been processed by Artemis (Figure 2A). Fortuitously, the untrimmed 3′-PG terminus is refractory to the 3′→5′ exonuclease of T4 pol, and thus resection or fill-in by T4 pol will be confined to DNA molecules whose 3′ ends had been trimmed by Artemis/DNA-PK. The substrate was finally treated with TaqI, which releases a short labeled fragment from the DNA end that can then be analyzed on a denaturing gel. TaqI cleaves both single- and double-stranded DNA, so that even substrate molecules that had undergone extensive 5′→3′ resection would be cleaved.
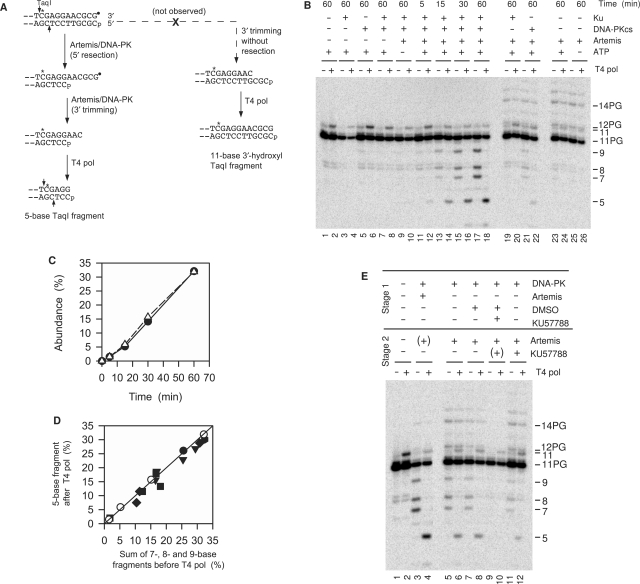
Figure 2.
Coordinate 5′ and 3′ trimming of a blunt 3′-PG (filled circle) 5′-phosphate substrate by Artemis/DNA-PK. (A) Experimental rationale. If 3′ trimming is accompanied by 5′ trimming (left pathway), subsequent treatment with T4 pol will resect or fill in the top strand to match the bottom strand. If 3′ trimming occurs without 5′ trimming (right pathway), T4 pol will fill in the gap, resulting in an increase in labeled 11-base 3′-hydroxyl fragments. (B) Time course and requirements for 3′ and concomitant 5′ trimming. Samples were treated for the indicated times with 25 nM Ku, 50 nM DNA-PKcs and/or 90 nM Artemis. Following deproteinization, half of each sample was treated with T4 pol, and then all samples were treated with TaqI. (C) Time course for accumulation of 3′-trimmed 7-, 8- and 9-base fragments without T4 pol treatment (filled circle) or 5-base fragment with T4 pol treatment (open triangle). (D) Comparison of trimming before and after T4 pol treatment; different symbols indicate six independent experiments similar to that shown in (B). (E) Effect of the DNA-PKcs inhibitor KU57788 on 3′ and 5′ trimming by Artemis in a two-stage reaction. In stage 1, samples were treated with Ku, DNA-PKcs and/or Artemis in the presence or absence of 1 μM KU57788 or 2% DMSO for 30 min. Artemis and/or KU57788 was then added and the sample incubated for an additional 45 min. A ‘(+)’ indicates that a component was present in stage 2 because it had been added in stage 1. Protein concentrations are the same as in (B).
In such experiments, T4 pol treatment consistently eliminated the 7-, 8- and 9-base TaqI fragments resulting from Artemis-mediated trimming, and produced a prominent 5-base fragment, indicating removal of 6 bases from the 5′ end (Figure 2B, lanes 11–18). Like the 7-, 8- and 9-base fragments, production of this 5-base fragment was completely dependent on Artemis (lanes 1–8) and DNA-PKcs (lanes 19 and 20), and largely dependent on Ku (lanes 21 and 22). Moreover, throughout the course of the reaction, the abundance of 5-base fragment after reaction with T4 pol was precisely equal to sum of the 7-, 8- and 9-base fragments prior to T4 pol treatment (Figure 2C). This equivalence was consistently observed over several experiments (Figure 2D). These results strongly suggest that trimming of 2–4 bases from the 3′-terminus by Artemis/DNA-PK was necessarily accompanied by, and probably preceded by, trimming of six bases from the 5′-terminus. Had 3′ trimming occurred without 5′ trimming, T4 pol would have filled in the resulting 5′ overhang to yield an 11-base 3′-hydroxyl fragment. On the contrary, there was no Artemis-dependent formation of such a fragment, implying that there was essentially no 3′ trimming without concomitant 5′ trimming. (The low level of 3′-hydroxyl 11-mer seen even in samples treated with T4 pol alone presumably results from fill-in of a small number of incidental breaks near the 3′ end; however, the abundance of 11-mer fragment was reduced rather than increased by Artemis treatment.)
As reported previously, 3′ trimming of the blunt substrate by Artemis/DNA-PK was completely dependent on ATP (Figure 2B, compare lanes 9–10 to lanes 17–18), suggesting a requirement for phosphorylation of DNA-PK and/or Artemis. In an attempt to distinguish the critical phosphorylation target, the reaction was performed in two stages (10). DNA-PK was preincubated with substrate in the presence of ATP, and then the DNA-PK inhibitor KU57788 was added prior to Artemis (Figure 2E). Under these conditions, qualitatively similar 3′ trimming was still detected (lanes 11 and 12), albeit at a lower level than when no inhibitor was added (lanes 7 and 8). When KU57788 was added at the same time as DNA-PK, no trimming was detected (lanes 9 and 10). Similar results were obtained with the less specific but irreversible DNA-PK inhibitor wortmannin (Supplementary Figure 1). These results suggest that autophosphorylation of DNA-PK is essential for such trimming, as reported previously for trimming of 5′ overhangs (10). However, because the level of trimming was reduced under these conditions and because Artemis may have been constitutively phosphorylated during production in insect cells, a role for Artemis phosphorylation in promoting trimming cannot be excluded.
Trimming of blunt ends by Artemis/DNA-PK does not require a 5′-phosphate
For single-stranded oligomeric substrates, it has been reported that the 5′→3′ exonuclease activity of Artemis requires a 5′-phosphate terminus (2), and we have confirmed this specificity for soluble recombinant Artemis (Figure 6A). Thus, if the 6-base 5′ resection that accompanies 3′ trimming is catalyzed by Artemis exonuclease, then it might be expected that removal of the 5′-phosphate would block 5′ as well as consequent 3′ trimming of blunt ends by Artemis/DNA-PK. To test this prediction, the blunt-ended 3′-PG substrate was dephosphorylated, treated with Artemis and/or DNA-PK and finally treated (or not) with T4 pol to produce blunt ends as above.
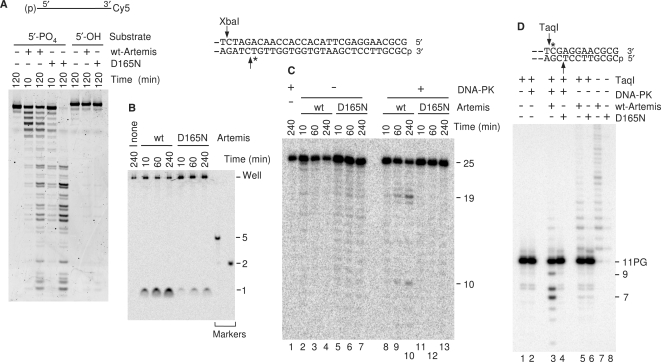
Figure 6.
Presence of 5′→3′ exonuclease activity and lack of 5′ and 3′ endonucleolytic trimming activities in Artemis mutant D165N. (A) A single-strand 3′-Cy5-labeled 25-mer (2 nM) with either a 5′-phosphate or 5′-hydroxyl terminus, was treated with 180 nM wild-type or D165N mutant Artemis for the indicated times and products were analyzed on a 20% sequencing gel. The mutant enzyme retains 5′→3′ exonuclease activity. (B) The substrate shown, labeled 25 bases from the 5′-terminus, was treated with Artemis alone for the indicated times, and then products were analyzed on a 36% gel without further treatment. (C) The same substrate was treated with Artemis or Artemis/DNA-PK for the indicated times, and then cut with XbaI. (D) The internally labeled 3′-PG 5′-phosphate substrate shown was treated with Artemis and/or DNA-PK for 2 h, and then cut with TaqI as in Figure 2. Protein concentrations in (B—D) are as in Figure 2. At least two replicate experiments were performed with each substrate, with essentially identical results.
Treatment of the blunt 3′-PG 5′-phosphate substrate with Artemis in the absence of DNA-PK resulted in TaqI-independent, ATP-independent release of labeled 12- to 15-base fragments, indicating endonucleolytic cleavage farther from the 3′-terminus than in the presence of DNA-PK (Figure 2B, lanes 23–26 and Figure 3A, lanes 5–8). These fragments were not affected by T4 pol treatment, suggesting that they retained a 3′-PG terminus (Figure 3A, lanes 5–8), nor were they cleaved by TaqI, probably because they extend only a few bases beyond the TaqI site and may bind poorly to the enzyme. However, 5′ dephosphorylation of the blunt 3′-PG substrate prior to Artemis treatment completely prevented generation of these fragments (Figure 3A, lanes 13 and 14). The simplest explanation of these data is that in the absence of DNA-PK, Artemis catalyzed extensive 5′→3′ exonucleolytic resection (Figure 1A), followed by endonucleolytic cleavage of the resulting 3′ overhang. The 5′-hydroxyl substrate would presumably be resistant to exonucleolytic 5′ resection (2), and thus no long 3′ overhangs that could be cleaved by Artemis would be produced.
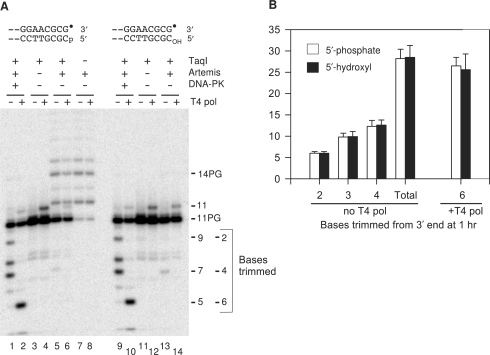
Figure 3.
Trimming of a 3′-PG 5′-hydroxyl substrate by Artemis/DNA-PK. (A) Blunt-ended 3′-PG substrates with either 5′-phosphate (lanes 1–8) or 5′-hydroxyl termini (lanes 9–14) were treated with Artemis and/or DNA-PK in the presence of ATP for 1 h, and then half of each sample was treated with T4 pol as in Figure 2. Dephosphorylation blocked trimming in the absence of DNA-PK but had no effect on trimming in the presence of DNA-PK. (B) Quantitation of trimming by Artemis/DNA-PK. The 5-, 7-, 8- and 9-base fragments in (A) correspond to trimming of 6, 4, 3 and 2 bases from the 3′ end, respectively. Error bars show standard errors of three replicate experiments including that shown in (A).
Unexpectedly, however, in the presence of DNA-PK, removal of the 5′-phosphate had no discernable effect on the extent or the specificity of trimming of the blunt 3′-PG substrate by Artemis; the 3′-PG 5′-hydroxyl substrate still showed the trimming of 2–4 bases from the 3′-terminus, and six bases from the 5′-terminus (Figure 3A, lanes 1–2 and 9–10; quantitative data in Figure 3B). Thus, either the observed 5′ trimming was endonucleolytic, or the specificity of the 5′→3′ exonuclease was altered in the presence of DNA-PK to accept a 5′-hydroxyl substrate.
Trimming of blunt ends by Artemis/DNA-PK does not require a blocked 3′-terminus
To determine whether a 3′-PG terminus was required for trimming, a similar internally labeled blunt-ended substrate with a 3′-hydroxyl terminus was constructed. Trimming of this substrate by Artemis/DNA-PK was qualitatively similar to that of the 3′-PG substrate, with 2–4 bases being removed from the 3′-terminus, generating Artemis-dependent 7-, 8- and 9-mer bands. As with the 3′-PG substrate, subsequent T4 pol treatment yielded a prominent 5-mer band, indicating trimming of six bases from the 5′-terminus (Figure 4A). In addition, all T4 pol-treated samples showed a minor Artemis-independent 10-mer band, probably reflecting equilibrium between the polymerase and exonuclease functions of T4 pol at the blunt end. However, the 3′-hydroxyl substrate was trimmed two to three times more slowly than the 3′-PG substrate (compare Figures 2C and 4C). The specificity of trimming was also slightly different, with more 2- and 4-base, but less 3-base trimming. As seen previously with a 6-base 3′ overhang substrate (8), there was a time-dependent progression from 2-base to 4-base trimming (Figure 4B), consistent with sequential removal of two bases at a time. As with the 3′-PG blunt end, the extent of 6-base 3′ trimming following T4 pol treatment was equal to the sum of 2-, 3- and 4-base 3′ trimming before T4 pol treatment (Figure 4C). However, with the 3′-hydroxyl blunt end, there was a significant fraction of fragments corresponding to 2- to 5-base 3′ trimming in the T4 pol-treated samples (Figure 4A, lanes 4 and 6). These fragments were not seen with the 3′-PG substrate (Figure 2A, lanes 14 and 16), suggesting that they represent substrates that were trimmed by Artemis in the 5′-terminal strand only. Thus, the sum of fragments corresponding to trimming of 2–6 bases was greater in the samples subsequently treated with T4 pol, particularly at early time points (Figure 4C). The simplest explanation of all these data is that intermediates, with trimming of 2–5 bases from the 5′-terminus but no trimming from the 3′-terminus, accumulate early in the reaction; but when 5′ trimming reaches six bases, the resulting 3′ overhang is rapidly trimmed as well.
Figure 4.
Trimming of a 3′-hydroxyl 5′-phosphate substrate by Artemis/DNA-PK. (A) The substrate shown was treated with 25 nM Ku, 50 nM DNA-PKcs and/or 90 nM Artemis for the times indicated, and then half of each sample was treated with T4 pol. Note that the 10-mer band was present in all T4 pol-treated samples even with Artemis (lane 2); it probably represents an equilibrium between the exonuclease and polymerase functions of the enzyme at the blunt end. (B) Quantitation of trimming. Abundance of 7- (filled triangle), 8- (filled square) and 9-base (open circle) fragments (corresponding to trimming of 4, 3 and 2 bases, respectively) were determined from three independent experiments similar to that shown in (A). Error bars indicate standard errors when larger than the symbols. (C) Comparison of 3′ trimming by Artemis/DNA-PK with and without subsequent treatment with T4 pol. The total of 2-, 3-, 4-, 5- and 6-base 3′ trimming with T4 pol (filled square), 6-base trimming alone with T4 pol (filled circle) and the total of 2-, 3- and 4-base trimming without T4 pol (open circle), were calculated from two independent experiments; error bars show the range of values. The Artemis-independent 10-mer was not included in the calculations. (D) A substrate with identical terminal sequence and structure, but labeled 25 bases from the 5′-terminus, was similarly treated with Artemis or Artemis/DNA-PK and then cut with XbaI to release terminal fragments. (E) Same as (D), except samples were directly loaded on a 36% gel without XbaI treatment.
Unlike the 3′-PG substrate, the 3′-hydroxyl substrate would be subject to 3′ overhang removal by T4 pol if it had undergone 5′ resection but not 3′ trimming by Artemis. However, T4 pol-mediated processing of the substrate treated with Artemis/DNA-PK without ATP was indistinguishable from that of the untreated substrate (Figure 4A, lanes 11 and 12), showing only a low level of single-nucleotide removal (10-mer). This result suggests that, when DNA-PK is present, Artemis-mediated 5′ resection requires DNA-PK catalytic activity.
Treatment of the blunt 3′-hydroxyl substrate with Artemis in the absence of DNA-PK produced fragments 12–15 bases in length (Figure 4A, lane 9). These fragments were almost completely digested by subsequent treatment with T4 pol (lane 10), suggesting that they were single-stranded. These data thus support the view that the fragments were formed by Artemis-mediated 5′ resection followed by cleavage of the resulting 3′ overhang, as proposed above for the similar but T4 pol-resistant fragments produced from the 3′-PG substrate (Figure 3A). The prominent 11-mer band in the sample treated with Artemis alone (lane 9) was also largely eliminated by T4 pol treatment (lane 10), suggesting that although a significant fraction of molecules with intact 3′-termini remained following Artemis treatment, most of them had already been 5′-resected.
To verify more directly the 5′ trimming inferred from T4 pol digestion experiments, a substrate was prepared with an identical terminal sequence, but with an internal label 25 bases from the 5′-terminus. Following reaction with Artemis, DNA was deproteinized and cut with XbaI to release terminal fragments (Figure 4D). Analysis of these fragments confirmed the predominant trimming of six bases from the 5′ end by Artemis plus DNA-PK (lanes 9–11), as well as the more extensive degradation of this strand by Artemis alone (lanes 3–5). As with other substrates, treatment with Artemis plus DNA-PKcs without Ku resulted in qualitatively similar but less robust 6-base 5′ trimming (Supplementary Figure 2).
When samples of the same substrate treated with Artemis alone were directly loaded on a 36% gel without XbaI treatment, radioactivity was found either in the well or at the position of a mononucleotide, indicating that digestion was exonucleolytic (Figure 4E). Qualitatively similar, but dramatically reduced mononucleotide release was seen in the presence of Ku (lanes 6–8), as expected from Figure 4D. For samples treated with Artemis in the presence of both Ku and DNA-PKcs, no mononucleotide release was seen, and instead traces of di-, tri- and tetranucleotides were detected after 4 h (Figure 4E, lane 11). This result suggests that DNA-PK promotes a switch from exo- to endo-nucleolytic digestion, and that in a small fraction of molecules such digestion had proceeded 25 bases from the DNA end by that time.
Trimming of blunt ends by Artemis/DNA-PK is at least partially endonucleolytic in both strands
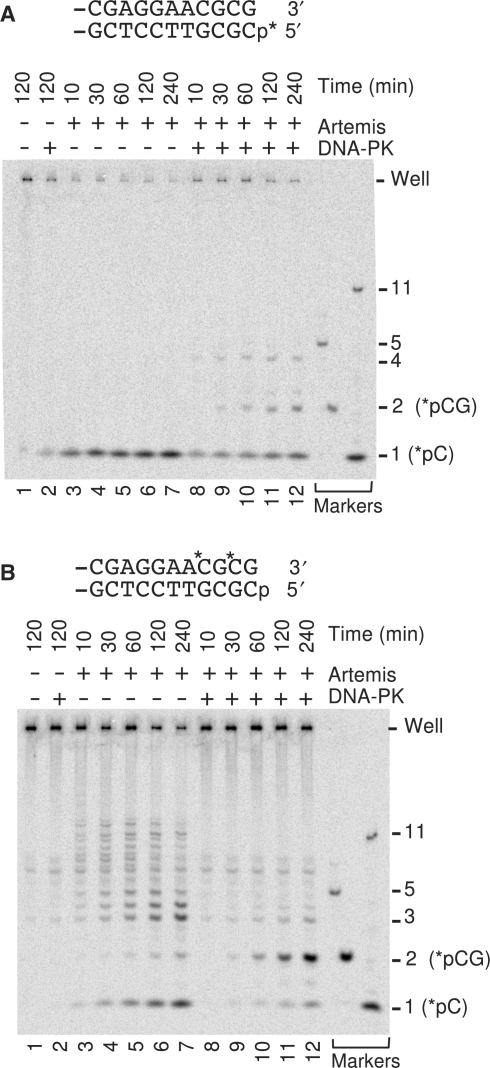
Artemis degrades single-stranded oligomers exonucleolytically, but in the presence of DNA-PK, Artemis trims 5′ overhangs endonucleolytically (2,10). To determine which of these activities was responsible for resection of blunt-ended DNA by Artemis/DNA-PK, blunt 5′-phosphate 3′-hydroxyl substrates were prepared that were identical to the internally labeled plasmid substrate, but were labeled either at the 5′-terminal phosphate or at the two C residues in the 3′-terminal CGCG sequence. Following treatment with Artemis in the presence or absence of DNA-PK, samples were analyzed on 36% polyacrylamide gels, which afforded resolution of mono-, di- and oligonucleotide products (Figure 5).
Figure 5.
Endonucleolytic trimming of both DNA strands by Artemis/DNA-PK. (A) The 5′-end-labeled plasmid substrate shown was treated with 90 nM Artemis in the presence or absence of DNA-PK (25 nM Ku plus 50 nM DNA-PKcs) for the indicated times, and the products analyzed on a 36% polyacrylamide gel. (B) An identical substrate labeled (asterisk) at the two C residues near the 3′-terminus was likewise treated with Artemis or Artemis/DNA-PK.
In the absence of DNA-PK, Artemis released the 5′-terminal label exclusively as a mononucleotide, consistent with exonucleolytic degradation of this strand (Figure 5A, lanes 3–7). Although small amounts of hydrolyzed mononucleotide were detected after 2 h of incubation without protein, and with DNA-PKcs alone (Figure 5A, lanes 1 and 2), the Artemis nuclease activity exceeds this background hydrolysis at even the earliest time point of 10 min (Figure 5A, compare lanes 1–3). In contrast to 5′ label, label near the 3′-terminus was released as a broad distribution of oligonucleotides from 3 to about 12 bases in length (Figure 5B, lanes 3–7), consistent with endonucleolytic cleavage of the 3′ overhangs generated by 5′ resection, as proposed above. At longer times, the distribution was shifted toward shorter oligomers (including monomers), suggesting that once released, the oligomers may have been shortened by the 5′→3′ exonuclease activity of Artemis.
In the presence of DNA-PK, the label near the 3′-terminus was released almost exclusively as dinucleotides (Figure 5B, lanes 8–12). If a large proportion of the DNA molecules were 5′-resected to leave a 6-base 3′ overhang (as suggested by the data above, Figures 2–4), then sequential removal of two dinucleotides from the 3′ end is the expected result, based on the known action of Artemis/DNA-PK on short overhangs (8). It is unlikely that the dinucleotide was derived primarily from longer fragments that were subsequently digested by 5′→3′ exonuclease activity, because in that case at least half of the label would have been released as mononucleotides; instead, in the presence of DNA-PK very little mononucleotide was produced.
The presence of DNA-PK altered the manner of 5′ trimming as well, in that 5′-end-label was released as dinucleotides and tetranucleotides in addition to mononucleotides (Figure 5A, lanes 8–12). This result indicates that 5′ trimming was at least partially endonucleolytic. It is possible that the 5′ trimming was exclusively endonucleolytic, and that the labeled mononucleotide was generated by Artemis-mediated digestion of oligonucleotides that had already been released from the 5′- terminus. Indeed, given the apparent requirement of the exonuclease activity for a 5′ phosphate (2), the finding that 5′-dephosphorylation has no effect at all on 5′ trimming of the blunt-ended substrate (Figure 3) seems more compatible with a wholly endonucleolytic mode of trimming in the presence of DNA-PK. However, the possibility that 5′ resection by Artemis/DNA-PK was a mixture of endo- and exo-nucleolytic trimming cannot be excluded.
Artemis-mediated endonucleolytic trimming is abrogated by treatment with antiserum to Artemis or by a D165N mutation
Despite the high purity of the Artemis preparation used in these experiments (8), the possibility must be considered that one or more of the observed nuclease activities might be due to some low-abundance, highly active contaminant. To address this question, aliquots of soluble recombinant Artemis were pretreated with rabbit antiserum raised against recombinant Artemis fragments produced in E. coli (Supplementary Figure 3). Whereas preimmune serum had little effect on Artemis activity, antiserum to Artemis completely blocked 5′ and 3′ trimming of the blunt 3′-PG and 3′-hydroxyl substrates by Artemis/DNA-PK. The same antiserum completely blocked exonucleolytic 5′ resection of an internally labeled blunt-ended substrate by Artemis alone (as in Figure 4E), as well as the characteristic 6-base 5′ trimming of this same substrate by Artemis/DNA-PK (as in Figure 4D).
In previous work with immobilized Artemis and oligomeric substrates, a D165N mutation in Artemis was found to abrogate endonucleolytic cleavage of 3′ and 5′ overhangs, but not exonucleolytic digestion of single-stranded DNA (2). To confirm that Artemis was responsible for the observed endonucleolytic trimming activities, soluble recombinant Artemis with a D165N mutation was prepared by methods identical to those used for the wild-type enzyme. As reported (2), D165N mutant Artemis retained 5′→3′ single-strand exonuclease activity, and this activity required a 5′-phosphate but did not require DNA-PK. Although the mutant protein showed somewhat altered digestion kinetics, it nevertheless produced a qualitatively similar ladder of shorter fragments from a 3′-Cy5-labeled 25-mer (Figure 6A), and released a single labeled nucleotide from a 5′-end-labeled 11-mer (data not shown). In addition, the mutant retained 5′→3′ exonucleolytic activity toward double-stranded DNA, releasing a labeled mononucleotide from an internally labeled substrate, although 6- to 10-fold less efficiently than wild-type (Figure 6B). However, in the presence of DNA-PK the D165N mutant did not induce any of the 6-base 5′ trimming characteristic of the wild-type enzyme (Figure 6C; 19-base fragment in lanes 9 and 10, absent in lanes 12 and 13), nor did it release traces di-, tri- and tetra-nucleotides when subsequent cleavage by XbaI was omitted (Supplementary Figure 4). The mutant also did not induce any 3′ trimming of the internally labeled blunt-ended 3′-PG substrate, either in the presence or in the absence of DNA-PK (Figure 6D, lanes 3–4 and 7–8). These results are consistent with the D165N mutation causing loss of endonuclease activity, while only moderately affecting the exonuclease activity.
Thus, experiments with both antibody-mediated inactivation and with the D165N mutant indicate that the observed exo- and endo-nucleolytic activities are attributable to Artemis and not to any contaminants. Based on homology to β-lactamase enzymes, the D165N mutation was predicted to inactivate Artemis (2), and although the mutant enzyme clearly retains exonuclease activity, its specificity and kinetics appear to be altered (Figure 6A and B). Given the absence of any other obvious exonuclease motifs in the Artemis protein, we speculate that perhaps the endonuclease and exonuclease share the same active site, but that the endonuclease activity has more stringent structural requirements for substrate binding and activity.
DISCUSSION
Experiments with oligonucleotide substrates have shown that, in the presence of active DNA-PK, Artemis opens hairpin ends, removes 5′ overhangs and shortens 3′ overhangs to a length 4–5 nucleotides (2,8,10). However, when longer (5.5 kb) internally labeled substrates were treated with DNA-PK plus soluble recombinant Artemis, additional processing activities were revealed. Very short 3′ overhangs of 4–5 bases were efficiently trimmed to 2–3 bases. Long duplexes bearing 3′ overhangs of 1–2 bases, and even blunt ends, were also subject to Artemis-mediated trimming, albeit much more slowly than longer overhangs (8). The apparent lack of trimming of analogous oligomeric substrates (2,8,10) may reflect inability of the Artemis/DNA-PK complex to assemble and properly align the terminus with the active site on short duplexes, or interference between complexes loading from opposite ends of the substrate.
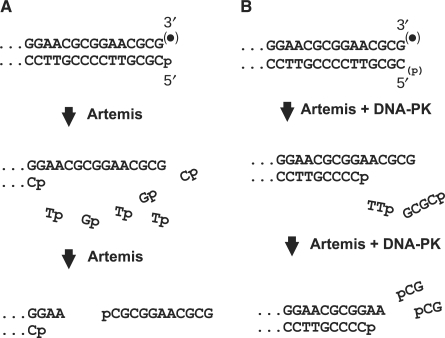
A more detailed examination of Artemis-mediated blunt-end trimming by Artemis/DNA-PK reveals that it is executed as a limited and tightly coordinated removal of very short oligonucleotides from both strands. For the particular blunt-ended substrate used in these experiments, the predominant products indicate trimming of six bases from the 5′-terminus, and two, three or four bases from the 3′-terminus, giving a 2-, 3-, or 4-base 3′ overhang. While the data do not unambiguously define the order of trimming, several experimental results suggest that 5′ trimming precedes and facilitates 3′ trimming. In particular, experiments with the 3′-PG substrate (Figure 2) indicate that 3′ trimming is always accompanied by 5′ trimming, whereas experiments with the 3′-hydroxyl substrate (Figure 4) indicate that there are some molecules trimmed in the 5′-terminal strand only. Such molecules dominate the product distribution early in the reaction, but do not continue to accumulate; rather, they rapidly reach a plateau, consistent with the view that they are transient intermediates in the reaction (Figure 4C). Moreover, previous work predicts that the proposed 6-base 3′ overhang intermediate would be rapidly shortened to four and then to two bases by Artemis/DNA-PK, by successive release of two dinucleotides (8) and as predicted, radiolabel near the 3′-terminus is released almost exclusively as dinucleotides (Figure 5). Taken together, data from all substrates examined are consistent with the model of Artemis activity at blunt ends shown in Figure 7. In the absence of DNA-PK, the Artemis exonuclease activity catalyzes extensive 5′→3′ resection, which is followed by endonucleolytic release of oligomers as long as 15 nucleotides from the resulting 3′ overhang. DNA-PK strongly suppresses the exonuclease, but upon autophosphorylation it promotes endonucleolytic release of short oligomers from the 5′ end, ultimately producing a 6-base overhang that is then trimmed to 2–4 bases. Unlike the exonuclease activity, which requires a 5′-phosphate, endonucleolytic trimming does not require a specific structure at either the 5′- or the 3′-terminus. Thus, this endonucleolytic activity could serve to remove chemically modified 3′- and 5′-termini such as 3′-PGs and 5′-aldehydes. Failure to efficiently process ends of DSBs induced by neocarzinostatin (bearing 3′-PG and 1-base-recessed 5′-aldehyde termini) and by bleomycin (typically 3′-PG/5′-phosphate blunt ends) could potentially explain the sensitivity of Artemis-deficient cells to these agents (6,8,17,18).
Figure 7.
Action of Artemis on blunt ends in the absence (A) or presence (B) of DNA-PK. DNA-PK induces a shift from exo- to endo-nucleolytic 5′ resection, eliminates the requirement for a 5′-phosphate, and limits the extent of trimming. Both exo- and endo-nucleolytic 5′ trimming can occur with either a 3′-PG or a 3′-hydroxyl terminus.
Previous work has shown that DNA-PK autophosphorylation at multiple sites relieves its total sequestration of DNA ends, initially rearranging to allow limited access to enzymatic processing and finally dissociating (19–21). In the case of Artemis, the effect of DNA-PK is to both confine nuclease activity to very near the DNA end and to alter its specificity, suggesting that Artemis acts while autophosphorylated DNA-PK is still bound to DNA.
Evidence that Artemis is involved in cell cycle signaling (22), and that Artemis is epistatic with ATM in terms of radiosensitivity (5), has raised the possibility that a signaling defect rather than an end processing defect may be primarily responsible for the chemo- and radio-sensitivity of Artemis-deficient cells. However, Artemis shows epistasis with ATM in direct DSB repair assays as well, even in growth-arrested cells that should not be subject to cell cycle effects (5). Artemis, ATM, 53BP1 and the MRN complex all appear to be required for repair of a specific subset of radiation-induced DSBs that in normal cells are rejoined relatively slowly (∼2–24 h after irradiation), but the criteria that define this fraction of presumably difficult-to-repair breaks are not known (4,5). One possibility is that DSBs with complex end structures, for example a combination of blocked termini and adjacent base damage, might only be resolvable by Artemis-mediated trimming of the damaged ends. Upon prolonged incubation, Artemis/DNA-PK-mediated trimming can proceed at least 25 bp from a DNA end (Figure 4E) in a small fraction of molecules. Thus, one can envision a process, wherein heavily damaged ends are subjected to successive rounds of Artemis-mediated trimming, interspersed with attempts to patch and ligate two juxtaposed ends (8,23). This end-digestion would presumably proceed until a segment of undamaged (or minimally damaged) DNA suitable for patching/ligation is exposed. However, the DSBs induced by radiomimetic drugs such as bleomycin and neocarzinostatin are much more homogeneous in structure than radiation-induced DSBs, and rarely if ever involve base lesions or clustered damage (17,18). Thus, while end structure may well be a major factor, the sensitivity of Artemis-deficient cells to the radiomimetic compounds, which is comparable to their radiosensitivity (6,8), suggest that there are additional factors that determine whether a given DSB will require Artemis for repair.
An alternative possibility is that any DSBs, which fail to rejoin within 1–2 h are channeled into the Artemis- and 53BP1-dependent rejoining pathway that is associated with repair foci. Such DSBs might include chemically complex breaks, but also breaks in highly condensed chromatin and/or breaks whose two ends have become physically separated. Once a DSB enters this pathway, the DNA ends may become inaccessible to enzymes such as Ape1 (24) or tyrosyl-DNA phosphodiesterase (25) that might otherwise be capable of removing blocked termini. It would thus be essential that this pathway include an enzyme that can resolve diverse end structures with minimal sequence loss, in what may be a final attempt at repair. Previous work has shown that Artemis/DNA-PK can resolve hairpin ends, as well as long and short 3′ and 5′ overhangs, including overhangs bearing 3′-PG and 3′-phosphotyrosyl termini (2,8). The present work shows that Artemis/DNA-PK can also convert terminally blocked blunt ends to ends with short 3′ overhangs and ligatable termini. Although the kinetics of this blunt end trimming are slow, they are consistent with the slow kinetics of Artemis-dependent DSB repair in cells (4,5). Moreover, annealing of adventitious complementarities in the resulting 3′ overhangs, followed by patching and ligation (16), would yield precisely the types of repair joints typically seen in nonhomologous end joining, with very short microhomologies and terminal deletions of a few to several base pairs (26).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Graeme Smith and KUDOS Pharmaceuticals for providing KU-57788. This work was supported by Grants CA40615 and CA72955 from the National Cancer Institute, and Grant AG023783 from the National Institute on Aging, DHSS. Work at LBNL was supported by the U.S. Department of Energy Office of Science, under contract no. DE-AC02-05CH11231 and US National Institutes of Health grant CA104660. Funding to pay the Open Access publication charges for this article was provided by CA40615.
Conflict of interest statement. None declared.
REFERENCES
- 1.Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 2.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 3.Li L, Moshous D, Zhou Y, Wang J, Xie G, Salido E, Hu D, de Villartay JP, Cowan MJ. A founder mutation in Artemis, an SNM1-like protein, causes SCID in Athabascan-speaking native Americans. J. Immunol. 2002;168:6323–6329. doi: 10.4049/jimmunol.168.12.6323. [DOI] [PubMed] [Google Scholar]
- 4.Wang J, Pluth JM, Cooper PK, Cowan MJ, Chen DJ, Yannone SM. Artemis deficiency confers a DNA double-strand break repair defect and Artemis phosphorylation status is altered by DNA damage and cell cycle progression. DNA Repair. 2005;4:556–570. doi: 10.1016/j.dnarep.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol. Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 6.Rooney S, Alt FW, Lombard D, Whitlow S, Eckersdorff M, Fleming J, Fugmann S, Ferguson DO, Schatz DG, Sekiguchi J. Defective DNA repair and increased genomic instability in Artemis-deficient murine cells. J. Exp. Med. 2003;197:553–565. doi: 10.1084/jem.20021891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L, Salido E, Zhou Y, Bhattacharyya S, Yannone SM, Dunn E, Meneses J, Feeney AJ, Cowan MJ. Targeted disruption of the Artemis murine counterpart results in SCID and defective V(D)J recombination that is partially corrected with bone marrow transplantation. J. Immunol. 2005;174:2420–2428. doi: 10.4049/jimmunol.174.4.2420. [DOI] [PubMed] [Google Scholar]
- 8.Povirk LF, Zhou T, Zhou R, Cowan MJ, Yannone SM. Processing of 3′-phosphoglycolate-terminated DNA double strand breaks by Artemis nuclease. J. Biol. Chem. 2007;282:3547–3558. doi: 10.1074/jbc.M607745200. [DOI] [PubMed] [Google Scholar]
- 9.Drouet J, Frit P, Delteil C, de Villartay JP, Salles B, Calsou P. Interplay between Ku, artemis and DNA-PKcs at DNA ends. J. Biol. Chem. 2006;281:27784–27793. doi: 10.1074/jbc.M603047200. [DOI] [PubMed] [Google Scholar]
- 10.Goodarzi AA, Yu Y, Riballo E, Douglas P, Walker SA, Ye R, Harer C, Marchetti C, Morrice N, Jeggo PA, et al. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J. 2006;25:3880–3889. doi: 10.1038/sj.emboj.7601255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Povirk LF, Han Y.-H, Steighner RJ. Structure of bleomycin-induced DNA double-strand breaks: predominance of blunt ends and single-base 5′ extensions. Biochemistry. 1989;28:8508–8514. doi: 10.1021/bi00440a016. [DOI] [PubMed] [Google Scholar]
- 12.Hsu H-L, Yannone SM, Chen DJ. Defining the interactions between DNA-PK and ligase IV/XRCC4. DNA Repair. 2001;1:225–235. doi: 10.1016/s1568-7864(01)00018-0. [DOI] [PubMed] [Google Scholar]
- 13.Yannone SM, Roy S, Chan DW, Murphy MB, Huang S, Campisi J, Chen DJ. Werner syndrome protein is regulated and phosphorylated by DNA-dependent protein kinase. J. Biol. Chem. 2001;276:38242–38248. doi: 10.1074/jbc.M101913200. [DOI] [PubMed] [Google Scholar]
- 14.Chen S, Hannis JC, Flora JW, Muddiman DC, Charles K, Yu Y, Povirk LF. Homogeneous preparations of 3′-phosphoglycolate-terminated oligodeoxynucleotides from bleomycin-treated DNA as verified by electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Anal. Biochem. 2001;289:274–280. doi: 10.1006/abio.2000.4936. [DOI] [PubMed] [Google Scholar]
- 15.Bennett RAO, Gu X-Y, Povirk LF. Construction of a vector containing a site-specific DNA double-strand break with 3′-phosphoglycolate termini and analysis of the products of end-joining in CV-1 cells. Intl. J. Radiat. Biol. 1996;70:623–636. doi: 10.1080/095530096144509. [DOI] [PubMed] [Google Scholar]
- 16.Chen S, Inamdar KV, Pfeiffer P, Feldmann E, Hannah MF, Yu Y, Lee JW, Zhou T, Lees-Miller SP, Povirk LF. Accurate in vitro end-joining of a DNA double-strand break with partially cohesive 3′-overhangs and 3′-phosphoglycolate termini: effect of Ku on repair fidelity. J. Biol. Chem. 2001;276:24323–24330. doi: 10.1074/jbc.M010544200. [DOI] [PubMed] [Google Scholar]
- 17.Dedon PC, Goldberg IH. Free-radical mechanisms involved in the formation of sequence-dependent bistranded DNA lesions by the antitumor antibiotics bleomycin, neocarzinostatin, and calicheamicin. Chem. Res. Toxicol. 1992;5:311–332. doi: 10.1021/tx00027a001. [DOI] [PubMed] [Google Scholar]
- 18.Povirk LF. DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: bleomycin, neocarzinostatin and other enediynes. Mutat. Res. 1996;355:71–89. doi: 10.1016/0027-5107(96)00023-1. [DOI] [PubMed] [Google Scholar]
- 19.Weterings E, Verkaik NS, Bruggenwirth HT, Hoeijmakers JH, van Gent DC. The role of DNA dependent protein kinase in synapsis of DNA ends. Nucleic Acids Res. 2003;31:7238–7246. doi: 10.1093/nar/gkg889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calsou P, Frit P, Humbert O, Muller C, Chen DJ, Salles B. The DNA-dependent protein kinase catalytic activity regulates DNA end processing by means of Ku entry into DNA. J. Biol. Chem. 1999;274:7848–7856. doi: 10.1074/jbc.274.12.7848. [DOI] [PubMed] [Google Scholar]
- 21.Block WD, Yu Y, Merkle D, Gifford JL, Ding Q, Meek K, Lees-Miller SP. Autophosphorylation-dependent remodeling of the DNA-dependent protein kinase catalytic subunit regulates ligation of DNA ends. Nucleic Acids Res. 2004;32:4351–4357. doi: 10.1093/nar/gkh761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, Succi J, Feng Z, Prithivirajsingh S, Story MD, Legerski RJ. Artemis is a phosphorylation target of ATM and ATR and is involved in the G2/M DNA damage checkpoint response. Mol. Cell Biol. 2004;24:9207–9220. doi: 10.1128/MCB.24.20.9207-9220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma Y, Lu H, Schwarz K, Lieber MR. Repair of double-strand DNA breaks by the human nonhomologous DNA end joining pathway: the iterative processing model. Cell Cycle. 2005;4:1193–1200. doi: 10.4161/cc.4.9.1977. [DOI] [PubMed] [Google Scholar]
- 24.Suh D, Wilson III DM, Povirk LF. 3′-Phosphodiesterase activity of human apurinic/apyrimidinic endonuclease at DNA double-strand break ends. Nucleic Acids Res. 1997;25:2495–2500. doi: 10.1093/nar/25.12.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inamdar KV, Pouliot JJ, Zhou T, Lees-Miller SP, Rasouli-Nia A, Povirk LF. Conversion of phosphoglycolate to phosphate termini on 3′ overhangs of DNA double-strand breaks by the human tyrosyl-DNA phosphodiesterase hTdp1. J. Biol. Chem. 2002;276:24323–24330. doi: 10.1074/jbc.M204688200. [DOI] [PubMed] [Google Scholar]
- 26.Povirk LF. Biochemical mechanisms of chromosomal translocations resulting from DNA double-strand breaks. DNA Repair. 2006;5:1199–1212. doi: 10.1016/j.dnarep.2006.05.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.