Abstract
DNA nonhomologous end-joining (NHEJ) and homologous recombination are two distinct pathways of DNA double-strand break repair in mammalian cells. Biochemical and genetic studies showed that DNA ends can also be joined via microhomology-mediated end joining (MHEJ), especially when proteins responsible for NHEJ, such as Ku, are reduced or absent. While it has been known that Ku-dependent NHEJ requires DNA ligase IV, it is unclear which DNA ligase(s) is required for Ku-independent MHEJ. In this study, we used a cell-free assay to determine the roles of DNA ligases I, III and IV in MHEJ and NHEJ. We found that siRNA mediated down-regulation of DNA ligase I or ligase III in human HTD114 cells led to impaired end joining that was mediated by 2-, 3- or 10-bp microhomology. In addition, nuclear extract from human fibroblasts harboring a mutation in DNA ligase I displayed reduced MHEJ activity. Furthermore, treatment of HTD114 nuclear extracts with an antibody against DNA ligase I or III also significantly reduced MHEJ. These data indicate that DNA ligases I and III are required in MHEJ. DNA ligase IV, on the contrary, is not required in MHEJ but facilitates Ku-dependent NHEJ. Therefore, MHEJ and NHEJ require different DNA ligases.
DNA double-strand breaks (DSBs) are the most serious form of DNA damage and a single unrepaired DSB can lead to cell death (1). In mammalian cells, there are at least two enzymatically distinct pathways for the repair of DSBs, homologous recombination (HR) and nonhomologous end joining (NHEJ). HR uses a homologous template (most frequently the sister chromatid) to carry out DSB repair, whereas NHEJ joins two ends without the requirement for extensive homology. Proteins known to be involved in NHEJ include DNA-PKCS (the catalytic subunit of DNA-dependent protein kinase), Ku70/Ku80 heterodimer, XRCC4 (X-ray Cross Complementing factor 4), and DNA ligase IV (2,3). However, DNA ends can also be joined via microhomologous sequences flanking the break point, especially when proteins responsible for NHEJ, such as Ku, are absent or limiting in mammalian cells (4–13). Microhomology-mediated end joining (MHEJ) is always accompanied by a deletion that spans one of the two homologous sequences and the intervening sequence, if any, and thus is a mutagenic repair pathway. Indeed, microhomologies were observed at deletion break points in the HPRT gene in primary human fibroblasts (14) and in the Aprt gene in hamster cells (15). Furthermore, translocations mediated by MHEJ were frequently detected in pre-B cell lymphomas in mouse models (16).
Little is known about the factors involved in MHEJ, but it can be assumed that the MHEJ pathway may consist of a series of steps, culminating in the sealing of DNA nicks by the action of a DNA ligase. DNA ligases catalyze the joining of nicked DNA in DNA replication, recombination and repair (17). Eukaryotic cells encode three well-characterized ATP-dependent DNA ligases, DNA ligases I, III and IV, each specializing in distinct pathways of DNA repair and replication (18). Although these DNA ligases vary in sequence and size, sequence and structural analyses have shown that they contain a common catalytic core (18). While the central core of the enzyme carries out the catalytic function of sealing nicked DNA, other domains may determine the specificity of the various ligases in different DNA metabolic reactions, e.g. by targeting ligases to different parts of the nucleus or by mediating interactions with different proteins. DNA ligase I is involved in at least two distinct processes within the nucleus: the joining of Okazaki fragments during DNA replication, and the ligation of a newly synthesized patch during base excision repair (BER) (17). DNA ligase I is recruited to sites of DNA replication by its interaction with proliferating cell nuclear antigen (PCNA) (19,20). It has been shown that the loss of its PCNA binding activity severely compromised the ability of DNA ligase I to join Okazaki fragments, and to participate in long-patch BER (21). Two isoforms of DNA ligase III resulting from alternately spliced mRNA variants have been characterized (22). Ligase IIIα is ubiquitously distributed, whereas ligase IIIβ has been detected only in testes, where it is believed to play a role in recombination during meiotic prophase (22). DNA ligase IIIα interacts with XRCC1 (X-ray Cross Complementing factor 1) via its carboxy-terminal BRCT (BRCA C-terminal) domains and functions in BER (23). DNA ligase IV is distinct from other DNA ligases in that it possesses two tandem C-terminal BRCT domains (18). This protein forms a complex with XRCC4, which appears to stabilize (24) and stimulate the overall activity of ligase IV (25). This complex further interacts with DNA-PKCS and the Ku70/Ku80 heterodimer to function in NHEJ (26).
Though many studies have revealed the role of DNA ligases in DNA repair pathways, such as NHEJ and BER, a systematic study of these ligases in the MHEJ pathway has been lacking. We recently developed a cell-free assay, with which factors modulating two end-joining pathways, i.e. Ku-dependent NHEJ and MHEJ, can be evaluated (9). We previously showed that Ku and histone H1 facilitate error-free NHEJ and inhibit MHEJ, and that FEN-1 is involved in MHEJ. In the present study, we have determined the roles of DNA ligases I, III and IV in these end-joining pathways. We found that while DNA ligase IV is required to join DNA ends in the Ku-dependent NHEJ pathway, DNA ligase I and ligase III are required in MHEJ.
MATERIALS AND METHODS
Cells and cell culture
Human cell line HTD114 was grown in DMEM containing 10% fetal bovine serum (FBS). The DNA ligase I-deficient human primary fibroblast cell line, GM16096, and the DNA ligase IV-deficient human primary fibroblast cell line, GM16088, were obtained from the NIGMS Human Genetic Mutant Cell Repository (Camden, NJ, USA), and were grown in Eagle's MEM with Earle's salts supplemented with 1× concentration of MEM nonessential amino acids, 2 mM glutamine and 10% FBS. WI38 normal diploid human fibroblasts were purchased from the American Type Culture Collection (Manassas, VA, USA) and grown in DMEM containing 10% FBS.
RNA interference
Twenty-one nucleotide RNA duplexes (most of them have a 3′-deoxy thymidine overhang) were purchased from Ambion, Inc. (Austin, TX, USA). The RNA sequences were as follows: DNA ligase I, siRNA121249, sense, 5′-GGC AUG AUC CUG AAG CAG Att-3′, antisense, 5′-UCU GCU UCA GGA UCA UGC Ctt-3′; DNA ligase III, siRNA121668, sense, 5′-CCA CAA AAA AAA UCG AGG Att-3′, antisense, 5′-UCC UCG AUU UUU UUU GUG Gtg-3′; siRNA121372, sense, 5′-CGA CCU UUU AGA CUC AAU Utt-3′, antisense, 5′-AAU UGA GUC UAA AAG GUC Gtt-3′. A nonsilencing siRNA duplex (sense, 5′-UUC UCC GAA CGU GUC ACG Utt-3′, antisense, 5′-ACG UGA CAC GUU CGG AGA Att-3′) (Qiagen, Valancia, CA, USA) was used as a nonsilencing control. Each siRNA (20 nM) was transfected into HTD114 cells with siPORT Amine reagent (Ambion, Inc.). Forty-eight hours after transfection, total RNA and nuclear proteins were extracted.
Real-time PCR for quantification of gene expression
Total RNA from cells was purified using RNeasy kits (Qiagen) and reverse transcribed using the Taqman Reverse Transcription Reagent kit (Applied Biosystems, Foster City, CA, USA). The expression of DNA ligase I, III and IV genes was measured with a quantitative real time PCR assay as previously reported (27). Expression levels were calculated as a ratio of the mRNA level for a given gene relative to the mRNA level for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in the same cDNA sample. Primer sequences are: Lig1-1F, 5′-GAA TTC TGA CGC CAA CAT GCA-3′, Lig1-1R, 5′-CCG TCT CTC TGC TGC TAT TGG A-3′; Lig3-1F, 5′-GAT CAC GTG CCA CCT ACC TTG T-3′, Lig3-1R, 5′-GGC ATA GTC CAC ACA GAA CCG T-3′; Lig4-F, 5′-CAC CTT GCG TTT TCC ACG AA-3′, Lig4-R, 5′-CAG ATG CCT TCC CCC TAA GTT G-3′. To determine whether or not a certain ligase siRNA treatment could also knock down other nontargeted ligases, we measured the expression levels of all DNA ligases in all collected RNA samples. For each RNA sample, the real-time PCR was performed in triplicate.
Nuclear extract preparation
Cells were harvested and washed with ice-cold phosphate buffered saline (PBS). Nuclear protein extracts were prepared as described (9,28). Protein concentration was determined by the Bradford assay (29).
Immunoblotting
Equivalent amount of nuclear extracts were separated with SDS–PAGE gels and transferred onto polyvinylidene difluoride membranes. Membranes were blocked at room temperature for 30 min in blocking buffer [3% nonfat dry milk in PBS containing 0.1% Tween 20 (T-PBS)] and incubated at 4°C overnight with primary antibody diluted in blocking buffer. After five washes with T-PBS, membranes were incubated at room temperature for 1 h with secondary antibody diluted in blocking buffer. Immunoblots were visualized by enhanced chemiluminescence. Primary antibodies used in this study were: mouse anti-DNA ligase I (Novus, Biologicals, Inc., Littleton, CO, USA), mouse anti-DNA ligase III (Novus, Biologicals, Inc.), goat anti-DNA ligase IV (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) and rabbit anti-β-actin (Abcam, Cambridge, MA, USA).
End-joining substrates
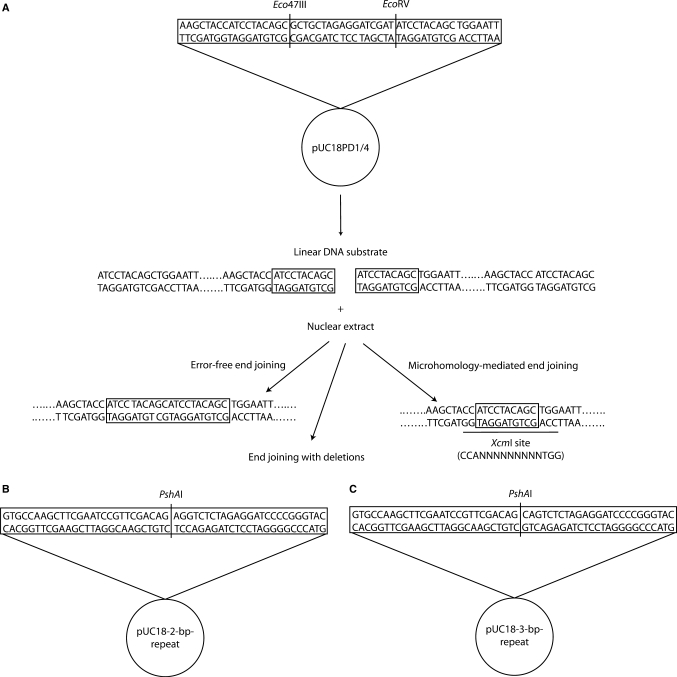
The construction of linear end-joining substrates with a 10-bp microhomology at both ends (pUC18PD1/4) was described by Liang et al. (9). After cleavage with the restriction enzymes Eco47III and EcoRV, the 2.7-kb linear DNA possesses a 10-bp direct repeat (ATCCTACAGC) at both ends (Figure 1A). Linear end-joining substrates with a 2- or 3-bp microhomology at both ends were also constructed. For the pUC18-2-bp-repeat substrates, a fragment, which was generated by annealing oligonucleotides (2-bp-repeat-F, 5′-AGC TTC GAA TCC GTT CGA CAG AGG TCT-3′, and 2-bp-repeat-R, 5′-CTA GAG ACC TCT GTC GAA CGG ATT CGA-3′), was ligated between the HindIII and XbaI sites of pUC18 (Invitrogen, Calsbad, CA, USA). After cleavage with the restriction enzyme PshAI, the 2.7-kb linear DNA possesses a 2-bp direct repeat (AG) at both ends (Figure 1B). For the pUC18-3-bp-repeat substrates, the fragment to be inserted between the HindIII and XbaI sites of pUC18 was generated by annealing oligonucleotides 3-bp repeat-F (5′-AGC TTC GAA TCC GTT CGA CAG CAG TCT-3′) and 3-bp repeat-R (5′-CTA GAG ACT GCT GTC GAA CGG ATT CGA-3′). After cleavage with the restriction enzyme PshAI, the 2.7-kb linear DNA carries a 3-bp direct repeat (CAG) at both ends (Figure 1C).
Figure 1.
DNA substrates of end-joining assays. (A) The plasmid pUC18PD1/4 was digested with Eco47III and EcoRV, resulting in a blunt-ended linear molecule with a 10-bp direct repeat (ATCCTACAGC) at each end. Deletion of one 10-bp repeat in joined DNA will produce an XcmI restriction site. This particular restriction site can not be created by other joining reactions. (B) The plasmids pUC18-2-bp-repeat and pUC18-3-bp-repeat were digested with PshAI, resulting in blunt-ended linear molecules with a 2-bp (AG) or 3-bp (CAG), respectively, direct repeat at each end.
DNA end-joining assay
The end-joining assay, which evaluates the relative contribution of MHEJ and NHEJ to DNA end joining, using nuclear extracts and DNA substrates containing direct repeats at the ends, was conducted as described (9). Briefly, linearized DNA substrates were incubated with nuclear protein extract at 14°C for 2 h. DNA products were deproteinized, purified and separated by electrophoresis through 0.6% agarose gels. The intensity of DNA bands was quantified by using Kodak Gel Logic 100 Imaging System and Kodak 1D Image Analysis Software. The overall end-joining efficiency was calculated as the percentage of end-joined product (dimer and multimers) as a fraction of total DNA in the reaction (monomer, dimer and multimers). All DNA end-joining experiments were repeated at least three times.
To study the involvement of DNA ligases I and III in MHEJ, we also treated nuclear extract from HTD114 cells (1.6 µg total protein) with dilutions of monoclonal antibody against ligase I or III (Novus Biologicals, Inc.) for 30 min on ice. Nuclear extract was then incubated with 380 ng linear DNA at 14°C for 1 h, and DNA end-joined products were analyzed with agarose gel electrophoresis.
Quantification of MHEJ
The MHEJ activity of individual protein extract was determined by two steps. First, a quantitative real-time PCR assay was used to measure the total amount of ‘head to tail’ end-joined products, including MHEJ products. Second, the fraction of end-joined products mediated by microhomology was determined by PCR amplification of the junction of rejoined ends, followed by XcmI digestion for DNA substrates with 10-bp repeats or by DNA sequencing for substrates with 2- and 3-bp repeats. The MHEJ activity of individual nuclear extracts was calculated by multiplying the values obtained in these two steps.
Step one
The primers utilized in the quantitative real-time PCR were designed using the Primer Express software (Applied Biosystems). One pair of primers amplifies a DNA fragment containing the rejoined ends. The sequences of the primers for amplifying the 10-bp-mediated products are: forward, 5′-GCC AGT GCC AAG CTA CCA TC -3′, and reverse, 5′-TGG AAT TGT GAG CGG ATA ACA AT-3′. The sequences of the primers for amplifying the 2- or 3-bp-mediated end-joined products are: forward, 5′-GTT GTA AAA CGA CGG CCA GTG-3′, and reverse, 5′-CAG CTA TGA CAT GAT TAC GAA TTC G-3′. As an internal loading control, a pair of primers were used to amplify a DNA fragment that is about 70 bp away from the junction of rejoined ends. All DNA in the end-joining reaction, including monomers and multimers, can be amplified by this pair of primers, forward, ATT CAG GCT GCG CAA CTG TT-3′, and reverse, 5′-CCC AAC TTA ATC GCC TTG CA-3′. An aliquot of the end-joined product was amplified for 40 cycles on a GeneAmp 7900HT Sequence Detection System. Syber Green dye was used for signal detection. All analyses were carried out in triplicate, and nontemplate controls and dissociation curves were used to ensure specific template amplification. For each primer pair, serial dilutions of a control DNA were used to determine a standard curve, and curves with R2 > 0.97 were then used to determine the DNA levels in individual samples.
Step two
The relative contribution of MHEJ using the 10-bp microhomology was determined by PCR amplification of the junction of rejoined ends, followed by XcmI digestion (9). If joining of the DNA ends is mediated by the 10-bp microhomology, resulting in an XcmI site (CCAN9TGG) at the junction, digestion of PCR products (∼600 bp) with XcmI would give rise to a 400- and 200-bp fragment. Restriction fragments were separated on a 1% agarose gel. The intensity of the undigested and XcmI-digested fragments was quantified using Kodak Gel Logic 100 Imaging System and Kodak 1D Image Analysis Software. The relative contribution of the 10-bp microhomology-mediated end-joining of the DNA substrate was calculated as the percentage of the XcmI-digested fragments of total PCR products (sum of the undigested and XcmI-digested fragments).
The relative contribution of MHEJ using the 2- or 3-bp microhomology was determined by sequencing the junction of rejoined ends. After PCR amplification of the junction, the PCR products were purified with the QIAquick PCR purification kit (Qiagen). The purified DNA was cloned into the TOPO TA cloning vector pCR4-TOPO (Invitrogen) according to the manufacturer's recommendations. Individual clones were then sequenced on a Beckman CEQ8000 DNA sequencer.
RESULTS
We previously established an in vitro assay to evaluate factors involved in MHEJ pathway by using nuclear extracts and linear DNA substrates containing 10-bp repeats at the ends (9). We found that the occurrence of MHEJ in our experimental setting is determined by the relative abundance of nuclear proteins. At low DNA/protein ratios, a Ku-dependent end-joining mechanism predominated over MHEJ. As the DNA/protein ratio increased, end-joining shifted toward MHEJ (9). The present study was conducted to determine the roles of DNA ligases in MHEJ, by using cell extracts that have reduced, defective or inactive DNA ligases.
Ligase I is required for 10-bp MHEJ
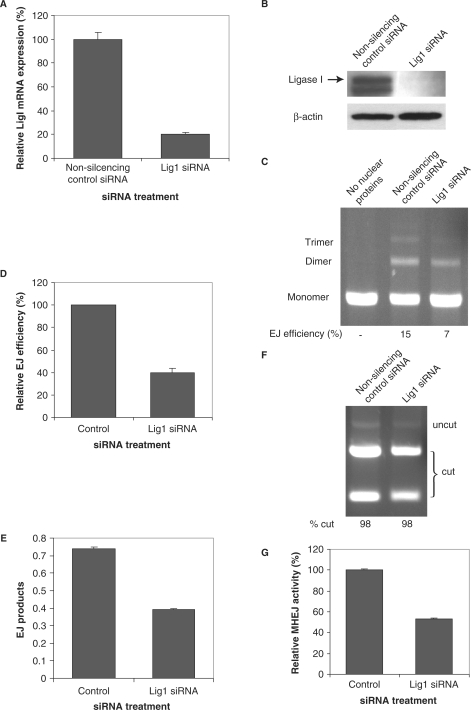
Using siRNA 121249, we were able to decrease the mRNA level of ligase I in HTD114 cells by 5-fold, as compared with a nonsilencing control siRNA (Figure 2A). Meanwhile, this siRNA did not change the mRNA level of DNA ligases III or ligase IV (data not shown). siRNA 121249 also resulted in no detectable ligase I protein (Figure 2B).
Figure 2.
Down-regulation of DNA ligase I leads to reduced MHEJ activity. HTD114 cells were transfected with siRNA oligonucleotides against DNA ligase I (siRNA 121249). Forty-eight hours after transfection, total RNA and nuclear extracts were prepared. RNA samples were reverse transcribed to cDNA, followed by real time-PCR, repeated three times for each RNA sample, to determine the expression level of ligase I (A). Data represent the mean and the SEM. Nuclear extracts were analyzed by immunoblotting with the indicated antibodies (B). (C) End-joining activity affected by the depletion of Lig1. Linearized pUC18PD1/4 DNA (300 ng) was incubated without (lane 1) or with 1 μg nuclear protein extracts from HTD114 transfected with the indicated siRNA oligonucleotides (lanes 2 and 3). The end-joined products were separated by electrophoresis in an agarose gel and the efficiency of end joining was calculated as the percentage of end-joined products (dimer and multimers) in total DNA in the reaction (monomer, dimer and multimers). (D) A summary of three independent end-joining experiments performed as in C (above). Data represent the mean and SEM. (E) The amount of end-joined products from individual reactions shown in C (above) was determined, in triplicate, with a quantitative real time-PCR assay as described in Materials and methods section. The bar represents the mean and the error bar is SEM. (F) End-joined products from reactions shown in C were PCR amplified and subsequently digested with XcmI. The relative contribution of 10-bp MHEJ to DNA end joining was calculated as the percentage of the XcmI-digested fragments in total PCR products (sum of the undigested and XcmI-digested fragments). (G) Quantification of MHEJ activity. The MHEJ activity was calculated by multiplying the amount of end-joined products measured from (E) with the relative contribution of MHEJ obtained in (F).
To test whether or not the decrease in DNA ligase I affects MHEJ, we incubated 300 ng linear DNA substrates containing 10-bp repeats at the ends with 1 μg nuclear extracts (high DNA/protein ratio) prepared from HTD114 cells that were transfected with siRNA 121249. As shown in Figure 2C and D, siRNA 121249 significantly decreased the overall end-joining efficiency to 40%—a 2.5-fold reduction, of the control (Figure 2D, end-joined products were the result of the ‘head to tail’, ‘head to head’ or ‘tail to tail’ end joining).
We performed a 2-step assay to quantify how much of the decreased end joining was attributed to MHEJ. Since the 10-bp microhomology only mediates the ‘head to tail’ end joining, we first measured the total amount of ‘head to tail’ end-joined products using quantitative real time PCR. As shown in Figure 2E, siRNA 121249 decreased the amount of the ‘head to tail’ end-joined products to about 53% of the control. To determine the fraction of the ‘head to tail’ end-joined products contributed by MHEJ using the 10-bp repeat, we PCR amplified the ligated DNA products, using primers flanking the end-joined junction, and subsequently digested the PCR products with XcmI. Rejoining of DNA substrates catalyzed by MHEJ using the 10-bp repeat generates an XcmI restriction enzyme site at the joint. No other joining reaction could create such a restriction site (9) (Figure 1A). As shown in Figure 2F, at the 300 ng DNA/1 μg protein ratio, the relative intensity of cleaved PCR fragments by XcmI digestion is 98% in both end-joining reactions. By multiplying the values obtained from step 1 (Figure 2E) with the relative contribution of MHEJ in the corresponding end-joining reaction as shown in Figure 2F, we were able to compare the relative MHEJ activity between individual extracts. As shown in Figure 2G, siRNA 121249 decreased the MHEJ activity to 53% of the control.
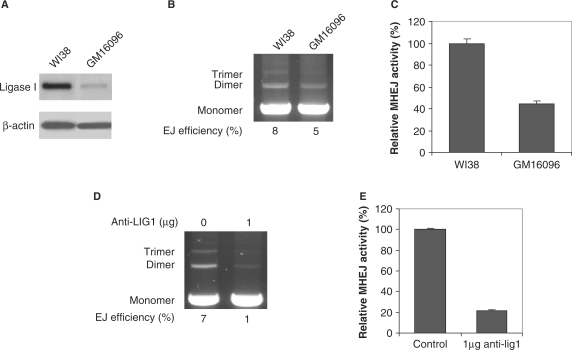
GM16096 fibroblast cells, derived from a compound human heterozygote for ligase I, carry two different missense mutations. The paternal allele showed a substitution of lysine for glutamic acid, resulting from G-to-A transition and the maternal allele carried an Arg771-to-Trp mutation, the result of a C-to-T transition (30). Western blot analysis shows that GM16096 has a significantly lower level of ligase I than normal human fibroblast WI38 cells (Figure 3A). We extracted nuclear proteins from GM16096 and WI38 and incubated 1 μg extract with 500 ng DNA substrate. As shown in Figure 3B, GM16096 had a reduced overall end-joining efficiency. MHEJ using the 10-bp repeat, the predominant pathway of end joining, was reduced by 2.5-fold in the extract prepared from GM16096 (Figure 3C), as compared to WI38. Preincubation of 1.6 μg nuclear protein extract prepared from HTD114 cells with 1 μg monoclonal antibody raised against ligase I before initiating the end-joining assay reduced the overall end-joining efficiency by 7-fold, 1% versus 7% in the control (Figure 3D), and the MHEJ activity by about 5-fold, as compared to the control, (Figure 3E). Taken together, these data clearly demonstrate that DNA ligase I is required for the 10-bp repeat-mediated end joining.
Figure 3.
Mutation in the ligase I gene results in decreased MHEJ. (A) Western blot analysis with the antibody against DNA ligase I. GM16096 cells are primary human fibroblast cells that carry two different missense mutations in the ligase I gene. WI38 cells are primary normal human fibroblasts. (B) End-joining activities. End-joining assays were carried out using 1 μg nuclear extract prepared from GM16096 or WI38 fibroblast cells. (C) Quantification of MHEJ activity. (D) Inhibition of end-joining by anti-Ligase 1 antibodies. Nuclear extract from HTD114 cells (1.6 µg total protein) was incubated for 30 min on ice with 1 µg antibody against human DNA ligase I, then incubated with 400 ng linear DNA at 14°C. After 1 h incubation, DNA products were analyzed by agarose gel electrophoresis. (E) Quantification of MHEJ affected by anti-ligase 1 antibodies.
DNA ligase III is also required in 10-bp MHEJ
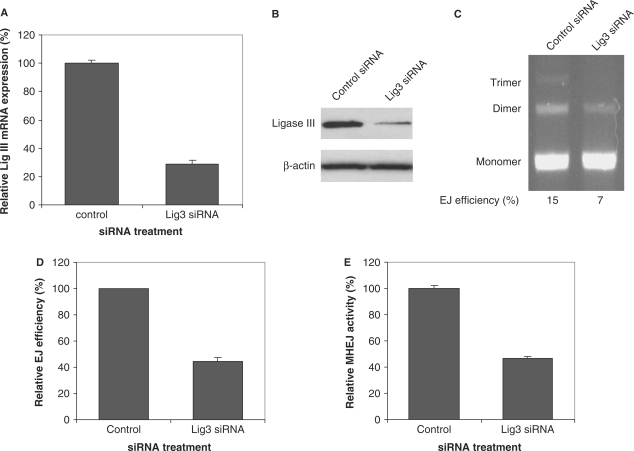
siRNA 121668, which targets against DNA ligase III, significantly reduced the mRNA level and the protein level of ligase III (Figure 4A and B). At high DNA/protein (300 ng DNA/1 μg protein) ratio, DNA end-joining activity was significantly decreased in the nuclear extract prepared from cells treated with siRNA 121668 (Figure 4C and D), indicating that reduction in ligase III compromises the end-joining activity. Further quantification of the 10-bp repeat-mediated MHEJ revealed that reduction in ligase III resulted in decreased MHEJ (Figure 4E), suggesting that the decrease in overall end-joining efficiency was caused by a decrease in MHEJ.
Figure 4.
Down-regulation of DNA ligase III results in decreased MHEJ activity. HTD114 cells were transfected with nontargeted siRNA (control) or siRNA oligonucleotides against ligase III (siRNA 121668). Forty-eight hours after transfection, total RNA and nuclear extracts were prepared. Expression of ligase III at mRNA level and protein level was determined by quantitative real time-PCR (A) and by immunoblotting analysis (B), respectively. (C) End-joining activity affected by the reduction of ligase III. Linearized pUC18PD1/4 DNA (300 ng) was incubated with 1 μg nuclear protein extracts from HTD114 transfected with nontargeted siRNA (lane 1) or the ligase III siRNA 121668 (lane 2). (D) A summary of three independent end-joining reactions as performed in (C). (E) Quantification of MHEJ.
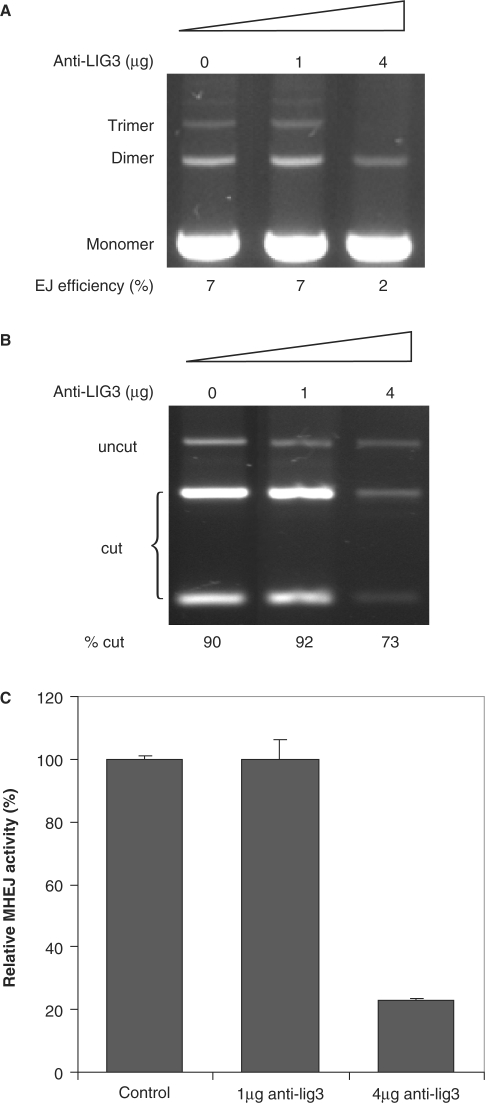
To substantiate the above finding, we preincubated 1.6 μg nuclear protein extract prepared from HTD114 cells with a monoclonal antibody raised against ligase III before initiating the end-joining assay. While the addition of 1 μg anti-ligase III antibody had no effect on the end-joining efficiency, addition of 4 μg antibody resulted in a 3.5-fold reduction, 2% versus 7% in the control, in the end-joining efficiency (Figure 5A). Addition of 4 μg antibody against mouse exonuclease 1, on the other hand, did not affect the end-joining efficiency. XcmI digestion of PCR products of joined DNA showed that the addition of 4 μg ligase III antibody reduced the relative activity of MHEJ using the 10-bp repeat, 73% versus 90% in control (Figure 5B). Together with the result of quantitative real-time PCR amplification of end-joined products, these data indicate that application of 4 μg anti-ligase III antibody caused a 5-fold decrease in 10-bp repeat-mediated MHEJ (Figure 5C). Collectively, these data demonstrate that DNA ligase III is required in MHEJ.
Figure 5.
Requirement of DNA ligase III in MHEJ. (A) The effect of anti-ligase III antibodies. Nuclear extract from HTD114 cells (1.6 µg total protein) was incubated for 30 min on ice with the antibody against human DNA ligase III, then incubated with 400 ng linear DNA at 14°C. After 1 h incubation, DNA products were analyzed by agarose gel electrophoresis. (B) PCR products of end-joined products shown in (A) were digested with XcmI. (C) Quantification of MHEJ.
DNA ligase IV is not required in 10-bp MHEJ but facilitates Ku-dependent NHEJ
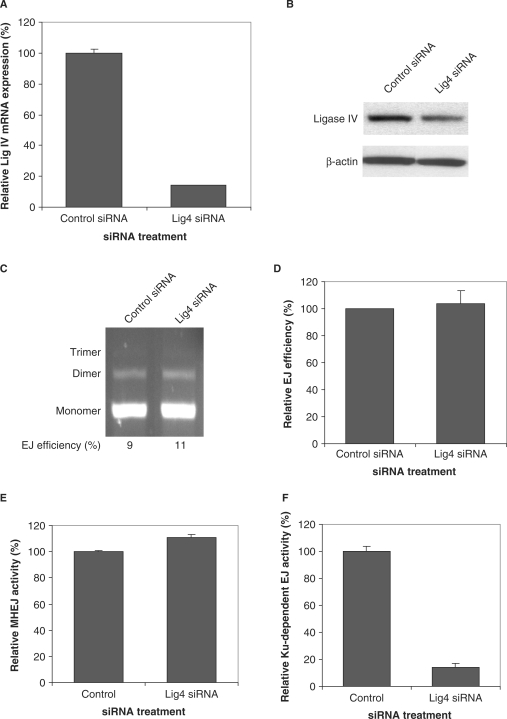
By using siRNA 121372 that targets ligase IV, we were able to reduce the expression of ligase IV at the mRNA level and at the protein level in HTD114 cells (Figure 6A and B). Incubation of 300 ng DNA substrate with 1 μg nuclear extract isolated from siRNA 121372-transfected HTD114 cells showed that knockdown of ligase IV slightly increased the MHEJ activity (Figure 6C–E).
Figure 6.
DNA ligase IV is not required for MHEJ. (A) siRNA-mediated down-regulation of DNA ligase IV in HTD114 cells. Expression of ligase IV at the mRNA level was determined by quantitative real time-PCR. (B) Expression of ligase IV at the protein level, as determined by the western blot assay. (C) End-joining activity in nuclear extract with reduced ligase 4. Linearized DNA substrate (300 ng) was incubated with 1 μg nuclear protein extracts from HTD114 transfected with nontargeted siRNA (lane 1) or the ligase IV siRNA 121372 (lane 2). (D) A summary of three independent end-joining reactions as performed in C. (E) Quantification of MHEJ. (F) DNA ligase IV is required for the Ku-dependent NHEJ pathway. End-joining reactions were carried out by incubating 10 ng DNA substrates with 8 μg nuclear extracts prepared from HTD114 cells treated with nontargeted siRNA, or siRNA 121372 against ligase IV. PCR products of end-joined products were digested with XcmI. The uncut band represents the Ku-dependent NHEJ pathway. NHEJ activity was calculated by multiplying the amount of end-joined products measured by real time PCR with the relative contribution of NHEJ obtained from (A).
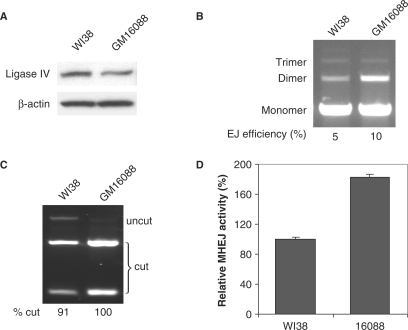
We also isolated nuclear extract from human fibroblast cells, GM16088, which harbor an Arg278 to His substitution in the ligase IV gene product, leading to a decreased level of ligase IV proteins (Figure 7A). The Arg278 residue is located within a highly conserved motif encompassing the active site, and the substitution was shown to significantly impair ligase IV function (31). Interestingly, end-joining assay showed that the nuclear extract (1 μg) from GM16088 cells has a 2-fold greater efficiency than that from WI38 cells to ligate DNA substrates (500 ng), 10 and 5%, respectively (Figure 7B). XcmI digestion of PCR products of joined DNA revealed that the MHEJ pathway mediated by the 10-bp repeat was more dominant in GM16088 than in WI38, 100% versus 91% (Figure 7C). Furthermore, the overall MHEJ activity in the nuclear extract of ligase IV-deficient GM16088 cells was measured to be 1.8-fold greater than that of WI38 cells (Figure 7D). Thus, two lines of evidence implicated DNA ligase IV in the negative regulation of the MHEJ pathway.
Figure 7.
Mutation in the ligase IV gene leads to increased MHEJ. (A) Western blot analysis with the antibody against DNA ligase IV. (B) End-joining activities. Nuclear extract (1 μg) from GM16088 cells, which carry a mutation in ligase IV gene, was incubated with the DNA substrate (500 ng). (C) PCR products of end-joined products shown in (A) were digested with XcmI. (D) Quantification of MHEJ.
DNA ligase IV is known to play an important role in DNA-PKcs/Ku-dependent NHEJ. Therefore, it is not unexpected that a decrease in ligase IV expression greatly decreased the Ku-dependent end-joining activity (Figure 6F), which was quantified by using our DNA end-joining at a low DNA/protein ratio (9). However, it is worth noting that when the function of ligase IV is compromised, MHEJ activity is increased (Figures 6 and 7). We can conclude that while DNA ligase IV may facilitate Ku-dependent end joining, it is not required in MHEJ.
Roles of DNA ligases I, III and IV in 2- and 3-bp MHEJ
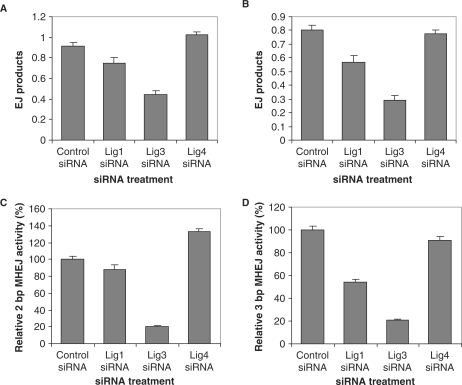
The results presented earlier showed that DNA ligases I and III, but not ligase IV, are required in 10-bp microhomology-mediated end joining. We further investigated their involvement in DNA end joining mediated by shorter repeat sequences, i.e. 2- or 3-bp repeats. We made two plasmid constructs and used PshAI to obtain linearized DNA substrates, which have 2- or 3-bp direct repeats at the ends (Figure 1B and C). At a high DNA/protein ratio (300/1 ng/μg), with nuclear extracts prepared from HTD114 cells, DNA substrates with 2- or 3-bp repeats were not rejoined as efficiently as those with the 10-bp repeats (data not shown). With the nuclear extracts from HTD114 cells in which DNA ligase I were depleted by siRNA 121249, the efficiency of rejoining the substrates with 2- or 3-bp repeats was decreased, as compared with the control (Figure 8A and B). A greater decrease was observed in HTD114 cells treated with siRNA 121668 against ligase III. With the siRNA 121668 treatment, the efficiencies of rejoining the substrates with 2- and 3-bp repeats were about 49 and 37%, respectively, of the control (Figure 8A and B). In contrast, the siRNA treatment against ligase IV (siRNA 121372) had little effect on the efficiencies of rejoining the substrates with 2- and 3-bp repeats, as compared with the control (Figure 8A and B).
Figure 8.
The involvement of DNA ligases I, III and IV in 2- and 3-bp MHEJ. End-joining reactions were carried out by incubating 300 ng linearized pUC18-2-bp-repeat (containing a 2-bp, AG repeat at each end, A) or pUC18-3-bp-repeat (containing a 3-bp, CAG repeat at each end, B) with nuclear extracts (1 μg) prepared from HTD114 cells treated with nontargeted siRNA, or siRNA against ligase I (121248), III (121668), or IV (121372). The amount of end-joined products from individual reactions was determined with a quantitative real time-PCR assay as described in Materials and methods section. The bar represents the mean end-joining efficiency of three independent reactions and the error bar is SEM. The activity of MHEJ using 2-bp (C) or 3-bp (D) microhomology was determined by multiplying the end joining efficiency (A and B) with the fraction of end-joined products mediated by 2- or 3-bp microhomology (Table 2).
DNA sequencing results of the end-joined junctions, shown in Table 1, indicated that majority of end-joining events were mediated by 2- or 3-bp repeats in the controls, accounting for 63% (12/19) or 62% (13/21), respectively, of overall end-joining events. Knockdown of ligase I or IV did not significantly change the fraction of such end-joining events (all P > 0.1, chi-squared test, Table 2). However, knockdown of ligase III, by treatment with siRNA 121668, resulted in a remarkable decline in the contribution of 2- and 3-bp MHEJ events, to 26 and 32%, respectively, of the total end-joining events (both P < 0.05, Table 2).
Table 1.
Sequences of junctions of end-joined productsa
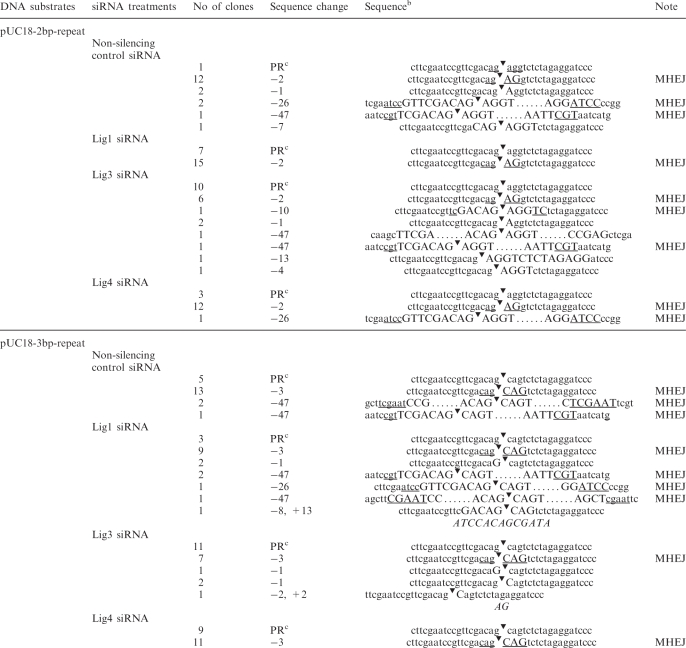 |
aNuclear extract was prepared from HTD114 cells treated with different siRNAs.
bThe deleted sequences are indicated by capital letters and the flanking sequences are in lower case. Direct repeats are underlined. The symbol, ‘▾’, denotes the ends of two joined DNA molecules. Italic letters represent the inserted sequence.
cPR, perfect rejoining.
Table 2.
Fractions of end-joined products mediated by 2-bp (AG) or 3-bp (CAG) repeats at the ends of substrates
| DNA substrates | siRNA treatments | No. of EJ products used repeats at the ends of substrates | No. of total EJ products sequenced | Percentage | P-valuea |
|---|---|---|---|---|---|
| pUC18-2bp-repeat | |||||
| Non-silencing control siRNA | 12 | 19 | 63 | ||
| Lig1 siRNA | 15 | 22 | 68 | 0.735 | |
| Lig3 siRNA | 6 | 23 | 26 | 0.016 | |
| Lig4 siRNA | 12 | 16 | 75 | 0.452 | |
| pUC18-3bp-repeat | |||||
| Non-silencing control siRNA | 13 | 21 | 62 | ||
| Lig1 siRNA | 9 | 19 | 47 | 0.356 | |
| Lig3 siRNA | 7 | 22 | 32 | 0.048 | |
| Lig4 siRNA | 11 | 20 | 55 | 0.654 | |
aComparisons of the fraction of end joined products mediated by 2- or 3-bp repeats at the ends of substrates in HTD114 nuclear extracts with siRNA treatments against different ligases to the control siRNA were done with contingency tables. Significant levels were determined with chi-squared test.
By multiplying the end-joining efficiency (Figure 8A and B) with the fraction of end-joined products mediated by 2- or 3-bp microhomology (Table 2), we were able to determine the effect of siRNA against DNA ligases I, III or IV on 2- and 3-bp MHEJ. As shown in Figure 8C, while siRNA 121249, which depletes ligase I, decreased the activity of 2-bp MHEJ to 88% of the control, it reduced the activity of 3-bp MHEJ to 54% of the control, suggesting that DNA ligase I plays a more important role in 3-bp MHEJ than in 2-bp MHEJ. The siRNA 121668, which reduces ligase III, had a greater inhibitory effect on 2- and 3-bp MHEJ than siRNA 121249. The activities of both 2- and 3-bp MHEJ were only about 20% of the control (Figure 8C and D). Interestingly, siRNA 121372, which reduces ligase IV, increased the activity of 2-bp MHEJ to 133% of the control (Figure 8C), consistent with its effect on 10-bp MHEJ (Figures 6 and 7).
DISCUSSION
The presence of multiple DNA ligases in eukaryotic cells suggests that these enzymes may each have specific functions. This notion is supported by findings showing that mammalian cell lines deficient in either DNA ligase I, III or IV exhibit different phenotypes (32). We showed here that human DNA ligases I and III contribute to microhomology-mediated end joining (MHEJ). The supporting evidence includes: first, siRNA-mediated down-regulation of DNA ligase I or III in human HTD114 cells leads to impaired joining of DNA ends mediated by a 10-, 2- or 3-bp microhomology (Figures 2, 4 and 8); second, GM16096, a cell line derived from a patient with immunodeficiency, increased sensitivity to DNA-damaging agents, and delayed development due to loss of DNA ligase I function, showed decreased level of MHEJ (Figure 3); third, treatment of HTD114 nuclear extracts with antiserum against DNA ligase I or III significantly reduced MHEJ (Figures 3 and 5). We have also shown that human DNA ligase IV is not required in MHEJ (Figures 6–8). In addition, reduction of ligase IV significantly decreased Ku-dependent NHEJ (Figure 6). Taken together, these findings demonstrate that human DNA ligases I, III and IV each play a different role in MHEJ and Ku-dependent NHEJ, the two end-joining pathways for DNA DSBs.
DNA ligase I functions in DNA replication to join Okazaki fragments during lagging-strand DNA synthesis and in DNA BER to seal single-strand nicks (30). Our data indicate that DNA ligase I is also involved in end joining of DNA DSBs, especially in MHEJ. Although ligase I interacts with several different proteins in DNA replication and BER (17,32), it remains unclear whether or not it acts alone in MHEJ. It has been shown that the amino terminus of the DNA ligase I protein has a proliferating cell nuclear antigen (PCNA)-binding motif that is required for the loading of the DNA ligase I protein to replication foci within the nuclei of S-phase cells (20). Moreover, the PCNA-binding motif is required for the efficient joining of Okazaki fragments and completion of long patch BER in cell extract assays (21). Accumulating evidence suggests that in addition to DNA ligase I, PCNA interacts with DNA polymerases δ ε and β replication factor C (RFC), and a DNA structure-specific endonuclease (FEN-1), and coordinates their activities by forming complexes that are involved in several DNA metabolic pathways, including the processing of Okazaki fragments, and the repair of DNA base damage and single-strand breaks (SSBs) (32–34). Therefore, it is reasonable to speculate that PCNA coordinates the involvement of DNA ligase I and other factors, such as FEN-1, in MHEJ. Indeed, it has been reported that FEN-1 is required for the MHEJ pathway (9,35). It is possible that FEN-1 is responsible for the removal of single-stranded flaps formed from broken DNA ends during MHEJ, and then ligase I seals the remaining nick. All these processes are probably mediated by the protein–protein interaction of FEN-1 and ligase I with PCNA.
Besides its critical role in the repair of damaged bases and SSBs (32), evidence from several studies suggests that DNA ligase III is also involved in DSB repair through an alternative route of DSB repair that complement the DNA-PK/XRCC4/ligase IV-dependent NHEJ (35–37). In line with these findings, we have shown that DNA ligase III is required for MHEJ. While ligase III exists in a tight complex with XRCC1 (23) in BER to repair DNA base damage and DNA SSBs (38,39), it was also reported that the Ku-independent end-joining, which may be mediated by microhomology sequences, required the synapsis activity of poly(ADP-ribose) polymerase 1 (PARP-1) and the ligation activity of the XRCC1-DNA ligase III complex (36), implying that ligase III may also work together with other factors in MHEJ. However, amongst DNA ligases, ligase III is unique in having a zinc-finger motif at the N-terminus (40), enabling its recruitment to DNA strand breaks (41). Furthermore, Taylor et al. (42) reported that the zinc-finger motif confers upon ligase III the ability to bind DNA duplexes and stimulates intermolecular end joining between DNA duplexes. Therefore, the possibility that ligase III functions alone in DNA end joining can not be excluded.
Like DNA ligase III that forms a complex with XRCC1, DNA ligase IV is tightly associated with XRCC4, upon which it depends for stability and activity (24,25,43). Strikingly, ligase IV–XRCC4 complex functions in coupling with Ku heterodimers and DNA PKCS in NHEJ, but not in other DNA repair pathways (44). In line with this notion, we here demonstrated that DNA ligase IV facilitated Ku-dependent end joining (Figure 6), but was not required for MHEJ (Figures 6 and 7). Furthermore, when ligase IV was absent, the activity of MHEJ in the nuclear extract was increased, suggesting that DNA ligase IV may negatively regulate the MHEJ pathway. It is not clear how ligase IV inhibits MHEJ. One possibility is that the Ku-ligase IV–XRCC4-dependent NHEJ pathway, which is initiated by binding of Ku proteins to DNA ends, may compete with the MHEJ pathway for binding to and processing DNA ends (9). Once the DNA ends are ligated by DNA ligase IV/XRCC4 complex, Ku proteins presumably become dissociated from DNA, and bind to other DNA ends to start another round of NHEJ reaction. Lack of DNA ligase IV would lead to an incomplete NHEJ reaction in which Ku proteins cannot be released from DNA ends. Due to lack of competition caused by a shortage of recycled Ku proteins, DNA ends would have a greater chance to be exposed to nuclease attack and to be joined by the alternative MHEJ pathway. In addition, there is growing evidence showing that DNA ligase IV may coordinate DNA processing activities that are critical for both NHEJ and MHEJ. For instance, Ma et al. (45) showed that there are conserved physical and functional interactions between DNA polymerase (pol) X family members (pol μ and pol λ) and DNA ligase IV that coordinate gap-filling DNA synthesis and DNA ligation in NHEJ. Notably, in yeast, DNA ligase IV physically and functionally interacts with Rad27 (FEN-1 in mammalian cells) to process and join DNA molecules (46). Therefore, while ligase IV, by interacting with other DNA processing proteins, may facilitate the Ku-dependent NHEJ pathway, it may inhibit MHEJ as a consequence of the competition for components common to both pathways.
Overall, our work showed that human DNA ligase I and ligase III, which are involved in long patch BER and short patch BER, respectively, to repair DNA base damage and SSBs, are also critical for the repair of DSBs by MHEJ. It should also be noted that the requirement of ligase I and III seems to vary depending on the length of microhomology sequences. While a reduction in ligase III affects MHEJ using both short and long microhomology, a reduction in ligase I had only slightly affected MHEJ using 2-bp repeats, though it greatly decreased the MHEJ using 10-bp repeats. Since the 2- and 3-bp substrates are much more likely to occur in vivo, ligase III is probably the primary ligase contributing to MHEJ. This notion is in accordance with previously published observations (35–37). Furthermore, our observations also suggest that DNA ligase I and ligase III may function in MHEJ through two independent pathways. In keeping up with this notion, protein partners of DNA ligases I and III, which are required for their stability, DNA-binding recruitment and catalytic activity, have never been shown to overlap. Although little is known about whether or not other factors may be involved in MHEJ, further studies of factors involved in the repair of DNA base damage and SSBs by using our end-joining assay may shed more light on the mechanisms of DNA DSB repair.
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health (ES011633 and P30ES05022), the National Aeronautics and Space Administration (NNG05GN24G), and New Jersey Stem Cell Research grants from New Jersey Commission on Science and Technology (06-2042-014-85 and 07-2042-014-90). Funding to pay the Open Access publication charges for this article was provided by the New Jersey Stem Cell Research grant (07-2042-014-90).
Conflict of interest statement. None declared.
REFERENCES
- 1.Rich T, Allen RL, Wyllie AH. Defying death after DNA damage. Nature. 2000;407:777–783. doi: 10.1038/35037717. [DOI] [PubMed] [Google Scholar]
- 2.Jackson SP. Sensing and repairing DNA double-strand breaks. Carcinogenesis. 2002;23:687–696. doi: 10.1093/carcin/23.5.687. [DOI] [PubMed] [Google Scholar]
- 3.Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell. Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 4.Chen S, Inamdar KV, Pfeiffer P, Feldmann E, Hannah MF, Yu Y, Lee JW, Zhou T, Lees-Miller SP, Povirk LF. Accurate in vitro end joining of a DNA double strand break with partially cohesive 3'-overhangs and 3'-phosphoglycolate termini: effect of Ku on repair fidelity. J. Biol. Chem. 2001;276:24323–24330. doi: 10.1074/jbc.M010544200. [DOI] [PubMed] [Google Scholar]
- 5.DiBiase SJ, Zeng ZC, Chen R, Hyslop T, Curran W.J., Jr, Iliakis G. DNA-dependent protein kinase stimulates an independently active, nonhomologous, end-joining apparatus. Cancer Res. 2000;60:1245–1253. [PubMed] [Google Scholar]
- 6.Feldmann E, Schmiemann V, Goedecke W, Reichenberger S, Pfeiffer P. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 2000;28:2585–2596. doi: 10.1093/nar/28.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kabotyanski EB, Gomelsky L, Han JO, Stamato TD, Roth DB. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 1998;26:5333–5342. doi: 10.1093/nar/26.23.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang F, Jasin M. Ku80-deficient cells exhibit excess degradation of extrachromosomal DNA. J. Biol. Chem. 1996;271:14405–14411. doi: 10.1074/jbc.271.24.14405. [DOI] [PubMed] [Google Scholar]
- 9.Liang L, Deng L, Chen Y, Li GC, Shao C, Tischfield JA. Modulation of DNA end joining by nuclear proteins. J. Biol. Chem. 2005;280:31442–31449. doi: 10.1074/jbc.M503776200. [DOI] [PubMed] [Google Scholar]
- 10.Perrault R, Wang H, Wang M, Rosidi B, Iliakis G. Backup pathways of NHEJ are suppressed by DNA-PK. J. Cell. Biochem. 2004;92:781–794. doi: 10.1002/jcb.20104. [DOI] [PubMed] [Google Scholar]
- 11.Smith J, Riballo E, Kysela B, Baldeyron C, Manolis K, Masson C, Lieber MR, Papadopoulo D, Jeggo P. Impact of DNA ligase IV on the fidelity of end joining in human cells. Nucleic Acids Res. 2003;31:2157–2167. doi: 10.1093/nar/gkg317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, Perrault AR, Takeda Y, Qin W, Wang H, Iliakis G. Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic Acids Res. 2003;31:5377–5388. doi: 10.1093/nar/gkg728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang HC, Zeng ZC, Perrault AR, Cheng XB, Qin W, Iliakis G. Genetic evidence for the involvement of DNA ligase IV in the DNA-PK-dependent pathway of non-homologous end joining in mammalian cells. Nucleic Acids Res. 2001;29:1653–1660. doi: 10.1093/nar/29.8.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris T, Thacker J. Formation of large deletions by illegitimate recombination in the HPRT gene of primary human fibroblasts. Proc. Natl Acad. Sci. USA. 1993;90:1392–1396. doi: 10.1073/pnas.90.4.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miles C, Meuth M. DNA sequence determination of gamma-radiation-induced mutations of the hamster aprt locus. Mutat. Res. 1989;227:97–102. doi: 10.1016/0165-7992(89)90004-3. [DOI] [PubMed] [Google Scholar]
- 16.Zhu C, Mills KD, Ferguson DO, Lee C, Manis J, Fleming J, Gao Y, Morton CC, Alt FW. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 17.Timson DJ, Singleton MR, Wigley DB. DNA ligases in the repair and replication of DNA. Mutat. Res. 2000;460:301–318. doi: 10.1016/s0921-8777(00)00033-1. [DOI] [PubMed] [Google Scholar]
- 18.Martin IV, MacNeill SA. ATP-dependent DNA ligases. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-4-reviews3005. REVIEWS3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levin DS, Bai W, Yao N, O’Donnell M, Tomkinson AE. An interaction between DNA ligase I and proliferating cell nuclear antigen: implications for Okazaki fragment synthesis and joining. Proc. Natl Acad. Sci. USA. 1997;94:12863–12868. doi: 10.1073/pnas.94.24.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montecucco A, Rossi R, Levin DS, Gary R, Park MS, Motycka TA, Ciarrocchi G, Villa A, Biamonti G, Tomkinson AE. DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. EMBO J. 1998;17:3786–3795. doi: 10.1093/emboj/17.13.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levin DS, McKenna AE, Motycka TA, Matsumoto Y, Tomkinson AE. Interaction between PCNA and DNA ligase I is critical for joining of Okazaki fragments and long-patch base-excision repair. Curr. Biol. 2000;10:919–922. doi: 10.1016/s0960-9822(00)00619-9. [DOI] [PubMed] [Google Scholar]
- 22.Mackey ZB, Ramos W, Levin DS, Walter CA, McCarrey JR, Tomkinson AE. An alternative splicing event which occurs in mouse pachytene spermatocytes generates a form of DNA ligase III with distinct biochemical properties that may function in meiotic recombination. Mol. Cell. Biol. 1997;17:989–998. doi: 10.1128/mcb.17.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caldecott KW, McKeown CK, Tucker JD, Ljungquist S, Thompson LH. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol. Cell. Biol. 1994;14:68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bryans M, Valenzano MC, Stamato TD. Absence of DNA ligase IV protein in XR-1 cells: evidence for stabilization by XRCC4. Mutat. Res. 1999;433:53–58. doi: 10.1016/s0921-8777(98)00063-9. [DOI] [PubMed] [Google Scholar]
- 25.Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 26.Featherstone C, Jackson SP. DNA double-strand break repair. Curr. Biol. 1999;9:R759–R761. doi: 10.1016/S0960-9822(00)80005-6. [DOI] [PubMed] [Google Scholar]
- 27.Liang L, Chen J, Vittal R, Selvanayagam ZE, McAteer JA, Deng L, Tischfield J, Chin KV, Sahota A. Expression profiling of crystal-induced injury in human kidney epithelial cells. Nephron Physiol. 2006;103:53–62. doi: 10.1159/000090503. [DOI] [PubMed] [Google Scholar]
- 28.Jessberger R, Berg P. Repair of deletions and double-strand gaps by homologous recombination in a mammalian in vitro system. Mol. Cell. Biol. 1991;11:445–457. doi: 10.1128/mcb.11.1.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 30.Barnes DE, Tomkinson AE, Lehmann AR, Webster AD, Lindahl T. Mutations in the DNA ligase I gene of an individual with immunodeficiencies and cellular hypersensitivity to DNA-damaging agents. Cell. 1992;69:495–503. doi: 10.1016/0092-8674(92)90450-q. [DOI] [PubMed] [Google Scholar]
- 31.Riballo E, Doherty AJ, Dai Y, Stiff T, Oettinger MA, Jeggo PA, Kysela B. Cellular and biochemical impact of a mutation in DNA ligase IV conferring clinical radiosensitivity. J. Biol. Chem. 2001;276:31124–31132. doi: 10.1074/jbc.M103866200. [DOI] [PubMed] [Google Scholar]
- 32.Tomkinson AE, Vijayakumar S, Pascal JM, Ellenberger T. DNA ligases: structure, reaction mechanism, and function. Chem. Rev. 2006;106:687–699. doi: 10.1021/cr040498d. [DOI] [PubMed] [Google Scholar]
- 33.Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair. 2003;2:955–969. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 34.Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J. Cell. Sci. 2003;116:3051–3060. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- 35.Gottlich B, Reichenberger S, Feldmann E, Pfeiffer P. Rejoining of DNA double-strand breaks in vitro by single-strand annealing. Eur. J. Biochem. 1998;258:387–395. doi: 10.1046/j.1432-1327.1998.2580387.x. [DOI] [PubMed] [Google Scholar]
- 36.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Rosidi B, Perrault R, Wang M, Zhang L, Windhofer F, Iliakis G. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005;65:4020–4030. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 38.Thompson LH, Brookman KW, Dillehay LE, Carrano AV, Mazrimas JA, Mooney CL, Minkler JL. A CHO-cell strain having hypersensitivity to mutagens, a defect in DNA strand-break repair, and an extraordinary baseline frequency of sister-chromatid exchange. Mutat. Res. 1982;95:427–440. doi: 10.1016/0027-5107(82)90276-7. [DOI] [PubMed] [Google Scholar]
- 39.Thompson LH, Brookman KW, Jones NJ, Allen SA, Carrano AV. Molecular cloning of the human XRCC1 gene, which corrects defective DNA strand break repair and sister chromatid exchange. Mol. Cell. Biol. 1990;10:6160–6171. doi: 10.1128/mcb.10.12.6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei YF, Robins P, Carter K, Caldecott K, Pappin DJ, Yu GL, Wang RP, Shell BK, Nash RA, Schar P, et al. Molecular cloning and expression of human cDNAs encoding a novel DNA ligase IV and DNA ligase III, an enzyme active in DNA repair and recombination. Mol. Cell. Biol. 1995;15:3206–3216. doi: 10.1128/mcb.15.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mackey ZB, Niedergang C, Murcia JM, Leppard J, Au K, Chen J, de Murcia G, Tomkinson AE. DNA ligase III is recruited to DNA strand breaks by a zinc finger motif homologous to that of poly(ADP-ribose) polymerase. Identification of two functionally distinct DNA binding regions within DNA ligase III. J. Biol. Chem. 1999;274:21679–21687. doi: 10.1074/jbc.274.31.21679. [DOI] [PubMed] [Google Scholar]
- 42.Taylor RM, Whitehouse CJ, Caldecott KW. The DNA ligase III zinc finger stimulates binding to DNA secondary structure and promotes end joining. Nucleic Acids Res. 2000;28:3558–3563. doi: 10.1093/nar/28.18.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Critchlow SE, Bowater RP, Jackson SP. Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr. Biol. 1997;7:588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- 44.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 45.Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh CL, Schwarz K, Lieber MR. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol. Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 46.Tseng HM, Tomkinson AE. Processing and joining of DNA ends coordinated by interactions among Dnl4/Lif1, Pol4, and FEN-1. J. Biol. Chem. 2004;279:47580–47588. doi: 10.1074/jbc.M404492200. [DOI] [PubMed] [Google Scholar]