Abstract
Microarrays and high-throughput sequencing methods can be used to measure the expression of thousands of genes in a biological sample in a few days, whereas PCR-based methods can be used to measure the expression of a few genes in thousands of samples in about the same amount of time. These methods become more costly as the number of biological samples increases or as the number of genes of interest increases, respectively, and these factors constrain experimental design. To address these issues, we introduced ‘vertical arrays’ in which RNA from each biological sample is converted into multiple, overlapping cDNA subsets and spotted on glass slides. These vertical arrays can be queried with single gene probes to assess the expression behavior in thousands of biological samples in a single hybridization reaction. The spotted subsets are less complex than the original RNA from which they derive, which improves signal-to-noise ratios. Here, we demonstrate the quantitative capabilities of vertical arrays, including the sensitivity and accuracy of the method and the number of subsets needed to achieve this accuracy for most expressed genes.
INTRODUCTION
Regulated gene expression plays important roles in almost every aspect of biology, including the differentiation and migration of cells, maintenance of homeostasis, responses to stress, damage or infection and aging. Evolutionary changes in gene expression account for many of the differences between species and between individuals within a species. Inappropriate expression can lead to disability, disease, and death, but can also serve as a sensitive indicator of disease. The immense implications of gene expression in basic biology and medicine have motivated the invention of a wide variety of analytical methods to track regulated changes, including microarrays (1–3), high-throughput sequencing (HTS) (4–6) and quantitative PCR (7,8). These methods apply to the two extremes of the experimental design spectrum: microarrays and HTS can be used to measure the expression of thousands of genes simultaneously in individual biological samples, whereas quantitative PCR can be used to measure the expression of individual genes in thousands of biological samples. These methods are very fast and economical relative to their predecessors. However, microarrays and HTS methods become less convenient and more costly as the number of biological samples increases, and quantitative PCR becomes more costly as the number of genes of interest increases. Which of these methods to use is a strategic decision based on experimental design factors, such as the number of samples, genes of interest, and replicates required, and on practical factors, such as the accessibility of the technology and cost. Experimental designs involving a few thousand biological samples (e.g. cells treated with thousands of different drugs) in which the behavior of a few hundred genes is of interest are impractical using these methods for most laboratories.
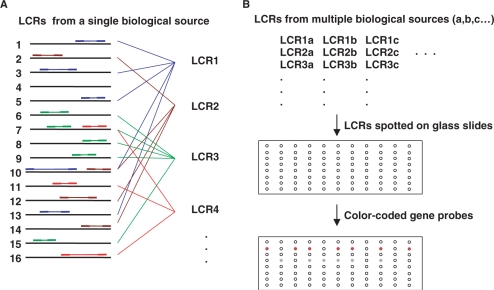
There are several methods that address this problematic neighborhood of experimental design, including a method by Kuhn et al. (9) that involves the capture of targets to a preassembled array of probe-bearing beads, a method by Yang et al. (10) involving target capture on encoded beads and a method by Geiss et al. (11) involving the use of color-coded probe pairs. Traditional dot blots (12), in which total cDNA is spotted on a membrane support and hybridized with single gene probes, have been used to measure the expression of single genes in multiple biological samples, but dot blots have poor performance characteristics and the membrane format is inconvenient. To remedy this, Rogler and colleagues (13) devised RNA expression microarrays (REM) that are essentially dot blots implemented in a glass slide microarray format, greatly improving both performance and convenience. A potential drawback of this direct approach is that rare transcripts remain rare in cDNA made using methods that seek to preserve representation. We previously introduced the idea of printing low complexity representations (LCRs) of mRNA population glass slide microarrays to monitor the expression of individual genes in many biological samples (14). The strategy is outlined in Figure 1. These LCRs comprise overlapping subsets of the RNA population. The representation of rare transcripts is enhanced in these subsets, leading to the surmise that their use might permit expression profiling of rare transcripts. However, the quantitative behavior of the method has not been established, either with regard to sensitivity to rare transcripts, or with regard to the number of LCRs needed to achieve high sensitivity for most expressed genes, and the method cannot be used effectively without this critical information. Here, we describe these quantitative aspects of vertical arrays in comparison to microarrays, real-time RT–PCR and ‘spike-in’ experiments.
Figure 1.
Production of a vertical array. (A) Multiple LCRs are produced using different arbitrary primers, indicated by different colors. The 16 horizontal bars represent a small number of the many different RNAs in a sample. Arbitrary primers match opposing sequences in the RNA population by chance, generating partially overlapping arbitrary sample sequences. (B) Several LCRs are prepared from each biological sample and these are spotted on a glass slide to form a vertical array. Fluorescently tagged single gene probes hybridize to LCRs that have sampled sequence from the mRNA corresponding to the gene.
MATERIALS AND METHODS
RNA sample preparation
Normal human diploid fibroblasts (cell line ATCC CRL 2091) were deprived of serum for 48 h and serum was reintroduced as described by Iyer et al. (15). Total RNA was isolated at 0, 20 and 240 min after reintroduction of serum using RNeasy Mini Kits (Qiagen, Valencia, CA, USA). RNA was treated with DNase I, and purified again using the RNeasy Mini Kit cleanup protocol. RNA concentration was determined by UV absorbance at 260 nm and adjusted to 25 ng/μl.
LCR preparation
Reverse transcription and RNA arbitrarily primed polymerase chain reaction (RAP-PCR) were performed in a single reaction mixture containing 1× M-MLV buffer (Promega, Madison, WI, USA), 0.2 mM each dNTP (ICN, Aurora, OH, USA), 1 μCi [α-P32] dCTP (ICN, Irvine, CA, USA), 5 μM arbitrary primer (Proligo, Boulder, CO, USA), 50 U M-MLV (Promega), 50 U AmpliTaq DNA polymerase Stoffel fragment (Applied Biosystems, Foster City, CA, USA) and 100 ng of RNA. The reaction was incubated at 37°C for 60 min, heated at 94°C for 3 min and temperature cycled through 94°C for 15 s, 35°C for 2 min and 72°C for 2 min for 35 cycles. Eleven different 10-mer arbitrary primers were used for RAP-PCR: c8(TCACCAGCCA), d8(ACGGGCCAGT), g8(CAAGGGCAGT), h8(GGCAGGCTGT), c9(GGGCACCAGG), d9(GGGGCACCAC), f9(CACCAGGGGC), g9(CTGACTGCCT), a10(ACCTGGGGAG), c10(ACAGCCCCCA) and OPN28(GCACCAGGGG). The reactions were assembled using a Biomek FX liquid handling workstation. Products were purified using the PSI Ψ Clone PCR 96 purification kit (Princeton Separations, Adelphia, NJ, USA), eluted with 80 μl of distilled water (pH 9.5), and DNA concentration was measured for one replicate of each RAP-PCR reaction type. RAP-PCR repeatability was assessed qualitatively by electrophoresis through 4% polyacrylamide, 8 M urea gels and autoradiography.
Standard microarray analysis of RAP-PCR products
To identify differentially regulated genes with which to characterize vertical arrays, standard microarray analysis using LCRs was done as shown previously (16). LCRs were labeled for hybridization to standard arrays as follows: ∼500 ng of LCR was mixed with 8 μg of random hexamer (final vol. 36 μl), boiled for 5 min and cooled on ice. Five mocroliters of a 10× reaction mixture was added, and the volume was adjusted to 50 μl, such that the final reaction contained 10 mM Tris–HCl (pH 7.5), 5 mM MgCl2, 7.5 mM DTT, 0.025 mM dGTP, dATP and dCTP, 0.009 mM dTTP, 0.04 mM Cy3- or Cy5-dUTP (Amersham Pharmacia Biotech, Buckinghamshire, England) and 10 U of DNA Polymerase I Klenow fragment (New England Biolabs, Beverly, MA, USA). The reaction was incubated at 37°C overnight and then heated at 70°C for 10 min. The samples were purified using QIAquick PCR purification kit (Qiagen) and eluted in 25 μl of distilled water. Each labeling reaction was done in duplicate, the purified products from duplicate reactions were pooled (50 μl) and incorporation was measured by spectrophotometery (Cy3: 550 nm, Cy5: 650 nm). Approximately 80 pmols of dye per microgram of DNA were incorporated. The t = 0 sample was mixed with t = 240 min sample and hybridized to arrays assembled on UltraGAPS coated slides (Corning, Corning, NY, USA) containing three replicates of PCR products from 3840 human cDNA clones (I.M.A.G.E) (Invitrogen, Carlsbad, CA, USA). Each slide contained three replicates, for six data points per gene. Prehybridization, hybridization and washes were performed following the manufacturer's instructions for UltraGAPS slides, except that the isopropanol wash step was omitted and probes were not allowed to cool to room temperature. Formamide was used at 25% final concentration and 0.1 mg/ml denatured salmon sperm DNA was used as blocking agent. Standard microarrays were scanned using a ScanArray 5000 Laser scanner using ScanArray version 2.1 software and were quantified using Quantarray version 2.0 software (PerkinElmer, Waltham, Massachusetts, USA). Reciprocal dye-swapping was performed for every experiment.
Vertical microarray printing
RAP-PCR reaction products were dried and resuspended in 22 μl of distilled water to achieve average DNA concentrations of 100 ng/ul for printing. A total of 4 μl of DNA was mixed with 4 μl of DMSO. Each of the eight replicate RAP-PCR reactions for every time-point were printed 12 times on the slide. The printing was done with an Omnigrid microarrayer (GeneMachines, San Carlos, CA, USA) on Ultra GAPS coated slides (Corning). After printing, the DNA was cross-linked to the slides by UV irradiation (300 mJ) using a UV StrataLinker (Stratagene, La Jolla, CA, USA) and baked for 2 h at 80°C. Slides were then washed in water and spin-dried for storage. Salmonella LT2 DNA digested with EcoRV and ClaI at 50 ng/μl was printed as negative controls. This digested Salmonella LT2 DNA was also used to produce the positive controls, wherein 12 of the sequences selected to be probed in the vertical arrays were PCR amplified and spiked in as serial dilutions at 3 ng/ul, 0.6 ng/ul, 0.12 ng/ul, 24 pg/ul and 4.8 pg/ul, with 50 ng/ul Salmonella LT2.
Vertical microarray probe synthesis and hybridization
Twenty-four genes that were differentially regulated were initially chosen for study on the vertical arrays, and six genes exhibiting no change in expression were selected as negative controls. Three of these were eventually excluded from the analysis due to the presence of repetitive elements, and one is an independent cDNA clone from the same Unigene. The corresponding I.M.A.G.E. clones were grown, and the inserts were amplified by PCR using the primers M13F (GTTTTCCCAGTCACG- ACGTTG) and M13R (TGAGCGGATAACAATTTCACACAG). The PCR products were purified using the QIAquick PCR purification kit (Qiagen), and concentrations were measured spectrophotometrically. Insert sizes were confirmed by electrophoresis. 25–50 ng of insert DNA was labeled by in vitro transcription (IVT) using a Megascript kit (Ambion, Austin, TX, USA), in a reaction containing 7.5 mM GTP, ATP and CTP, 2.5 mM UTP, 1.75 mM Cy5-UTP; (Amersham Pharmacia Biotech) and 1 μl of enzyme mix, with a final volume of 10 μl. The reaction was incubated at 37°C for 3 h. A total of 7.5 U of T7 RNA Polymerase (Promega) were then added, and after 3 h at 37°C, 1 U of RNase-free DNase 1 was added and tubes were incubated at 37°C for 15 min. RNA was purified using an RNeasy Mini Kit (Qiagen). IVT produced 8–10 μg of RNA labeled with 400–800 pmols of dye.
The vertical arrays were prehybridized, hybridized and washed following the manufacturer's protocol for Ultra GAPS slides with a few modifications. 0.1 mg/ml of Poly dT (Amersham Pharmacia Biotech), 0.1 mg/ml of human Cot-1 DNA (Invitrogen) and 25% formamide were used. A probe consisted of 1–1.2 μg of Cy5-labeled RNA (∼50–80 pmols of dye) corresponding to a gene was mixed with 10–12 ng of a Cy3-labeled (0.7–1 pmols of dye) pool of all LCRs, blocking agents, formamide and buffer. Cy3-dUTP labeling of this pool followed the protocol described above for labeling LCRs.
Real-time RT-PCR
Transcript abundances for 20 genes studied using vertical arrays were also quantified by real-time RT–PCR (Table 1S, a and b). The primers were designed with Primer Express software version 2.0.0 (Applied Biosystems), and chosen to span splice junctions to avoid amplification of possible contaminating genomic DNA or unspliced transcript. Primers used can be found in Table 1Sb. First-strand cDNA synthesis was performed using oligo (dT)15 and real-time RT–PCR was carried out in the presence of SYBR Green using the ABI Prism 7900HT sequence detector (Applied Biosystems). A melting curve was used to identify a temperature where only the amplicon, and not primer dimers, accounted for SYBR Green-bound fluorescence. Standard curves for candidate cDNAs were prepared from a four-point 1/10 serial dilution and were run in duplicate, as were all the samples and the nontemplate control. cDNA quantities were normalized to an internal glyceraldehyde-3-phosphate dehydrogenase mRNA control.
Spike-in experiments
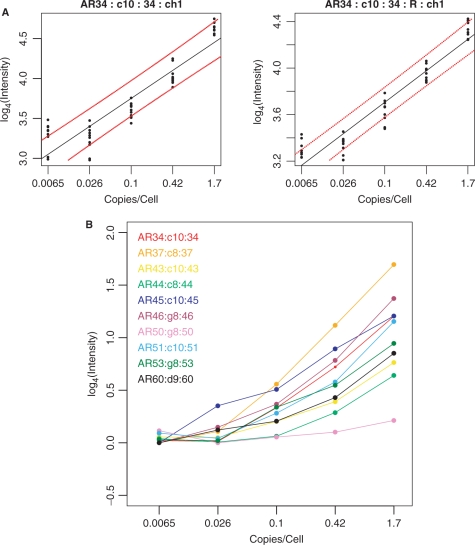
Sequences from 10 Arabidopsis thaliana cDNA clones were amplified by PCR using specific primer pairs, with one of each pair having a 5′ T7 promoter sequence extension (AR34, AF325016.2, 545 bp; AR37, AF325019.2, 817 bp; AR43, AF325025.2, 874; AR44, AF325026.2, 647 bp; AR45, AF325027.2, 600 bp; AR46, AF325028.2, 767 bp; AR50, AF325032.2, 1056 bp; AR51, AF325033.2, 1224 bp; AR53, AF325035.2, 1170 bp; AR60, AF325042.2, 634 bp). T7 RNA polymerase was then used to synthesize the spike-in transcripts. The transcripts were purified and added to human fibroblast total RNA at the proportions discussed in the text. LCRs were prepared as described above for arbitrary primers c8, c9, c10, d9, g8 and h8, and these were each spotted five times on glass slide arrays, as described previously. Fluorescent probes for each of these spike-in transcripts were prepared from the same PCR products using Cy3 or Cy5 and hybridized to the arrayed LCRs. The mean intensity values at each of the five dilutions were calculated for each LCR, and averaged between dye-swap chips, log-transformed and plotted. Figure 5a shows an example, and Figure 3S, shows results for each spike-in and the corresponding probe. To select the best LCR for each gene, the correlation between log4 measured intensities and log4 spike-in concentrations, excluding the lowest concentration (i.e. zero spike-in) was determined, and rp > 0.95 was used as the first selection criterion. The second criterion for the best LCR was to choose the best Student's pairwise t-test P-value indicating a measurable difference between the lowest nonzero spike-in concentration and the highest. These criteria resulted in the selections in Figure 5b. The zero spike-in concentrations were excluded because these samples were often in different physical locations on the microarrays, resulting in greater variance due to background issues. Student's t-tests were then calculated for every pairwise difference in spike-in concentrations (Table 2S).
Figure 5.
(A) Example of vertical array measurements of four of the 10 spiked-in A. thaliana transcripts. Names are as follows. AR34:c10:34:ch1:0.95 corresponds to A. thaliana sequence 34, LCR from primer c10, probe from sequence 34, channel 1. The ‘R’ signifies the dye-swap replicate. The dashed lines correspond to 95% linear calibration confidence limits. (B) Results for all 10 spiked-in transcripts. Data for each transcript was averaged within each vertical array, then normalized and scaled between forward and dye-swap arrays. Student's t-tests for each spike-in concentration transition are in Table 2S.
RESULTS
In these experiments, LCRs were prepared using RAP-PCR (17), in which arbitrarily chosen oligonucleotide primers are used in low stringency reverse transcription and PCR (Figure 1a). LCRs made in this way are similar to multiplex PCR products, except that single short oligonucleotide primers are used, and these primers find frequent matches, or approximate matches, in the RNA due to their short length. Regions of the RNA that are flanked by sequences with partial matches to the arbitrary primers succeed in reverse transcription and PCR amplification. The sequence complexity of LCRs is lower than that of the RNA from which they are derived because only a subset of the sequences in the template molecules amplifies. Successful sequences amplify reproducibly but with different efficiencies, such that rare mRNAs can be abundantly represented in an LCR, while abundant mRNAs can be represented at low levels. Thus, any individual sequence in an LCR can have higher representation than in the mRNA population from which the LCR was derived. While the relative abundances of different sequences within a sample can be highly distorted, relative abundances any particular sequence between samples are maintained, as in multiplex PCR, and consequently, LCRs can be used to infer relative transcript abundances between samples.
When printed on microarrays (Figure 1b), the higher representation of rare transcript sequences in LCRs leads to better signal-to-noise behavior in hybridization experiments (16–18), and multiple LCRs can be prepared such that most RNAs have enhanced representation in at least one LCR. Arrays prepared in this manner can be queried with gene-specific probes to explore differential expression in potentially thousands of biological samples with very high sensitivity. In the discussion that follows, single sequences will be referred to as ‘probes’, and complex mixtures will be referred to as ‘targets’. In standard microarrays, the probes are affixed to the array surface and the target is used in solution. In vertical arrays, it is the other way around: the complex targets are spotted and the simple probe is in solution. The LCRs prepared using RAP-PCR were used in these two different capacities, first as solution-phase targets for standard microarrays and then as spotted targets on vertical arrays.
Detection of LCR-specific differentially regulated genes
To initiate these experiments, we used LCRs as hybridization targets for standard cDNA expression arrays. This procedure revealed genes that were differentially regulated in response to serum-starvation and refeeding of fibroblasts, and also identified the LCRs in which these genes are represented. Total RNA from a serum starvation-refeeding treatment in fibroblasts performed according to Iyer et al. (15) was purified at 0, 20 and 240 min after the reintroduction of serum. This RNA was converted to LCRs using RAP-PCR and 11 different arbitrary primers. There are two different RAP-PCR procedures, one of which generates LCRs from an initial oligo(dT)-primed first strand cDNA template (19), and the other of which generates LCRs directly from RNA using arbitrary priming of reverse transcription to make first strand cDNA (17,20,21). The latter was used in these experiments because it can be done in a single well, with fewer pipetting steps, and without cDNA purification, facilitating preparation using a pipetting robot. The LCRs were radioactively labeled and reproducibility was assessed qualitatively by gel electrophoresis and autoradiography. The 0 and 240 min LCRs were fluorescently labeled and used as targets against standard cDNA microarrays containing about 4000 human cDNA probes (16). Reciprocal dye swap experiments were also performed. This procedure identified differentially regulated genes and the LCR in which each differentially regulated gene was represented. Analysis involved print-tip loess normalization and scaling between arrays using the limma package in BioConductor and the R programming environment (22–24). A modified t-statistic was used to estimate the probability that a gene was differentially regulated (22), with P-values adjusted for multiple testing to predict the false discovery rate (25). Ten transcripts having a modified t-statistic with P ≤ 0.05 from at least one LCR target, four with P-values in the range 0.05 ≤ P ≤ 0.33, and four having larger P-values (P ≥ 0.5) were tested using real-time RT–PCR to confirm differential gene expression. RT–PCR spanned splice junctions to avoid possible interference from unspliced transcripts or residual genomic DNA. Table 1S, contains real-time PCR results, gene names, accession numbers and the associated LCR. All of these genes met the additional criterion that their average signal intensities exceeded the mean intensity of 96 control probe sequences derived from the rat by three standard deviations. The real-time RT–PCR measurements correlated well (rp = 0.92) with the corresponding microarray measurements for those transcripts that had modified t-statistics with P ≤ 0.20 in the standard microarrays (Figure 1S).
Detection of differential gene expression using vertical arrays
Vertical arrays were then assembled by spotting LCRs prepared from the same RNA preparations and arbitrary primers on glass slides using a microarray printer. Eleven different LCRs for each of the three time-points (t = 0, 20, 240 min) after reintroduction of serum were prepared in eight replicates. Eight replicate oligo(dT)-primed cDNAs from the same RNAs were prepared in parallel. Each LCR and oligo(dT)-primed cDNA was spotted four times in each of three subarrays, for a total 3168 spots from the LCRs and 288 spots from oligo(dT)-primed cDNA. In addition, a complete replicate of the LCRs prepared using arbitrary primer OPN28 was included, for an additional 288 spots. The final array contained 3456 LCR spots and 288 oligo(dT)-primed cDNA spots. Spots were also included comprising serial dilutions of the anticipated probe sequences diluted in restriction digested Salmonella genomic DNA as positive controls for hybridization. Additional spots containing Salmonella sequences were included as controls for cross-hybridization and other ill-defined foreground nuisance problems.
Fluorescently labeled probes corresponding to 28 genes selected from standard microarrays were made by reverse transcription with incorporation of Cy5-labeled nucleotides. A control comprising equal masses of all 11 LCRs from each time point was labeled by random primed synthesis with Cy3-labeled nucleotides and these were hybridized to the vertical arrays simultaneously with the gene-specific probes to allow normalization for spotted DNA mass and probe availability to hybridization. This control mixture is sufficiently complex that individual gene expression differences do not contribute significantly to variance in the hybridization signal.
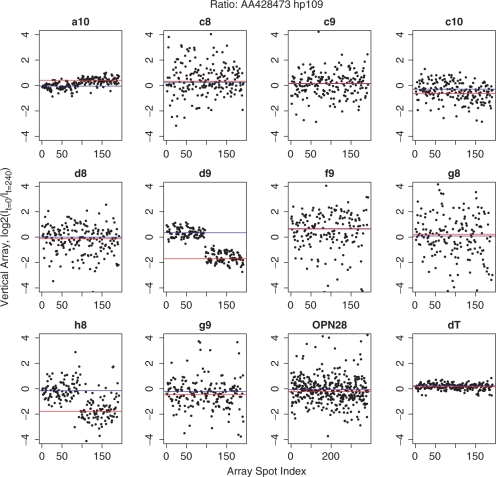
LCRs from all three time points were spotted adjacently in small groups throughout the chip. Ratios of measurements from the time points t = 20 and t = 240 were generated by dividing by a measurement from adjacent t = 0 spots on the chip after normalization to the mixed LCR control signal, and these ratios were plotted for each gene and each LCR. The t = 0 adjacent spots were used to help correct for local variation in background. Figure 2 shows one such graph for gene AA428473, which maps to Nuclear receptor subfamily 1, group D, member 2 (NR1D2). Four-fold down-regulation is implied at t = 240 by two different LCRs, d9 and h8. Quantitative RT–PCR (real-time RT–PCR) indicated 5.3-fold down-regulation of this gene. The other nine LCRs do not report a change, nor is the change reflected in vertical array data acquired from the oligo(dT)-primed first strand cDNA targets. The signals from the oligo(dT)-primed targets were typically 20-fold or more larger than the LCR signals, and differential regulation detected in LCRs were not detected in the oligo(dT) targets. However, we did not attempt to optimize for detection in the oligo(dT) targets. Consistent with this observation, in a standard microarray experiment using an oligo(dT)-primed probe and four replicate arrays, only two among these 28 genes (AA251800 and H77766) had changes with P-values of P ≤ 0.05. For all LCRs where standard arrays implied a change in transcript abundance with P ≤ 0.20, the corresponding LCR on the vertical array implied a similar change.
Figure 2.
Scatter plots for gene AA428473, which maps to Nuclear receptor subfamily 1, group D, member 2 (NR1D2). On the x-axis, 1–96 correspond to replicate measurements made at t = 20 min after re-feeding serum-starved fibroblasts, and 97–192 correspond to measurements made at t = 240 min, except for OPN28, for which twice as many measurements were made at all time points. The vertical axes are the log2(It/It= 0) and the blue and red lines pass through the median values for the 20 and 240 min, respectively. Four-fold down-regulation of NR1D2 is implied at t = 240 by two different LCRs, d9 and h8.
More than one LCR might report a change for any gene, and several examples of this are shown in Figure 2S. To decide which LCR was the best reporter of differential expression for that gene, we employed a prescreen of data falling outside two standard deviations on the log2 scale to exclude outliers, which can usually be attributed to defects in the microarray such as high-local background, and followed this with t-tests. Boxplots of vertical array results for 27 of the 28 transcripts are shown in Figure 3. One gene failed quality control and was omitted. Plotted for each gene are the data from the LCR having the largest t-statistic and P ≤ 10−5. The log2-transformed data is approximately normally distributed for each gene.
Figure 3.
Boxplot summary of results from vertical array measurements for all genes tested, showing the log2 of the ratio of intensities measured at t = 0 and t = 240 min after re-feeding serum to serum-starved fibroblasts. The vertical lines near the center of each box correspond to the median. The whiskers extend to the most extreme data point, which is not more than 1.5 times the interquartile range from the box. The red dotted vertical lines show the position of a 2-fold change.
Figure 4a shows strong Pearson's correlation (rp = 0.94) between measurements made using standard arrays and vertical arrays, and Figure 4b shows the corresponding comparison between vertical arrays and those that were tested using real-time RT–PCR (rp = 0.92). The standard error of the estimate calculated from the data in Figure 4b was sest = 0.62 on the log2 scale, and assumes that the real-time RT–PCR measurements were error-free. These studies indicate that vertical arrays measure changes in transcription abundances quite accurately. Correlation between real-time PCR and the oligo(dT)-primed targets was rp = 0.16, indicating that the direct oligo(dT)-priming approach did not provide useful information for most transcripts. Vertical arrays did not show changes after 20 min of re-exposure to serum, with the possible exception of H79778 (Histone deacetylase 3) (Figure 2S).
Figure 4.
(A) Correlation between measurements of change made using standard arrays and vertical arrays. (B) Correlation between measurements of change using real-time RT–PCR and vertical arrays.
The genes used in this study were chosen without specific reference to their biological functions. Their expression profiles were largely in accord with results of Chang et al. (26) deposited in GEO (http://www.ncbi.nlm.nih.gov/geo/), with the exceptions of CTSB (AA598950), which is down-regulated after 4 h in our data but only slightly upregulated in (26) NR1D2 (AA428473), which is very strongly down-regulated in our data, but not so in (26) LRRFIP1 (AA085597), which is strongly up-regulated in our data, but very modestly up-regulated in (26) and HDAC3 (H79778), which is not regulated in our data, but is moderately upregulated in (26).
Detection sensitivity
We performed ‘spike-in’ experiments to determine the number of LCRs needed to detect changes in most transcripts in the neighborhood of one transcript per cell. Ten different in vitro synthesized transcripts from A. thaliana were prepared and added to total mRNA equivalent to 30 000 fibroblasts at 4-fold dilutions comprising 1.67, 0.42, 0.10, 0.026 A. thaliana transcripts per cell-equivalent of RNA. Vertical arrays were prepared from the RNA containing the spike-in transcripts using 6 of the 11 arbitrary primers described above, and each LCR was spotted five times on each array. These arrays were hybridized in duplicate with fluorescently tagged probes for each of the spike-in sequences. Figure 5a shows an example of signal intensity versus spike-in concentration for one of the spike-in sequences, Figure 3S, shows the corresponding graphs for all of the spike-in sequences and corresponding probes, and Figure 5b shows a summary for all 10 spike-in sequences. Student's t-tests indicated that the transitions between 0 and 1.67 transcripts per cell could be detected for 8 out of 10 transcripts with P ≤ 0.05. The transitions between 0 and 0.42 transcripts per cell could be detected for 7 out of 10 transcripts with P ≤ 0.05, and the transitions between 0 and 0.1 transcripts per cell could be detected for 6 out of 10 transcripts with P ≤ 0.05. Differences between larger spike-in concentrations were robust, for example, transitions between 0.1 and 0.42 transcripts per cell could be detected for 6 out of 10 transcripts with P ≤ 0.05. The results are summarized in Table 2S. Detection of 8 out of 10 transcripts with six LCRs suggests that detection of similar changes in 95% of all transcripts could be achieved using 11 LCRs assuming a Poisson model. The number of LCRs required per sample is important because it determines the number of different biological samples that can be surveyed on a single glass slide array.
DISCUSSION
Previously, we presented qualitative evidence that vertical arrays prepared using LCRs spotted on glass slides could be used to assess differential gene expression. In the experiments presented here, we determined the sensitivity of the method with respect to transcript abundance and the number of LCRs required to achieve comprehensive coverage. The vertical arrays in this demonstration had only three variables (i.e. time points) represented in over 300 redundant features for each LCR for the purpose of evaluation of the approach. However, in practice, thousands of experimental variables could be tested on a single vertical array with far lower redundancy. For example, a vertical array with 22 000 spots and 11 LCRs would yield expression information for 2000 experimental conditions. The variant of RAP-PCR used in these experiments can be performed in a single well with two pipetting steps, without any intervening purification step, making it simple to automate LCR synthesis. Final purification of LCRs and robotic spotting were also automated. Therefore, the throughput of vertical array analysis is potentially very high.
The use of vertical arrays rather than standard microarrays or quantitative RT–PCR is a strategic decision based on experimental design, time lines and cost. In experiments in which there is interest in the behavior of a preselected set of genes in a large number of biological samples, vertical arrays can be more efficient and cost-effective than standard microarrays or quantitative RT–PCR. (See Supplement A for a discussion of cost estimates and other considerations.) With vertical arrays, the number of hybridizations is proportional to the number of genes to be explored rather than to the number of biological samples, and unlike methods that rely on multiplex detection of a preselected set of gene targets (27), vertical arrays can be hybridized with any gene probe. This relaxes the requirement for appropriate selection of genes at the beginning of an experiment. Once the LCRs have been generated and the arrays printed, any number of genes can be examined at the small marginal cost of hybridizing a probe to an array, without returning to the original biological samples. This allows for post hoc selection of additional genes as the biological story unfolds, which is advantageous when compared to the usual arrangement of spotted select probes, where thousands of additional hybridizations have to be performed to accommodate additional genes. At the present time, the cost of examining the expression of 200 genes in 2000 biological samples using vertical arrays is about 5-fold lower than the most competitive alternative.
Finally, a potential advantage of the vertical arrays and REM is that microarrays representing thousands of samples are easy to replicate and ship relative to sample libraries arrayed in plates.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was funded by a National Cancer Institute IMAT program grant [CA116214], a National Cancer Institute grant [CA068822], and generous gifts to the Sidney Kimmel Cancer Center by Mr Sidney Kimmel and Mr Ira Lechner. Funding to pay the Open Access publication charges for this article was provided by NCI grants CA116214 and CA068822.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 2.Fodor SP, Rava RP, Huang XC, Pease AC, Holmes CP, Adams CL. Multiplexed biochemical assays with biological chips. Nature. 1993;364:555–556. doi: 10.1038/364555a0. [DOI] [PubMed] [Google Scholar]
- 3.Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat. Genet. 1999;21:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- 4.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 5.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- 7.Delidow BC, Peluso JJ, White BA. Quantitative measurement of mRNAs by polymerase chain reaction. Gene Anal. Tech. 1989;6:120–124. doi: 10.1016/0735-0651(89)90002-2. [DOI] [PubMed] [Google Scholar]
- 8.Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology. 1993;11:1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- 9.Kuhn K, Baker SC, Chudin E, Lieu MH, Oeser S, Bennett H, Rigault P, Barker D, McDaniel TK, Chee MS. A novel, high-performance random array platform for quantitative gene expression profiling. Genome Res. 2004;14:2347–2356. doi: 10.1101/gr.2739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang L, Tran DK, Wang X. BADGE, beads array for the detection of gene expression, a high-throughput diagnostic bioassay. Genome Res. 2001;11:1888–1898. doi: 10.1101/gr.190901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 12.Kafatos FC, Jones CW, Efstratiadis A. Determination of nucleic acid sequence homologies and relative concentrations by a dot hybridization procedure. Nucleic Acids Res. 1979;7:1541–1552. doi: 10.1093/nar/7.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rogler CE, Tchaikovskaya T, Norel R, Massimi A, Plescia C, Rubashevsky E, Siebert P, Rogler LE. RNA expression microarrays (REMs), a high-throughput method to measure differences in gene expression in diverse biological samples. Nucleic Acids Res. 2004;32:e120. doi: 10.1093/nar/gnh116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Risques R, Rondeau G, Judex M, McClelland M, Welsh J. Vertical arrays: microarrays of complex mixtures of nucleic acids. Methods Mol. Biol. 2006;317:99–109. doi: 10.1385/1-59259-968-0:099. [DOI] [PubMed] [Google Scholar]
- 15.Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JC, Trent JM, Staudt LM, Hudson J., Jr., Boguski MS, et al. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 16.Rondeau G, McClelland M, Nguyen T, Risques R, Wang Y, Judex M, Cho AH, Welsh J. Enhanced microarray performance using low complexity representations of the transcriptome. Nucleic Acids Res. 2005;33 doi: 10.1093/nar/gni095. e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welsh J, Chada K, Dalal SS, Cheng R, Ralph D, McClelland M. Arbitrarily primed PCR fingerprinting of RNA. Nucleic Acids Res. 1992;20:4965–4970. doi: 10.1093/nar/20.19.4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 19.Trenkle T, Welsh J, Jung B, Mathieu-Daude F, McClelland M. Non-stoichiometric reduced complexity probes for cDNA arrays. Nucleic Acids Res. 1998;26:3883–3891. doi: 10.1093/nar/26.17.3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ralph D, McClelland M, Welsh J. RNA fingerprinting using arbitrarily primed PCR identifies differentially regulated RNAs in mink lung (Mv1Lu) cells growth arrested by transforming growth factor beta 1. Proc. Natl Acad. Sci. USA. 1993;90:10710–10714. doi: 10.1073/pnas.90.22.10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McClelland M, Ralph D, Cheng R, Welsh J. Interactions among regulators of RNA abundance characterized using RNA fingerprinting by arbitrarily primed PCR. Nucleic Acids Res. 1994;22:4419–4431. doi: 10.1093/nar/22.21.4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 23.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 24.Smyth GK, Yang YH, Speed T. Statistical issues in cDNA microarray data analysis. Methods Mol. Biol. 2003;224:111–136. doi: 10.1385/1-59259-364-X:111. [DOI] [PubMed] [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. Ser B. 1995;57:289–300. [Google Scholar]
- 26.Chang HY, Sneddon JB, Alizadeh AA, Sood R, West RB, Montgomery K, Chi JT, van de Rijn M, Botstein D, Brown PO. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol. 2004;2:E7. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan JB, Yeakley JM, Bibikova M, Chudin E, Wickham E, Chen J, Doucet D, Rigault P, Zhang B, Shen R, et al. A versatile assay for high-throughput gene expression profiling on universal array matrices. Genome Res. 2004;14:878–885. doi: 10.1101/gr.2167504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.