Abstract
Modification of the N-terminal tail of histones is required for various nuclear processes. Here, we show that fission yeast Clr6-HDAC (histone deacetylase) regulates the checkpoint kinase Cds1 when DNA replication encounters a stressful condition. We found that the global level of acetylation of histone H4 was constant throughout the normal cell cycle, but was reduced significantly when the cell recovered from the HU-induced cell cycle arrest (or slow DNA replication). We identified the Clr6-HDAC as a component responsible for the reduction in the level of the H4 acetylation. Although DNA replication was completed, the HU-induced cell cycle arrest could not be released even after removal of HU in the clr6-1 mutant. Under this experimental condition, Cds1 kinase was maintained active and remained bound tightly to chromatin. We also demonstrated that Cds1 was active even after treatment with caffeine, an inhibitor for ATM/ATR that is an activator of Cds1. These results indicate that inactivation of Cds1 requires functional Clr6-HDAC independently of the conventional DNA replication checkpoint. When DNA replication is impeded, Clr6-HDAC activity may monitor damage on chromatin structure/environment, which is required for inactivation of Cds1.
INTRODUCTION
Posttranslational modification of histones plays important roles in maintenance of the genome integrity. Phosphorylation of histone H2AX, which occurs shortly after introduction of double-strand break (DSB) of DNA, is required for DNA repair as well as regulation of the damage checkpoint (1). It was also shown that acetylation of lysine residues at the N-terminal tail of H4 is required for DSB repair in budding yeast (2). A recent study demonstrated an important role of acetylation of histone H3 lysine 56 in generation of a favorable chromatin structure for DNA repair (3). Not only enzymes to modify the histone tail, but also enzymes to catalyze the reverse reaction play roles in coping with DNA damage. The multisubunit histone deacetylase (HDAC) complexes catalyze the deacetylation of highly conserved acetylated lysine residues of core histones in the N-terminal tails. The conditional mutation of Clr6-HDAC results in chromosome mis-segregation and sensitivity to DNA-damaging agents in fission yeast (4). Cells lacking Pst2, a homolog of Sin3 in budding yeast, which are stably associated with Clr6, are also sensitive to DNA-damaging agents (4). It has also been shown that the histone modification is required for regulation of DNA replication during the normal cell cycle. In S phase, acetylation of histone H4 tails seems to be coupled with DNA replication, since increased and unscheduled replication was observed in cells deficient in a Rpd3-HDAC (5) or cells treated with a HDAC inhibitor TSA (6). In addition, histone H4 tails are hyperacetylated at the active origins (7). It is generally believed that modification of the histone tail regulates accessibility of various proteins to chromatin/DNA and thereby plays important roles in biological processes involving DNA.
Stress on DNA replication causes a harmful lesion to the genome integrity and various biological pathways would be activated to cope with them. Upon inhibition of DNA replication in fission yeast, Rad3/Tel1 phosphorylate the effector kinase Cds1, which is mediated by the adaptor protein Mrc1 (8). In contrast, Rad3/Tel1 phosphorylate the effector kinase Chk1 when DNA is damaged during S/G2 phase (9).
The Cds1 kinase, in turn, prevents the onset of mitosis (10). In this study, we attempted to investigate regulation of the Cds1 kinase through deacetylation of histone H4 by Clr6-HDAC.
MATERIAL AND METHODS
Strains and media
The strains used in this study are derivative of Schizosaccharomyces pombe h− 972 and h+ 975 (11). The Δrad1, Δrad3 (our laboratory stock), cdc10-M17 (12), clr6-1, Δpst2 (13) and histone H4 mutants (14) were described previously. Standard fission yeast techniques and media were employed (15). To repress the transcription from the nmt1 promoter, cells were grown either in YES complete or EMM2 minimal medium with 4 µM thiamine. For determination of hydroxyurea (HU) sensitivity, HU was supplied to YES or EMM2 medium at 12 mM or otherwise indicated. For spot assays, exponentially growing cultures were 5-fold diluted serially, and each dilution was spotted onto agar plates containing HU and/or thiamine. For synchronization, cdc10-M17 mutant cells were first cultured at permissive temperature (26°C) before being shifted up to the restriction temperature (37°C) for 4 h. The clr6-1 mutant is temperature-sensitive for growth, but its HDAC activity is significantly reduced from 25°C to 36°C. The experiments with the clr6-1 mutant were performed at 32°C.
Gene disruption and myc8 tagging
The procedure of gene disruption was reported previously (16). For myc8 tagging, about 1-kb fragment of C-terminus region of cds1+ and mrc1+ genes were inserted into NotI site of pYC11-myc8 (generously provided by Dr Mitsuhiro Yanagida), respectively, and resultant plasmids were introduced into wild-type strains. The gene disruption and gene replacement were confirmed by Southern blotting and immunoblotting.
Cell extracts and immunoblot
Cells were collected, washed twice with distilled water, rapidly frozen by liquid nitrogen, and kept at –20°C. Cells were thawed and suspended at a concentration of 1 × 109 cells/ml in Yeast Buster Protein Extraction Reagent (Novagen), containing 1 mM PMSF and protease inhibitor cocktails (Nacalai tesque). Cells were disrupted by glass beads and centrifuged at 14 000 r.p.m. for 30 min. Surpernatants were used for immunoblot. Especially, for histone H4, cells were suspended in YBPER containing Laemmli sample buffer and boiled for 5 min before cell disruption. As primary and secondary antibodies, anti-c-myc (1:7500; Covance), anti-tetra acetylated histone H4 (1:5000; Serotec), total histone H4 (1:1000; Upstate biotechnology), anti-TAT-1 (1:5000; generously provided by Dr Andrea Baines) and goat HRP-conjugated anti-mouse (1:25000; Biosource) and antirabbit antibody (1:25000; NEN) were employed, respectively.
Immunofluorescence microscopy
Immunofluorescence was conducted as described (17). For staining acetylated histone H4 by using antitetra acetylated histone H4 (1:70; Serotec), cells were fixed with 3% formaldehyde alone for 5 min as described (18). As secondary antibody, goat Alexa 488-conjugated antirabbit (1:1000; Molecular Probes) was employed. Images were acquired with Axioplan2 (Carl Zeiss) and processed with Axiovision (Carl Zeiss). Triton extraction and immunostaining of Cds1-myc8 was performed as described (8).
Pulsed field gel electrophoresis (PFGE) analysis
PFGE analysis was conducted as described (19).
RESULTS
Acetylation in histone H4 during HU treatment
It was previously shown that when HU is added to an asynchronous culture of the wild-type fission yeast at 30°C, progression of the cell cycle is delayed in interphase for ∼6 h. Septated cells reappear after 7 h (20), indicating that slow DNA replication is eventually completed and the cells enter into mitosis under this condition. We performed a similar experiment with a temperature-sensitive allele of cdc10 (cdc10-M17), in which the cell cycle restarts from G1 phase upon the shift from the restrictive temperature to the permissive one (12). Following incubation at the restrictive temperature (36°C), the cdc10-M17 cells were released to the permissive temperature (26°C). As shown in Figure 1A, the septation index at 4 h after the shift down was ∼55% when grown in the absence of HU, indicating that the cell cycle proceeded synchronously. In contrast, the septation index reached only 15% at 8 h after the shift in the presence of HU. The result indicated that DNA replication proceeded very slowly and is completed between 6 and 8 h after the shift. This notion was supported by the measurement of the mobility of Cds1, which migrates slowly on SDS–PAGE due to phosphorylation when the DNA replication checkpoint is activated (21). In the presence of HU, Cds1 migrated slowly between 1 and 6 h after the shift (Figure 1B, upper panel). Mrc1 associates with Cds1 and is required for regulation of Cds1 by the checkpoint kinase Rad3. It is phosphorylated and accumurates upon HU treatment (8,22). As shown in Figure 1B, Mrc1 accumulated and its mobility decreased between 2 and 6 h after the shift. Examination of the two proteins indicated that the DNA replication checkpoint was activated by 6 h after the shift in most of the cells and that the cell cycle proceeded into G2/mitosis thereafter. In parallel, we monitored the level of acetylation in histone H4 by indirect immunofluorescent staining as well as western blot. While the level of the H4-acetylation was nearly constant in the cells released into the media containing no HU (Figure 1C and E), it was reduced gradually and was lowest at 6 h after the shift in the presence of HU (Figure 1D and E). Eight hours after the shift, the level of the H4-acetylation went up. We thereby concluded that acetylation of histone H4 reaches a minimum level at or around when the cell recovers from slow DNA replication under the stress imposed by HU. We also examined the level of H4 acetylation in a temperature-sensitive mutant of cdc22-M45. The cdc22+ gene encodes a ribonucleotide reductase, a major target of HU (23). As shown in Figure 1F, the level of H4 acetylation dropped in the cdc22 mutant upon the shift to the restrictive temperature. The reduction of the H4 acetylation is thus a cellular response to depletion of nucleotides, but not a side effect caused by HU. Taken together, these results would suggest that deacetylation of histone H4 may be involved in cell cycle regulation, in particular, when DNA replication slows down.
Figure 1.
Level of H4 acetylation during treatment with hydroxyurea (HU). (A) The cdc10-M17 mutant cells were released into the permissive temperature (26°C) and grown in the absence of HU (open circle) or in the presence of HU (closed square). The septation index was determined at the indicated time points. (B) The samples were prepared as in (A) and immunoblot was performed to detect Cds1 (upper panel) and Mrc1 (lower panels). The level of α-tubulin (loading control) was also determined by an antibody, TAT-1. (C–E) The samples were prepared as in (A) and indirect immunofluorescent staining (C and D) or immunoblot (E) was performed with an antitetra AcH4 antibody, which recognizes acetylated lysine residues, Lys5, -Lys8, -Lys12 and -Lys16 in histone H4 tail. (C) and (D) are images of the cdc10-M17 mutant cells grown in the absence and presence of HU, respectively. Signal intensity for each band in (E) was measured by NIH image. Relative band intensity of acetylated H4 was calculated with that of α-tubulin as a standard and shown below each lane. (F) Immunoblot was performed with the antitetra AcH4 antibody. Extracts were prepared from the cdc22 mutant cells grown at 26°C (permissive temperature) or at 36°C (restrictive temperature) for 4 h. Relative band intensity was calculated and shown as (E).
Involvement of Clr6-HDAC system
Fission yeast genome encodes three classical HDACs, namely Clr3, Clr6 and Hos2 (24). To examine which HDAC is involved in deacetylation of histone H4 in response to HU treatment, wild-type cells, in which each of the classical HDACs was overexpressed under the control of thiamine repressible nmt1 promoter, were plated on media containing HU. Overexpression of Clr6, but not Clr3 or Hos2, caused severe sensitivity to HU as well as a reduction in the level of H4-acetylation (Figure 2A). Importantly Clr3 is a histone H3 specific deacetylase, whereas Clr6 deacetylates both H4 and H3 (24). Taken together, modulation of acetylation in histone H4 tails but not H3 appears to be responsible for the cellular response to HU treatment.
Figure 2.
Overexpression of clr6+. (A) The wild-type cells transformed with the indicated plasmids were serially diluted (5-fold) and spotted on EMM2 containing HU and/or thiamine, and incubated at 32°C for 3 days (left panel). Each gene was placed under control of nmt1 promoter, which represses expression in the presence of thiamine. Cell extracts were prepared from cells transformed with the vector or a plasmid for overexpression of Clr6 and immunoblot was performed with the antitetra AcH4 antibody (right panel). Relative band intensity was calculated and shown as Figure 1E. (B) The septation index of cells overexpressing Clr6 (closed square) and the control cells (open circle) at indicated time points after addition of HU (left panel). The cells incubated with HU for 6 h were stained with DAPI and shown in the right panels. (C and D) Immunoblot was performed to detect Cds1-myc8 (C) and Mrc1-myc8 (D). Cell extracts were prepared from the wild-type cells transformed with pREP2 or pREP2-clr6. Expression of Clr6 was de-repressed when grown without thiamine (−Thi) and repressed with thiamine (+Thi). The level of α-tubulin (loading control) was monitored by immunoblot with an antibody, TAT-1.
The cells overexpressing Clr6 started septation after incubation for 5 h at 32°C in the presence of HU and the septation index reached a peak (27%) after incubation for 6 h (Figure 2B). In contrast, the septation index of the control cells was only 8% even after incubation for 7 h (Figure 2B). The two proteins, Cds1 and Mrc1, involved in the DNA-replication checkpoint, were also affected by overexpression of Clr6. Both the mobility shift of Cds1 and the increase in the level of Mrc1, which normally accompany HU treatment, were not observed in the cells overexpressing Clr6 (Figure 2C and D). These results suggested that overexpression of Clr6 results in premature entry into G2/mitosis. Microscopic observation revealed that the cells overexpressing Clr6 septated with segregated nuclei (Figure 2B, right panel). In contrast, mutants deficient in the DNA replication checkpoint septate with unsegregated nuclei, exhibiting the ‘cut’ phenotype (25), which is characteristic of the onset of mitosis before the completion of DNA replication. Although overexpression of Clr6 allows septation earlier, it seems that DNA replication is completed.
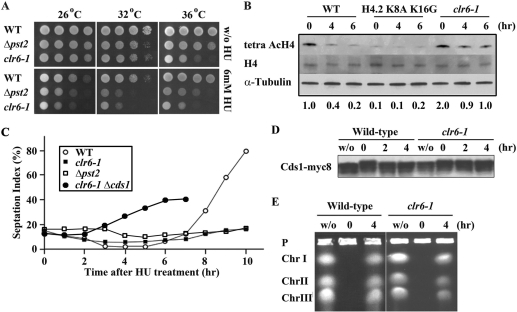
Having demonstrated that enhancement of the Clr6-HDAC activity shortens the delay induced by HU treatment, we speculated that a deficiency of the Clr6-HDAC system might result in a prolonged delay. To test this possibility, we examined response of mutants defective in Clr6-HDAC system to HU treatment. Pst2 forms a complex with Clr6 and is required for deacetylation of histone H3 and H4 (4). The spot test (Figure 3A) indicated that both the clr6-1 mutant and a strain lacking the pst2+ gene could grow at a semi-permissive temperature (32°C) comparatively with the wild-type strain, but exhibited hypersensitivity to HU. The level of H4 acetylation, measured by western blot, dramatically decreased in the wild-type strain in response to HU treatment (Figure 3B), but not in the clr6-1 mutant. Judged by the septation index (Figure 3C), the clr6-1 and Δpst2 mutants, in contrast to the wild-type strain, could not enter G2/mitosis in the continued presence of HU at 32°C. It appeared that the delay in entry into G2/mitosis was due to activated Cds1 because deletion of the cds1+ gene rescued this defect (Figure 3C). The delayed cell cycle progression was possibly because that (i) DNA replication was still incomplete in the clr6-1 mutant or (ii) the DNA replication checkpoint was prolongedly activated though DNA replication was completed. In order to further examine the defect of the clr6-1 mutant, we monitored the status of the DNA replication checkpoint after HU was removed. Following incubation in the media containing HU for 4 h, cells were released into the HU-free media. In the wild-type strain, Cds1, whose mobility was slow immediately after the release, migrated faster after incubation for 2 h in the HU-free media (Figure 3D). In a sharp contrast, the mobility of Cds1 was kept slow in the clr6-1 mutant even after incubation for 4 h in the HU-free media (Figure 3D). Under this experimental condition, DNA replication was completed in both the wild-type strain and the clr6-1 mutant. Chromosomal-sized DNAs with replicating structures, such as replication bubbles and forks, are retained in the gel loading wells during PFGE (26). As shown in Figure 3E, while chromosomal-sized DNAs could not migrate on the PFGE at 0 time (immediately after the release into the HU-free media), they could run after incubation for 4 h in the HU-free media. These results indicate that the checkpoint kinase Cds1 is maintained active in the clr6-1 mutant even after DNA replication is completed.
Figure 3.
Phenotype of the clr6-1 mutant. (A) The wild type, a strain lacking pst2+ and the clr6-1 mutant cells were serially diluted (5-fold) and spotted on YES plates with HU (upper three panels) or without HU (lower three panels) and incubated at the indicated temperature for 3 days. (B) Immunoblot was performed to detect tetra AcH4 and total H4. Cell extracts were prepared from the wild-type strain, H4.2 K8A K16G mutant, and the clr6-1 mutant grown with HU for the indicated time (0, 4 and 6 h). Intensity of each band was determined by NIH image. Relative band intensity of acetylated H4 was calculated with that of total H4 as a standard and shown below each lane. (C) The septation index of the wild-type strain (open circle), the clr6-1 (closed square) and Δpst2 (open square) and clr6-1 Δcds1 double-mutants (closed circle) at indicated time points after addition of HU at 32°C. (D) Immunoblot was performed to detect Cds1-myc8. Cell extracts were prepared from the wild-type strain and the clr6-1 mutant, which were first treated with HU for 4 h, and then washed and incubated without HU for the indicated time (0, 2 and 4 h). Extracts were also prepared from an asynchronous culture (w/o). (E) The wild-type strain and the clr6-1 mutant were grown as (D) and processed for PFGE.
Target of Clr6-HDAC system
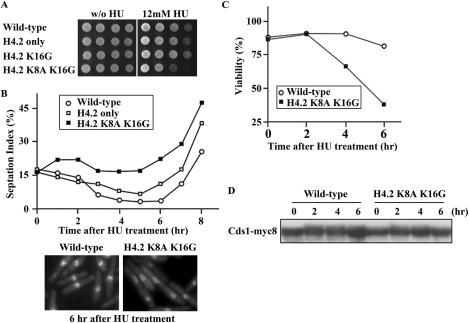
The results described above strongly suggested that the Clr6-HDAC system targets histone H4 and regulates cell cycle progression. To further confirm this notion, we examined mutants of histone H4 for the response to HU. As overexpression of Clr6-HDAC caused both reduction in the level of the H4 acetylation and premature inactivation of the DNA replication checkpoint, we anticipated that removal of acetylation sites from histone H4 tail could mimic the effect by overexpression of Clr6-HDAC. Fission yeast genome contains three genes encoding histone H4.1, H4.2 and H4.3. As shown in Figure 4A, a strain expressing only H4.2 or expressing only H4.2 in which an acetylation site (lys16) was replaced with glycine (H4.2 K16G) grew normally. However, additional replacement of an acetylation site (lys8) with alanine (H4.2 K8A K16G) caused hypersensitivity to HU. The replacement, indeed, greatly affected the level of the H4-acetylation (Figure 3B). In the continued presence of HU, the septation index of H4.2 K8A K16G was much higher than the wild-type strain (Figure 4B, upper panel). Consistently with this, the viability of H4.2 K8A K16G dropped rapidly (Figure 4C) in the media containing HU. The mobility of Cds1 also indicated that Cds1 was not phosphorylated normally in H4.2 K8A K16G (Figure 4D). Microscopic observation of H4.2 K8A K16G cells (Figure 4B, lower panel) indicated that their nuclei did not exhibit ‘cut’ phenotype. We thereby speculate that the DNA replication checkpoint is maintained normally in H4.2 K8A K16G though they septated earlier than the wild-type cells.
Figure 4.
Phenotype of H4.2 K8A K16G. (A) The wild-type strain and histone H4 mutants H4.2 only (lacking H4.1 and H4.3), H4.2 K16G (carrying Gly16 instead of Lys16 of H4.2 only), H4.2 K8A K16G (carrying Ala8 instead of Lys8 of H4.2 K16G) were serially diluted (5-fold) and spotted on YES plates without HU (left panel) or with HU (right panel) and incubated at 32°C for 3 days. (B) The septation index of histone H4 mutants, H4.2 only (open square) and H4.2 K8A K16G (closed square), and wild-type cells (open circle) at the indicated time points after incubation with HU at 32°C. The cells incubated with HU for 6 h were stained with DAPI and shown in the lower panels. (C) The viability of the wild-type strain (open circle) and H4.2 K8A K16G (closed square) at the indicated time points after incubation with HU at 32°C. (D) Immunoblot was performed to detect Cds1-myc8. Extracts were prepared from cells treated as in (B).
These results suggested that histone H4 is, at least in part, a target of Clr6-HDAC and involved in cell cycle regulation. The phenotype of H4.2 K8A K16G is, however, moderate in comparison to that observed in the cells overexpressing Clr6-HDAC. This is probably because that H4.2 K8A K16G can still be acetylated at the other two sites, lys5 and lys12, or another molecule could also be regulated by Clr6-HDAC in response to HU treatment.
Independent operation of Clr6-HDAC system
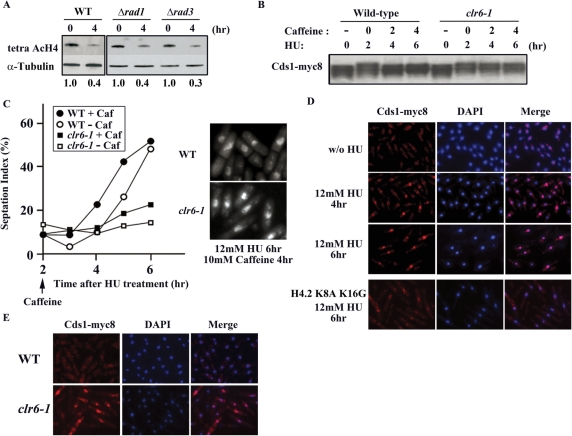
Activation of the DNA replication checkpoint is a major response to HU treatment. We addressed the question of whether the Clr6-HDAC system is operated under the control of the DNA replication checkpoint. We examined the level of the H4-acetylation in mutants, rad1 and rad3, both of which are defective in the DNA replication checkpoint. As shown in Figure 5A, the level of the H4-acetylation dropped in these mutants in response to HU treatment, just like the wild-type strains. The Clr6-HDAC system, therefore, responds to HU independently of the replication checkpoint. As shown in Figure 3E, Cds1 was still activated in the clr6-1 mutant even when HU was removed. We tested if prolonged activation of Cds1 in the clr6-1 mutant is dependent on the DNA replication checkpoint. Numerous studies demonstrated that caffeine, a potent inhibitor of ATM/ATR (fission yeast Tel1/Rad3) kinases, could override an arrest induced by the DNA replication checkpoint (25,27). We first treated fission yeast cells with HU for 2 h to activate Cds1. Caffeine was added subsequently in the continued presence of HU. After incubation with caffeine for 2 h, the mobility of Cds1 on SDS–PAGE increased in the wild-type strain (Figure 5B), indicating that caffeine abrogated the checkpoint and Cds1 was inactivated. Consistently with this, the septation index increased upon addition of caffeine in the wild-type strain (Figure 5C). In the clr6-1 mutant, on the other hand, the mobility of Cds1 remained slow even after incubation with caffeine for 6 h (Figure 5B). The septation index of the clr6-1 mutant also remained low (Figure 5C). Microscopic observation indicated that caffeine induced premature onset of mitosis and allowed septation with single nucleus in the wild-type cells, but not in the clr6-1 mutant (Figure 5C, right panel). These results indicated that inhibition of the Tel1/Rad3 kinase by caffeine was not sufficient for inactivation of Cds1 in the clr6-1 mutant. The functional Clr6-HDAC system, which is operated independently of the DNA replication checkpoint, is required for inactivation of Cds1.
Figure 5.
Effect of caffeine. (A) Immunoblot was performed to detect tetra AcH4. Extracts were prepared from cells lacking rad1+ or rad3+ after incubation with HU for the indicated time (0 and 4 h). Relative band intensities of acetylated H4 were calculated as Figure 1E. (B) Immunoblot was performed to detect Cds1-myc8. Extracts were prepared from the wild-type and the clr6-1 mutant cells. HU was added to the culture and caffeine was subsequently added after 2-h incubation with HU. (C) The septation index of the wild-type and the clr6-1 mutant cells. They were first grown with HU for 2 h and then incubated with or without caffeine. The cells incubated with HU for 6 h (and with caffeine for the last 4 h) were stained with DAPI and shown in the right panels. (D) Triton extraction and immunostaining of Cds1-myc8. The wild-type and H4.2 K8A K16G mutant cells were harvested at indicated time points after addition of HU, treated with 1% Triton, fixed with formaldehyde and subjected to immunostaining with anti-myc antibody to detect Cds1-myc8. (E) Four hours after washout of HU, the wild-type and the clr6-1 mutant cells were harvested, treated with 1% Triton, fixed with formaldehyde and subjected to immunostaining with anti-myc antibody to detect Cds1-myc8.
Status of Cds1
As our experiments demonstrated that Cds1 is still active in the clr6-1 mutant even after the completion of DNA replication (Figure 3D and E) or after treatment of caffeine (Figure 5B), we attempted to further examine the status of Cds1. It was previously shown that Mrc1, a component of the replication checkpoint that activates Cds1, remains bound to chromatin after extraction with Triton X-100 when the checkpoint is activated (8). We thereby speculated that Cds1 might also behave in a similar manner. As shown in Figure 5D, when the replication checkpoint was activated by HU, Cds1 was resistant to the Triton-extraction and remained bound to chromatin, because the level of Cds1 did not change after treatment with HU (Figure 1B). In contrast, Cds1 was washed out from chromatin after 6 h in H4.2 K8A K16G mutant (Figure 5D). These results indicated that Cds1 in an active form has an increased affinity to chromatin. We next characterized the chromatin affinity of Cds1 in the clr6-1 mutant. Cells were first treated with HU for 4 h and subsequently grown for 4 h in the HU-free media. In the wild-type strain, the Triton-extraction removed most of Cds1 from the nucleus (Figure 5E). In contrast, Cds1 remained in the nucleus in the clr6-1 mutant. Taken together, these results suggested that Cds1, which is activated by the replication checkpoint, remains bound to chromatin if Clr6-HDAC is not fully functional.
DISCUSSION
In this study, we investigated the level of H4-acetylation during the HU-induced cell cycle arrest and found a close link between the Clr6-HDAC system and regulation of the checkpoint kinase, Cds1.
Judged by indirect immuno-fluorescence staining of the nucleus in fission yeast, the global level of acetylation of histone H4 was maintained throughout the normal cell cycle, but reduced significantly when DNA replication was impeded by HU or a mutation in the cdc22+ gene encoding a ribonucleotide reductase. The H4 -acetylation is reduced to a minimum level when the cell recovers from the HU-induced cell cycle arrest (or slow DNA replication). We identified the Clr6-HDAC as a component responsible for the reduction in the level of the H4 acetylation. Analysis of the phenotypes of a clr6-deficient mutant, clr6-1, indicated that the HU-induced cell cycle arrest could not be released. In the clr6-1 mutant, although DNA replication was completed after removal of HU, the checkpoint kinase Cds1 was still activated. We also demonstrated that Cds1 was active even after treatment with caffeine, an inhibitor for ATM/ATR that is an activator of Cds1. These results would suggest that once the Cds1 kinase is activated in the clr6-1 mutant, it cannot be inactivated. The Clr6 protein forms two distinctive HDAC-complexes with Pst1 or Pst2 (4). Although we showed in this study that Clr6 plays a role in regulation of Cds1, it remains to be elusive which of the components, Pst1 or Pst2, is required for this process. As supportive evidence, we showed that cells lacking Pst2 were defective in recovery from a HU-induced arrest just like the clr6-1 mutant. Further biochemical analysis is obviously required to examine the involvement of Pst2 and/or Pst1, in regulation of Cds1.
In order to find a clue as to why Cds1 cannot be inactivated in the clr6-1 mutant, we examined localization of Cds1. Prior to incubation with the antibody for indirect immunofluorescent staining, cells were extracted with Triton X-100. If a nuclear protein is tightly bound to chromatin, it remains bound after extraction with Triton X-100 (28). Our analysis indicated that Cds1, when the replication checkpoint was activated by HU in the wild-type strain, persisted in the nucleus after the Triton-extraction. Upon removal of HU, it disappeared from the nucleus. The clr6-1 mutant, treated with HU, was subsequently grown in HU-free media for 4 h to allow DNA replication. Under this experimental condition, Cds1 still persisted in the nucleus as if the DNA replication checkpoint were active. These results would suggest that H4-deacetylation by Clr6-HDAC might, directly or indirectly, be required for removal of the Cds1 kinase from chromatin. Mrc1, which associates with Cds1 and has a DNA-binding activity (29), may possibly bridge Cds1 and chromatin. As Mrc1 did not accumulate in cells overexpressing Clr6, it is plausible that its stability and/or binding activity to DNA may be regulated depending on the level of H4-acetylation. The role of Mrc1 in localization of Cds1 should be defined in the future study.
On the basis of these results we propose several models for the role of Clr6-HDAC in regulation of Cds1. As shown in many studies (30,31) histone acetylation regulates gene expression. It is thereby possible that modification by Clr6-HDAC is required for gene expression of a component necessary for inactivation of Cds1. We, however, are not in favor of this model. The level of H4 acetylation changes globally in the nucleus during treatment with HU and is unlikely to regulate expression of a specific gene locus.
When DNA replication is impeded by HU, homologous recombination is a major pathway to overcome the replication blockage (32). Various protein complexes would be loaded on chromatin and play roles in recombination. Clr6-HDAC may be responsible for removing these complexes by deacetylating H4 before the onset of mitosis. It is also plausible that loading of newly synthesized histones on replicated DNA may require a specialized mechanism when DNA replication is impeded. The Clr6-HDAC might be involved in such a histone metabolism. In addition to the status of DNA replication, chromatin structure/environment may be an important element to regulate the checkpoint kinase, Cds1. The p38 MAP kinase pathway in fission yeast involving Spc1/Sty1 is known to be activated following addition of HU to slow DNA replication or in response to agents that cause DNA damage (33,34). We showed in this study that Clr6-HDAC is operated independently of the conventional DNA replication checkpoint. It is thus an interesting possibility that Clr6-HDAC may be under control of the MAP kinase signal cascade.
ACKNOWLEDGEMENTS
We thank O. Niwa, J. Nakayama, T. Shimura and all members of Radiation Biology Center for reagents, procedures and helpful discussions. We are particularly grateful to M. Yanagida, C. Shimoda, I. M. Hagan, K. Ekwall, R. C. Allshire, A. Baines and E. Sonoda for strains, plasmids, antibodies and procedures. This work was supported by the 21st Century COE grant from the Ministry of Education, Sports, Culture, and Technology, Japan to T.M. T.K. was partly supported by a grant to O. Niwa from Nuclear Safety Research Association, Tokyo. Funding to pay the Open Access publication charges for this article was provided by XXX.
Conflict of interest statement. None declared.
REFERENCES
- 1.Thiriet C, Hayes JJ. Chromatin in need of a fix: phosphorylation of H2AX connects chromatin to DNA repair. Mol. Cell. 2005;18:617–622. doi: 10.1016/j.molcel.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 2.Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- 3.Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 4.Nakayama J, Xiao G, Noma K, Malikzay A, Bjerling P, Ekwall K, Kobayashi R, Grewal SI. Alp13, an MRG family protein, is a component of fission yeast Clr6 histone deacetylase required for genomic integrity. EMBO J. 2003;22:2776–2787. doi: 10.1093/emboj/cdg248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M. Histone acetylation regulates the time of replication origin firing. Mol. Cell. 2002;10:1223–1233. doi: 10.1016/s1097-2765(02)00702-5. [DOI] [PubMed] [Google Scholar]
- 6.Kemp MG, Ghosh M, Liu G, Leffak M. The histone deacetylase inhibitor trichostatin A alters the pattern of DNA replication origin activity in human cells. Nucleic Acids Res. 2005;33:325–336. doi: 10.1093/nar/gki177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aggarwal BD, Calvi BR. Chromatin regulates origin activity in Drosophila follicle cells. Nature. 2004;430:372–376. doi: 10.1038/nature02694. [DOI] [PubMed] [Google Scholar]
- 8.Zhao H, Tanaka K, Nogochi E, Nogochi C, Russell P. Replication checkpoint protein Mrc1 is regulated by Rad3 and Tel1 in fission yeast. Mol. Cell. Biol. 2003;23:8395–8403. doi: 10.1128/MCB.23.22.8395-8403.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walworth NC, Bernards R. rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science. 1996;271:353–356. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- 10.Boddy MN, Furnari B, Mondesert O, Russell P. Replication checkpoint enforced by kinases Cds1 and Chk1. Science. 1998;280:909–912. doi: 10.1126/science.280.5365.909. [DOI] [PubMed] [Google Scholar]
- 11.Beach D, Rodgers L, Gould J. ran1+ controls the transition from mitotic division to meiosis in fission yeast. Curr. Genet. 1985;10:297–311. doi: 10.1007/BF00365626. [DOI] [PubMed] [Google Scholar]
- 12.Kim SM, Huberman JA. Regulation of replication timing in fission yeast. EMBO J. 2001;20:6115–6126. doi: 10.1093/emboj/20.21.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silverstein RA, Richardson W, Levin H, Allshire RC, Ekwall K. A new role for the transcriptional corepressor SIN3; regulation of centromeres. Curr. Biol. 2003;13:68–72. doi: 10.1016/s0960-9822(02)01401-x. [DOI] [PubMed] [Google Scholar]
- 14.Mellone BG, Ball L, Suka N, Grunstein MR, Partridge JF, Allshire RC. Centromere silencing and function in fission yeast is governed by the amino terminus of histone H3. Curr. Biol. 2003;13:1748–1757. doi: 10.1016/j.cub.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 15.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 16.Rothstein RJ. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 17.Ikui AE, Furuya K, Yanagida M, Matsumoto T. Control of localization of a spindle checkpoint protein, Mad2, in fission yeast. J. Cell Sci. 2002;115:1603–1610. doi: 10.1242/jcs.115.8.1603. [DOI] [PubMed] [Google Scholar]
- 18.Petersen J, Paris J, Willer M, Philippe M, Hagan IM. The S. pombe aurora-related kinase Ark1 associates with mitotic structures in a stage dependent manner and is required for chromosome segregation. J. Cell Sci. 2001;114:4371–4384. doi: 10.1242/jcs.114.24.4371. [DOI] [PubMed] [Google Scholar]
- 19.Yoshida SH, Al-Amodi H, Nakamura T, McInerny CJ, Shimoda C. The Schizosaccharomyces pombe cdt2(+) gene, a target of G1-S phase-specific transcription factor complex DSC1, is required for mitotic and premeiotic DNA replication. Genetics. 2003;164:881–893. doi: 10.1093/genetics/164.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brondello JM, Boddy MN, Furnari B, Russell P. Basis for the checkpoint signal specificity that regulates Chk1 and Cds1 protein kinases. Mol. Cell. Biol. 1999;19:4262–4269. doi: 10.1128/mcb.19.6.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka K, Boddy MN, Chen XB, McGowan CH, Russell P. Threonine-11, phosphorylated by Rad3 and atm in vitro, is required for activation of fission yeast checkpoint kinase Cds1. Mol. Cell. Biol. 2001;21:3398–3404. doi: 10.1128/MCB.21.10.3398-3404.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka K, Russell P. Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nat. Cell Biol. 2001;3:966–972. doi: 10.1038/ncb1101-966. [DOI] [PubMed] [Google Scholar]
- 23.Moynihan EB, Enoch T. Liz1p, a novel fission yeast membrane protein, is required for normal cell division when ribonucleotide reductase is inhibited. Mol. Biol. Cell. 1999;10:245–257. doi: 10.1091/mbc.10.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bjerling P, Silverstein RA, Thon G, Caudy A, Grewal S, Ekwall K. Functional divergence between histone deacetylases in fission yeast by distinct cellular localization and in vivo specificity. Mol. Cell. Biol. 2002;22:2170–2181. doi: 10.1128/MCB.22.7.2170-2181.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moser BA, Brondello JM, Baber-Furnari B, Russell P. Mechanism of caffeine-induced checkpoint override in fission yeast. Mol. Cell. Biol. 2000;20:4288–4299. doi: 10.1128/mcb.20.12.4288-4294.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hennessy KM, Lee A, Chen E, Botstein D. A group of interacting yeast DNA replication genes. Genes Dev. 1991;5:958–969. doi: 10.1101/gad.5.6.958. [DOI] [PubMed] [Google Scholar]
- 27.Wang SW, Norbury C, Harris AL, Toda T. Caffeine can override the S-M checkpoint in fission yeast. J. Cell Sci. 1999;112:927–937. doi: 10.1242/jcs.112.6.927. [DOI] [PubMed] [Google Scholar]
- 28.Kearsey SE, Montgomery S, Labib K, Lindner K. Chromatin binding of the fission yeast replication factor mcm4 occurs during anaphase and requires ORC and cdc18. EMBO J. 2000;19:1681–1690. doi: 10.1093/emboj/19.7.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao H, Russell P. DNA binding domain in the replication checkpoint protein Mrc1 of Schizosaccharomyces pombe. J. Biol. Chem. 2004;279:53023–53027. doi: 10.1074/jbc.M410449200. [DOI] [PubMed] [Google Scholar]
- 30.Durant M, Pugh BF. Genome-wide relationships between TAF1 and histone acetyltransferases in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006;26:2791–2802. doi: 10.1128/MCB.26.7.2791-2802.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabet N, Volo S, Yu C, Madigan JP, Morse RH. Genome-wide analysis of the relationship between transcriptional regulation by Rpd3p and the histone H3 and H4 amino termini in budding yeast. Mol. Cell. Biol. 2004;24:8823–8833. doi: 10.1128/MCB.24.20.8823-8833.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnaudeau C, Lundin C, Helleday T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 2001;307:1235–1245. doi: 10.1006/jmbi.2001.4564. [DOI] [PubMed] [Google Scholar]
- 33.Taricani L, Feilotter HE, Weaver C, Young PG. Expression of hsp16 in response to nucleotide depletion is regulated via the spc1 MAPK pathway in Schizosaccharomyces pombe. Nucleic Acids Res. 2001;29:3030–3040. doi: 10.1093/nar/29.14.3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taricani L, Wang TS. Rad4TopBP1 associates with Srr2, an Spc1 MAPK-regulated protein, in response to environmental stress. J. Biol. Chem. 2007;282:8793–8800. doi: 10.1074/jbc.M609282200. [DOI] [PubMed] [Google Scholar]