Abstract
UniPrime is an open-source software (http://uniprime.batlab.eu), which automatically designs large sets of universal primers by simply inputting a gene ID reference. UniPrime automatically retrieves and aligns homologous sequences from GenBank, identifies regions of conservation within the alignment and generates suitable primers that can amplify variable genomic regions. UniPrime differs from previous automatic primer design programs in that all steps of primer design are automated, saved and are phylogenetically limited. We have experimentally verified the efficiency and success of this program by amplifying and sequencing four diverse genes (AOF2, EFEMP1, LRP6 and OAZ1) across multiple Orders of mammals. UniPrime is an experimentally validated, fully automated program that generates successful cross-species primers that take into account the biological aspects of the PCR.
INTRODUCTION
Comparing the genomic structure and content of phylogenetic and ecologically diverse taxa will act as a ‘Rosetta’ stone allowing us to annotate and interpret our own genome (1–4). Evolutionary analyses of whole genomes and targeted genes sequenced in divergent species have advanced our understanding of the patterns of human disease mutations in many inherited diseases and cancers (5–7). Comparative genomics is a powerful and expanding field, which is evident from the exponential increase in the number of non-human sequence entries in GenBank and EMBL within the past decade. GenBank has doubled in size about every 18 months. It currently contains over 65 billion nucleotide bases from more than 61 million individual sequences, with 15 million new sequences added in the past year (8). Since the completion of the human sequence project in 2001 (9,10), the number of whole genomes sequenced or in the process of being sequenced is also increasing. Currently, over 617 genomes are completed, 531 are in assembly stage, 652 are in process and 421 have been approved for whole genome sequence (11). Researchers are faced with vast quantities of molecular data that can only be stored, analysed and mined with appropriate computer-based algorithms and programs. Subsequently, the bioinformatics software used to mine these data are also increasing exponentially (4).
Although many species will be sequenced at the whole genome level, this number still only represents a small fraction of the diversity of life: e.g. 107/5000 mammals either sequenced, in the process or accepted to be sequenced (11,12). Therefore, smaller comparative genomic projects that target key genes in key taxa (4) will still play a large role in future comparative studies. Good primer design is a crucial step in any comparative genomics project and ensures specificity and efficiency of target amplification, necessary to achieve reliable PCR results. Traditionally, universal primers were designed by first generating a multispecies alignment then, manually identifying conserved regions in that alignment, finally an algorithm was used to estimate the melting temperature of candidate primers sequences within the conserved regions. This is a laborious process with many defined user steps, including downloading and aligning sequences of phylogenetic interest.
Recent advances in bioinformatics primer design software have increased the speed of some steps within this process. Most programs will automatically design compatible forward and reverse PCR primers from an inputted sequence [e.g. Primer3 (13), Oligo 6 (http://www.oligo.net), AutoPrime (14), CODEHOP (15), ExonPrimer (http://ihg.gsf.de/ihg/ExonPrimer.html)], or an inputted user defined multiple alignment [e.g. PrimaClade (16), Primer Premier (http://www.premierbiosoft.com)]. A recent program QPRIMER (17) generates universal primers within exons by automatically creating multi-genome alignments of human, mouse, rat, chicken, dog, zebrafish and fruit fly. Although, all of these programs enhance the speed and accuracy of primer design, none automate all steps in the process for all regions within the genome. To our knowledge, no automatic primer design program uses phylogenetic information to retrieve and align all homologous sequences from GenBank. They do not automatically assess levels of variation across the alignment, design non-degenerate primers, nor estimate the possibility of false positive amplifications.
To address these problems, we have designed the UniPrime program, an open-source suite where users can automatically design sets of universal primers to amplify regions of suitable inter-specific variation across divergent taxa, by simply inputting a reference sequence or accession number. UniPrime uniquely allows all steps of the process to be saved, including initial data retrieval, multispecies alignment and primer design sites. We have experimentally verified the efficiency and success of this program by amplifying and sequencing four diverse genes (AOF2, EFEMP1, LRP6 and OAZ1) across multiple Orders of mammals. The program is available at http://uniprime.batlab.eu and is licensed under the Creative Commons GNU GPL.
METHODS
Algorithm
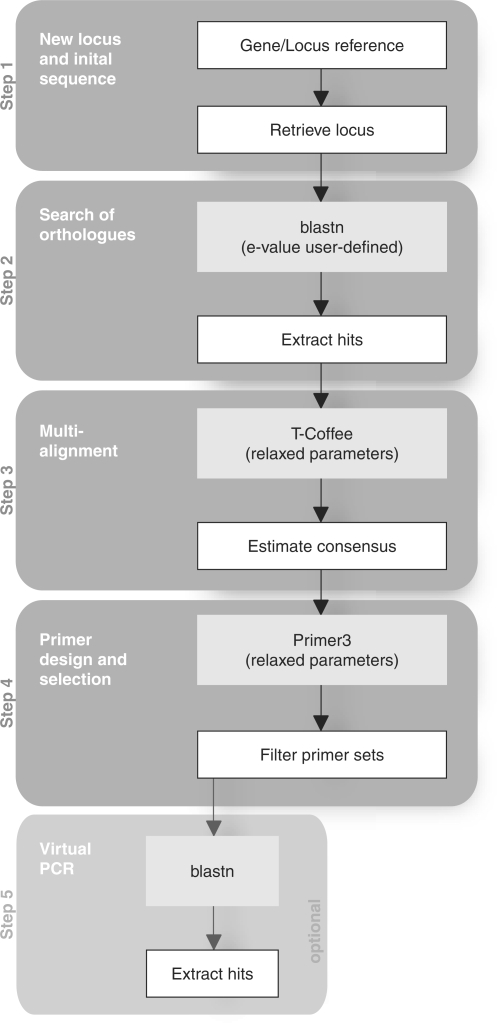
We designed an automated method to: (i) design successful universal primers for any given locus; (ii) maximize the number of variable positions in the fragment amplified given a user defined similarity threshold; (iii) evaluate the ‘mis-priming’ potential of the primers generated. This process is defined in five steps and shown schematically in Figure 1. Each step can be reviewed by the companion web interface to assess potential challenges resulting from sequencing error, incorrect annotation or an artifactual duplication of the locus. The main module of UniPrime uses a command line interface. The commands are simple, they are detailed in the ‘readme’ file and the number of parameters requested is small. Each step is described subsequently.
Figure 1.
Schematic diagram of the UniPrime algorithm. Steps carried out by UniPrime are shown in white boxes. Steps performed by external programs, are shown in grey boxes. The five main functions of UniPrime are highlighted.
Step 1—Initial sequence
Initially, the GenBank GeneID (unique identifiers for genes provided by Entrez Gene) of the target locus (protein coding gene, snRNA, etc.) is input by the user. This reference code is used to retrieve the sequences related to this target locus (the genomic DNA and the mRNA sequences). The program then selects a single nucleotide sequence (proto-type sequence), which is usually the reference mRNA sequence of the longest isoform of the gene. If no mRNA sequence is known for the gene, the reference genomic DNA sequence is then used. If required, the user can enforce the use of the DNA rather than mRNA sequence. In such cases, the quality of the alignment is less accurate with multiple insertions/deletions introduced due to introns, but the overall scheme of the primer design method is unchanged.
Step 2—Search for orthologues
The prototype sequence is used as a ‘query’ for a Blastn (18) search of the NCBI database to identify highly similar homologous sequences. The user can delimit the search at varying phylogenetic levels by incorporating the Entrez Query BLAST option. The NCBI ‘Reference genomic sequence’ (RefSeq) database (19) is used as the default search database. An e-value threshold of 10−100 is incorporated as the default cut-off point for valid hits. Both parameters are user-defined and can be modified. Due to problems with variable intron length and number within divergent taxa, this step preferentially uses an mRNA prototype sequence when available; otherwise, the DNA sequence of target locus is used. If the prototype sequence is mRNA, then only mRNA sequences will be retrieved. Likewise, if the prototype sequence is DNA then only DNA sequences will be retrieved. The sequences with the best e-value score for each species within the database are retrieved, and stored in the database. Using this method, we hope to retrieve orthologous sequences; however, as our retrieval method is based only on a high blast score, this step may also gather closely conserved paralogous sequences. UniPrime allows the user to review the retrieved sequences and remove any sequences that are considered paralogous before primer design. Also, users can further modify the phylogenetic distance between the taxa they want incorporated in the primer design step by including or removing the required taxa.
Step 3—multi-species alignment
The stored sequences are concatenated into a single file, which is then imported into the alignment program T-Coffee (20). Users can restrict the taxa they want to be included in this input file. The sequences are aligned using the default parameters in T-Coffee (20). From this alignment, a consensus sequence is inferred with a conservative default threshold of 60% (i.e. only the nucleotide which occurs >60% at a single position in the alignment is represented in the consensus sequence, otherwise an N is reported). Only A, T, C, G and N are used in the consensus sequence. As the number of non-N-sites in the consensus sequence is limited by the threshold, increasing this threshold should lower the number of primers designed but increase their specificity. Likewise, decreasing this threshold should increase the number of primers designed but lower their specificity. If required the user can manually incorporate their own alignment.
Step 4—primer design
All possible primers along the consensus sequence are generated by Primer3 (13) using the following parameters: melting temperature (Tm) 55–65°C; primer length 20–25 bp; GC clamp (G or C at 3′ end); GC content 40–60%; and, an optimum product size of 600 bp. The user can vary the product size. To ensure that all possible primers are defined along the consensus sequence, they are generated using a sliding window approach. The sliding window of 250 bp moves along each nucleotide position in the consensus sequence and primers are designed where possible. By default, sites that are over 40% variable within the alignment are defined as Ns; therefore, primers cannot be generated within these regions. Users can change this threshold, thereby controlling the level of expected variability within the resulting amplicon. To select the optimal primer pairs each primer sequence is examined against all input sequences using the following filters: (i) the entire primer sequence must be found in all input sequences but can have a mis-match of 20% with each sequence; (ii) the last 5 bp at the 3′ end of each primer must be 100% conserved in all input sequences. This is an essential filter, as DNA polymerase cannot bind efficiently to a template if there are mismatches at the 3′ end of a primer, regardless of the specificity in 5′ end region. If the last two residues of the primer do not match the template, then no amplification can occur (21).
Step 5—virtual PCR
The possibility of amplifying non-target sequences using the proposed primer pairs is assessed in an optional last step by completing a ‘virtual PCR’. The primer sequences are submitted for a Blastn search within the ‘Whole-genome shotgun reads’ (wgs) databases of GenBank to identify sequences that match the forward and reverse primer sequence within a compatible size range for PCR amplification (primers <10 kb apart).
All steps and results are stored in the UniPrime database and their details can be viewed using the web-based interface.
Computer implementation
We implemented this algorithm in a software suite called UniPrime. The basic algorithm requires Bioperl 1.4 (22), T-Coffee 3.2 (20), PostgreSQL 7 (http://www.postgresql.org) and Primer3 (13) version 1.0. Newer versions of these programs can also be used. Primer3 version 1.1.1 is used as it includes an advanced Tm calculation method (23).
Web interface
The intermediate results for each step of the UniPrime suite can also be viewed, accessed and modified through the web companion interface. This is implemented in PHP script using PHP 4.3 or above (http://www.php.net) and any PHP compliant web server.
Laboratory verification
We used UniPrime to generate primers from four diverse genes. We validated the primers designed by amplifying and sequencing five fragments from these genes in five divergent Orders across Class Mammalia. The sequences retrieved and used by UniPrime are available in the Supplementary data and belong to Bos taurus (cow), Canis lupus familiaris (dog), Homo sapiens (man), Macaca mulatta (macaque), Monodelphis domestica (opossum), Mus musculus (mouse), Pan troglodytes (chimp) and Rattus norvegicus (rat).
Taxa and genes
The genomic DNA from six divergent mammal species was used (Table 1). The four genes selected (Table 2) have diverse functions and show different variability levels as estimated by DnaSP 4.10.9 (24). The five selected primer sets are shown in Table 2.
Table 1.
Mammalian species used
| Species name | Abbrev. | Common name | Order |
|---|---|---|---|
| Myrmecophaga tridactyla | Mtri | Giant anteater | Xenarthra |
| Ornithorhynchus anatinus | Oana | Platypus | Monotremata |
| Rousettus lanosus | Rlan | Long-haired rousette | Chiroptera |
| Tragelaphus eurycerus | Teur | Bongo | Cetartiodactyla |
| Tupaia minor | Tmin | Pygmy tree shrew | Scandentia |
| Euphractus sexcinctus | Esex | Six-banded armadillo | Xenarthra |
For each species the common name and the Order name are indicated (12).
Table 2.
Primers used and estimated product length
| Gene | GeneID | Primers (5′ → 3′) | CORE index | Size | Location (human) | |
|---|---|---|---|---|---|---|
| AOF2 | 23028 | F | ATGCAGTTCTCTGTACCCTTCC | 41 | 550 | Exon 15–Exon 16 |
| R | AACATGCCCNAACAAATTGAC | |||||
| EFEMP1 | 2202 | F | GCATTGCAAAACTCTGTATGG | 37 | 650 | Intron 6–Exon 7 |
| R | TACCTTCACAGTTGAGCCTGTC | |||||
| LRP6 (1) | 4040 | F | ATCAGNTCCCTCAGTATCATGG | 37 | 800 | Exon 21–Exon 22 |
| R | TAATGTGATCGCTCTGTGG | |||||
| LRP6 (2) | 4040 | F | GAACTCAATTGTCCTGTNTGCTC | 37 | 1200 | Exon 18–Exon 19 |
| R | CAGTTCATCTGANTTGTCACTGC | |||||
| OAZ1 | 4946 | R | TCCCTNCACTGCTGTAGTAACC | 33 | 550 | Exon 2–Exon 3 |
| F | CNGGGATCTCGATGTAGAGG |
For each gene/locus the forward (F) and reverse (R) primers are indicated. The ‘GeneID’ was the entry used for the initial step. The ‘CORE index’ has been provided by T-coffee, and is the reliability score of the alignments. The expected product length is an average value inferred from the multiple alignment but varies between species. Two sets of primers were used for the LRP6 gene.
PCR and DNA sequencing
PCR was performed with 2 nM of each primer, 1.5 mM MgCl2, 1 U of Platinum Taq DNA polymerase (Invitrogen Corporation, Carlsbad, California, USA), and 10 ng of genomic DNA. Touchdown conditions of amplification were used for all species, as follows: 10 cycles of denaturation at 95°C for 30 s, annealing at 65°C for 30 s −1°C per cycle, extension at 72°C for 60 s; followed by 35 cycles with 95°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 60 s. The initial denaturation step and the last extension step were 3 min each. The PCR products were separated and visualized in 1% agarose gel. Sequencing reactions were performed in both directions on PCR products, using the same primer set as for amplification.
Sequence validation
Newly generated sequences were concatenated and aligned, using T-Coffee (20), with the original sequences used to generate the primers. Maximum likelihood (ML) analyses were performed for each data set with PAUP 4.0b10 (25) using the parameters settings (Table 3) for the optimal model of sequence evolution as estimated by Modeltest (26). Starting trees were obtained via neighbor-joining (NJ). 100 ML bootstrap analyses were performed using tree-bisection and recombination branch swapping. Ornithorhynchus anatinus (platypus) was used as an outgroup in all analyses apart from EFEMP1, where Monodelphis domestica (opossum) was used.
Table 3.
Model and parameters
| Model | ti/tv ratio | α-shape | p-inv | |
|---|---|---|---|---|
| AOF2 | HKY + I + G | 2.2047 | 4.1884 | 0.1939 |
| EFEMP1 | HKY + I + G | 1.8285 | 1.6702 | 0 |
| LRP6 (1) | HKY + I + G | 1.6993 | 5.5588 | 0.0800 |
| LRP6 (2) | HKY + I + G | 1.3773 | 2.9999 | 0.0905 |
| OAZ1 | HKY + I + G | 2.0732 | 0.5506 | 0.1105 |
Optimal model and parameters settings of sequence evolution estimated by Modeltest and used to establish the maximum likelihood bootstrap consensus trees. ti/tv, Transition/transversion ratio; α-shape, shape of the distribution; p-inv, proportion of invariable sites.
Supporting information
The generated sequences were deposited in GenBank (Table 4).
Table 4.
Genbank Accession numbers
| OAZ1 | LPR6 (1) | LPR6 (2) | EFEMP1 | AOF2 | |
|---|---|---|---|---|---|
| Euphractus sexcinctus | EF674548 | EF674524 | na | EF674539 | EF674533 |
| Rousettus lanosus | EF674547 | EF674526 | EF674529 | EF674542 | EF674535 |
| Myrmecophaga tridactyla | EF674546 | EF674525 | EF674528 | EF674541 | EF674534 |
| Tragelaphus eurycerus | EF674545 | EF674522 | EF674530 | EF674543 | EF674536 |
| Ornithorhynchus anatinus | EF674544 | EF674527 | EF674531 | na | EF674538 |
| Tupaia minor | na | EF674523 | EF674532 | EF674540 | EF674537 |
RESULTS AND DISCUSSION
Empirical evaluation of the optimal number of input sequences required to successfully design universal primers
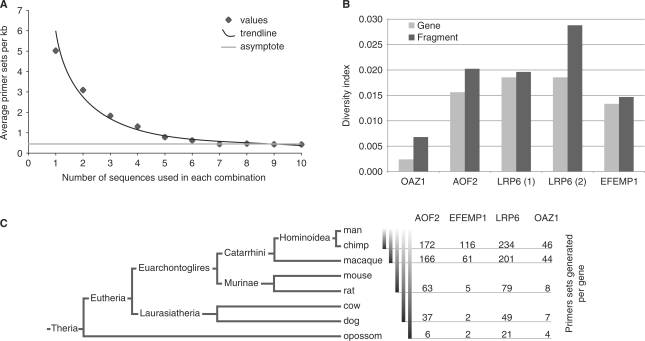
The design of optimal primers relies on the quality of the alignment and directly depends on the number of homologous sequences initially retrieved. We assessed how many optimal primer pairs UniPrime could design when the number of input sequences was varied. From 200 randomly selected mammalian genes, we identified and retrieved their homologous genes using UniPrime. The maximum number of species retrieved was 10 as only 10 mammalian genomes were annotated in the default RefSeq database at time of analyses. All possible combinations of retrieved sequences were generated for each gene and every combination was used to design primers: i.e. for each gene, regardless of phylogenetic affinity, we systematically jack-knifed the number of species present from 10 to one and each iteration was used to design primers. Figure 2A shows the correlation between the number of input sequences and the average number of primers identified per kilobase pair. The data fits with an inverse function (R2 = 0.97): above five sequences, the number of primer sets reached an asymptotical value of 0.5. Therefore, only five to six initial sequences are necessary to design optimal primers using UniPrime.
Figure 2.
(A) Impact of the number of sequences used for the alignment on the primer selection. The number of sequence used for the multi-alignment step varied from 1 to 10. The number of primer sets selected is expressed in primer sets per kb. The 200 random mammalian genes were used from 1 to 300 kb-long. The curve is asymptotic to a value of 0.5 primer set per kilobase (grey line). (B) Sequence diversity. Diversity indices (average number of variation per site) across the alignment of the entire gene (light grey) and of the amplified fragment (dark grey). The amplified regions are more variable than the complete gene as is expected due to the primer design process. (C) Primers sets generated per gene. For each gene, the number of primer sets generated is assessed across phylogenetic distances.
This result has only been established for mammals [last shared a common ancestor ∼220 MYA; (12)] and for the default consensus threshold of 60%. This result will vary at different Linnaean ranks and among phylogenetically diverse organisms. (Figure 2C). UniPrime will work with any data set regardless of genetic diversity as long as it is possible to create an alignment and thus a consensus sequence. The quality of the alignment will dictate the number of primers found, and this quality is dependent on both phylogenetic similarity and genetic diversity of the taxa or genes being used (Figure 2). UniPrime was consistently able to generate a similar amount of primers per kilobase pair regardless of gene function and structure, indicating that this is a robust and reliable primer design method. Interestingly, there is no difference in the number of primers designed when more than five input sequences are used. As at least seven whole mammalian genomes are available, it appears that it is possible to create reliable mammalian primers for coding regions using this method. Therefore, this is an invaluable tool for future phylogenetic and comparative studies.
Laboratory benchmark
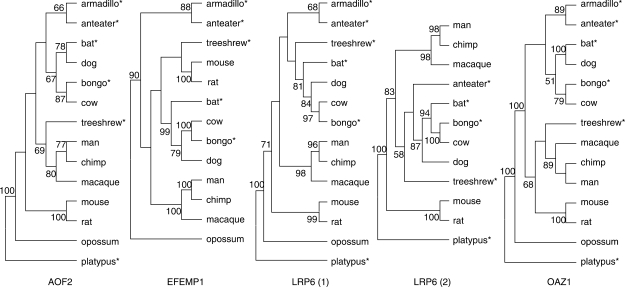
We randomly selected a total of four genes that are well studied, found throughout the genome and show a variable degree of diversity (Figure 2B) within the human population and among mammals: (i) AOF2 (also called LSD1) is a component of several histone deacetylase complexes and silences genes by functioning as a histone demethylase (27); (ii) EFEMP1, an extracellular matrix protein expressed in retina (28); (iii) LRP6, a low density lipoprotein receptor protein and putative tumour suppressor of leukaemia (29) and (iv) OAZ1, the ornithine decarboxylase antizymes, that regulates polyamine synthesis (30). Among the Class Mammalia, we applied our algorithm to these four genes to design five primer pairs providing amplification products of about 600 bp (Table 2 and Supplementary material for the alignments used). Despite the high levels of evolutionary divergence (∼220 MYA) among our input sequences, all genes were successfully amplified, sequenced and verified using phylogenetic analyses (Figure 3). The gene trees obtained for these sequences were congruent with the established species phylogeny (Figure 3).
Figure 3.
Benchmark ML bootstrap consensus trees based on our amplified fragments (indicated by *) and retrieved aligned sequences. Bootstrap support over 50% is shown.
Test design
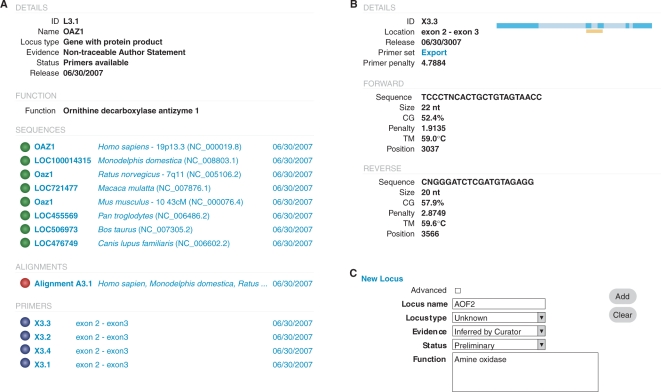
For OAZ1, the sequences retrieved and primers generated as an output by UniPrime are shown in Figure 4. Detailed information including related references for each sequence, alignment or primer pair are available, and can be reviewed through the web companion interface Figure 4A. Users can update, add or remove a locus, sequence alignment or primers through the web-based interface (Figure 4B and C). The response time of the web companion interface is nearly instantaneous, regardless of the quantity of information stored in the database or the number of target loci analysed. The primer generation time depends on the response time of the NCBI server but on average takes <10 min per locus (not including step 5—‘virtual PCR’).
Figure 4.
Screenshot of UniPrime web interface. (A) Detail of the output for the OAZ1 locus, including the sequences retrieved, the alignment and the primers available. (B) Details of one the primer sets generated. (C) Screenshot of the manual entry of a new locus (step 1).
To date, UniPrime has been implemented in four phylogenetically diverse projects at University College Dublin, Ireland. UniPrime is user friendly and has generated over 100 primer sets among which 90% have successfully amplified the target locus (data not shown). One of unique qualities of UniPrime is that only the Gene reference ID is required to enable the user generate a full suite of universal primers. UniPrime also stores and allows easy access to wealth of information via the web interface. UniPrime is an attractive alternative to the long and troublesome steps required for manual retrieval and alignment of homologous sequence from databases. UniPrime represents a new generation of primer design programs that builds on previous programs, automates all steps, enables great user versatility and efficiently mines the ever-expanding genomic databases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Alisha Goodbla for her assistance with laboratory work, William J. Murphy and two anonymous reviewers for their constructive comments. This work was supported by a Science Foundation Ireland PIYRA 06/YI3/B932, award to ECT. Funding to pay the Open Access publication charges for this article was provided by University College Dublin, Seed Funding Scheme.
Conflict of interest statement. None declared.
REFERENCES
- 1.Margulies EH, Blanchette M, Haussler D, Green ED. Identification and characterization of multi-species conserved sequences. Genome Res. 2003;13:2507–2518. doi: 10.1101/gr.1602203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy WJ, Pevzner PA, O’Brien SJ. Mammalian phylogenomics comes of age. Trends Genet. 2004;20:631–639. doi: 10.1016/j.tig.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 3.O’Brien SJ, Fraser CM. Genomes and evolution: the power of comparative genomics. Curr. Opin. Genet. Dev. 2005;15:569–571. doi: 10.1016/j.gde.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Tuggle CK, Dekkers JC, Reecy JM. Integration of structural and functional genomics. Animal Genet. 2006;37(Suppl. 1):1–6. doi: 10.1111/j.1365-2052.2006.01471.x. [DOI] [PubMed] [Google Scholar]
- 5.Fleming MA, Potter JD, Ramirez CJ, Ostrander GK, Ostrander EA. Understanding missense mutations in the BRCA1 gene: an evolutionary approach. Proc. Natl Acad. Sci. USA. 2003;100:1151–1156. doi: 10.1073/pnas.0237285100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burk-Herrick A, Scally M, Amrine-Madsen H, Stanhope MJ, Springer MS. Natural selection and mammalian BRCA1 sequences: elucidating functionally important sites relevant to breast cancer susceptibility in humans. Mamm. Genome. 2006;17:257–270. doi: 10.1007/s00335-005-0067-2. [DOI] [PubMed] [Google Scholar]
- 7.Thomas JW, Touchman JW, Blakesley RW, Bouffard GG, Beckstrom-Sternberg SM, Margulies EH, Blanchette M, Siepel AC, Thomas PJ, McDowell JC, et al. Comparative analyses of multi-species sequences from targeted genomic regions. Nature. 2003;424:788–793. doi: 10.1038/nature01858. [DOI] [PubMed] [Google Scholar]
- 8.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2007;35:D21–D25. doi: 10.1093/nar/gkl986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 10.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 11.NCBI. 2007. Genome sequencing projects statistics. [ http://www.ncbi.nlm.nih.gov/genomes/static/gpstat.html]. [Google Scholar]
- 12.Springer MS, Murphy WJ. Mammalian evolution and biomedicine: new views from phylogeny. Biol. Rev. Camb. Philos. Soc. 2007;82:375–392. doi: 10.1111/j.1469-185X.2007.00016.x. [DOI] [PubMed] [Google Scholar]
- 13.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 14.Wrobel G, Kokocinski F, Lichter P. AutoPrime: selecting primers for expressed sequences. Genome Biol. 2004;5:P11. [Google Scholar]
- 15.Rose TM, Henikoff JG, Henikoff S. CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Res. 2003;31:3763–3766. doi: 10.1093/nar/gkg524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gadberry MD, Malcomber ST, Doust AN, Kellogg EA. Primaclade—a flexible tool to find conserved PCR primers across multiple species. Bioinformatics. 2005;21:1263–1264. doi: 10.1093/bioinformatics/bti134. [DOI] [PubMed] [Google Scholar]
- 17.Kim N, Lee C. QPRIMER: a quick web-based application for designing conserved PCR primers from multigenome alignments. Bioinformatics. 2007;23:2331–2333. doi: 10.1093/bioinformatics/btm343. [DOI] [PubMed] [Google Scholar]
- 18.Ye J, McGinnis S, Madden TL. BLAST: improvements for better sequence analysis. Nucleic Acids Res. 2006;34:W6–W9. doi: 10.1093/nar/gkl164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 21.Drenkard E, Richter BG, Rozen S, Stutius LM, Angell NA, Mindrinos M, Cho RJ, Oefner PJ, Davis RW, Ausubel FM. A simple procedure for the analysis of single nucleotide polymorphisms facilitates map-based cloning in Arabidopsis. Plant Physiol. 2000;124:1483–1492. doi: 10.1104/pp.124.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 24.Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 25.Swofford DL. Sunderland, Massachusetts: Sinauer Associates; 2003. PAUP* 4.0. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. [Google Scholar]
- 26.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 27.Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 28.Downs K, Zacks DN, Caruso R, Karoukis AJ, Branham K, Yashar BM, Haimann MH, Trzupek K, Meltzer M, Blain D, et al. Molecular testing for hereditary retinal disease as part of clinical care. Arch. Ophthalmol. 2007;125:252–258. doi: 10.1001/archopht.125.2.252. [DOI] [PubMed] [Google Scholar]
- 29.Mani A, Radhakrishnan J, Wang H, Mani A, Mani MA, Nelson-Williams C, Carew KS, Mane S, Najmabadi H, Wu D, et al. LRP6 mutation in a family with early coronary disease and metabolic risk factors. Science. 2007;315:1278–1282. doi: 10.1126/science.1136370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ivanov IP, Atkins JF. Ribosomal frameshifting in decoding antizyme mRNAs from yeast and protists to humans: close to 300 cases reveal remarkable diversity despite underlying conservation. Nucleic Acids Res. 2007;35:1842–1858. doi: 10.1093/nar/gkm035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.