Abstract
DNA sequences capable of forming unusual secondary structures can be a source of genomic instability. In some cases that instability might be affected by transcription, as recently shown for the Z-DNA forming sequence (CG)14, which causes genomic instability both in mammalian cells and in bacteria, and this effect increases with its transcription. We have investigated the effect of this (CG)14 sequence on transcription with T7 RNA polymerase in vitro. We detected partial transcription blockage within the sequence; the blockage increased with negative supercoiling of the template DNA. This effect was not observed in a control self-complementary sequence of identical length and base composition as the (CG)14 sequence, when the purine–pyrimidine alternation required for Z-DNA formation was disrupted. These findings suggest that the inhibitory effect on T7 transcription results from Z-DNA formation in the (CG)14 sequence rather than from an effect of the sequence composition or from hairpin formation in either the DNA or the RNA product.
INTRODUCTION
DNA can assume a variety of unusual secondary structures in addition to the canonical B-form double helix. Among these structures, the Z-DNA conformation deviates strikingly from B-form DNA. Z-DNA is a left-handed helix, with about 12 bp/turn, in contrast to the B-form right-handed helix characterized by 10.5 bp/helical turn (1,2).
The transition from B to Z-DNA generally occurs at sequences with alternating pyrimidines and purines, preferably (CG)n. In addition, several factors are required for Z-DNA to form under physiological ionic conditions, where Z-DNA is less energetically favorable than the canonical B-DNA structure. One of these factors is negative supercoiling, which facilitates formation of DNA structures that are topologically equivalent to B-DNA unwinding, because they partially absorb or ‘relax’ negative superhelical stress. Because Z-DNA is a left-handed helix, its formation within a DNA region is topologically equivalent to unwinding more than the length of that region. Thus, the B–Z transition is strongly facilitated by negative supercoiling, and at a sufficient degree of negative supercoiling Z-DNA can be formed under physiological ionic conditions (3–9). Moreover, at a sufficiently high negative supercoiling, sequences with strong deviations from the purine–pyrimidine alternation can also form Z-DNA (10–12). Typically, a sequence forming Z-DNA is imbedded within DNA sequences which remain in B-form. Because of the very significant structural difference between Z-DNA and B-DNA, there are distortions in DNA base pairing at both junctions between Z-DNA and adjacent B-DNA (13,14). These energetically unfavorable distortions, called B–Z junctions, result in a relationship between the supercoiling required for the B–Z transition and the length of the Z-DNA forming region: the shorter the Z-DNA forming region, the larger the relative contribution of the B–Z junctions to the total energy of the B–Z transition, and therefore, a higher degree of negative supercoiling is required to induce the transition (7,9).
It has been documented that B–Z transitions can occur in living cells and that they affect mutagenesis, transcription initiation and recombination. It was hypothesized that Z-DNA formation in naturally occurring sequences could play a significant role in many biological processes, including regulation of gene expression. This hypothesis is strongly supported by the discovery of proteins that specifically bind Z-DNA (15–17).
One of the important properties of Z-DNA is its ability to interfere with DNA transcription elongation. This interference was shown with in vitro transcription systems both for Escherichia coli (18) and T7 (19) RNA polymerases (RNAP) for the Z-DNA forming sequence (CG)16. However, the mode of interference was evidently different for these two enzymes: E. coli RNAP was completely stalled at the B–Z junction proximal to the promoter (18),while most of the T7 RNAP passed through the sequence, some of the polymerases terminated transcription at either one of two B–Z junctions (19). Wheat Germ RNAP II was shown to transcribe through a poly(CG) sequence when it was in the Z-conformation, although somewhat less efficiently than when the same sequence was in the B-conformation (20). As (CG)n is self-complementary, its effect on transcription might also result from hairpin or cruciform structures. In protein-free supercoiled DNA under near-physiological ionic conditions (CG)n sequences shorter than 60–70 bp are predicted to form Z-DNA rather than cruciform structures (21). However, because a self-complementary sequence could also form a hairpin in the nascent transcript or within the exposed segment of the nontranscribed strand, such structures could potentially be the cause of interference with transcription. This question can be addressed directly by testing self-complementary sequences of the same length and G/C content, but which lack the ability to adopt the Z-DNA structure.
The effect of Z-DNA on transcription elongation is of particular interest from the point of view of the gratuitous transcription-coupled repair (TCR) hypothesis (22). TCR is a special pathway for repair of lesions located on the transcribed DNA strand of expressed genes (23,24). According to the generally accepted model, TCR is initiated by blockage of RNAP at lesions on the template DNA strand (25). The gratuitous TCR hypothesis suggests that stalling of RNAP by other factors, such as unusual DNA structures, might also lead to ‘futile’ DNA repair events that could be mutagenic. The dependence of DNA triplex induced mutagenesis on the specific TCR factor, CSB is consistent with this hypothesis (26). We have shown that an H-DNA triplex forming sequence from the human c-Myc promoter, which is mutagenic in mammalian cells (27), interferes with T7 RNAP transcription elongation in vitro (28). Recently it was shown that the sequence (CG)14, which potentially could form Z-DNA, is also mutagenic in mammalian cells, and that this effect is increased by transcription through that sequence (29). Therefore, we have investigated the effect of this sequence upon transcription in a model in vitro system using the T7 RNAP.
MATERIALS AND METHODS
Reagents
T7 RNAP, RNasin and BSA were purchased from Promega (Madison, WI, USA). T4 polynucleotide kinase, calf thymus DNA topoisomerase I, yeast tRNA, proteinase K and ethidium bromide were purchased from Invitrogen (Carlsbad, CA, USA).
Transcription substrates
The DNA templates used for transcription were closed circular plasmids. Sequences of interest (Figure 1) were cloned into the pUCGTG-TS plasmid (30) 252-bp downstream of the T7 promoter. For the plasmids containing a self-cleaving transcript, a ribozyme sequence (31) was cloned downstream of the inserts of interest with the cleavage site localized 564-bp downstream of the T7 promoter. Plasmids were purified using a HiSpeed Midi Plasmid Purification kit from Qiagen Sciences (MD).
Figure 1.
Sequences used in this study. Only the nontemplate strand is shown (5′-3′). The sequence CG14 is a Z-DNA forming sequence. The palindromic sequence PD was obtained from the sequence CG14 by three G-C permutations (bold, underlined) symmetrical relative to the center of the sequence (shown by a small arrow). The irregular sequence CON was used previously as a control in studies of Z-DNA induced mutagenesis (29), and the sequence TRANS was previously shown to have no effect on transcription elongation (unpublished data) and thus was used as a negative control.
Plasmids with varying levels of negative supercoiling were generated by incubation with calf thymus topoisomerase I in the presence of varying amounts of ethidium bromide (EtBr) in reaction mixtures of 0.25 ml containing 5 µg of template DNA, ∼50 U of topoisomerase, 50 mM Tris–HCl (pH 7.5), 50 mM KCl, 10 mM MgCl2, 0.5 mM DTT, 0.1 mM EDTA and 30 µg/ml BSA for 2 h at 37°C. The reaction mixtures were extracted twice with phenol:chloroform and once with chloroform. DNA in the aqueous phase was recovered by ethanol precipitation and analyzed by electrophoresis on 1% agarose gels in TAE buffer, either in the absence of EtBr and stained later, or run at a concentration of 0.015 µg/ml EtBr in both the gel and the running buffer.
BssHII digestion to monitor Z-DNA formation
One hundred nanograms of template DNA were incubated with 9.6 U of BssHII in a volume of 30 µl. BssHII was diluted with 1X NEBuffer3 (New England Biolabs) to 2 U/µl immediately before the reaction. Reactions were incubated at 37°C for 15 min and placed on ice until gel electrophoresis.
T7 RNAP single-round transcription reactions
Total 10–30 ng of DNA template were incubated at 37°C for 5 min in a mixture of 20 U of T7 RNAP, 40 mM Tris–HCl (pH 7.9), 6 mM MgCl2, 2 mM spermidine, 10 mM NaCl, 10 µCi [α-32P] GTP (specific activity 400 Ci/mM), 10 mM DTT, 16 U of RNasin, 200 µM UTP in a volume of 10 µl. Elongation proceeded until the end of the sequence GGGU immediately downstream from the promoter, at which point the first CTP was required for insertion. Heparin was added at 50 ng/µl to inactivate any unbound T7 RNAP. Resumed elongation by the stalled T7 RNAP complexes was induced by adding ATP, GTP and CTP to final concentrations 170 µM in a final volume of 12 µl. After 30 min, or less as indicated, reactions were stopped by the addition of 94 µl of mixture containing10 µg of proteinase K, 25 µg of tRNA, 1.1% SDS, 106 mM Tris–HCl (pH 7.5), 13.2 mM EDTA and 160 mM NaCl and incubated at room temperature for 15 min. The nucleic acids were precipitated with ethanol and dried under vacuum. The samples were resuspended in formamide dye and denatured at 90°C for 4 min prior to loading gels for electrophoresis. The transcription products were resolved on 5% denaturing polyacrylamide gels in Tris–borate EDTA containing 8.3 M urea. The gels were dried and exposed either to X-ray film or to a phosphoimager screen.
T7 RNAP multiple-round transcription reactions
Reaction conditions were the same as for the T7 RNAP single-round transcription reactions except that the nonradioactive nucleotide concentrations were 170 µM UTP, 170 µM CTP, 170 µM ATP and 17 µM GTP in a final volume of 1 µl. The initial incubation used for the single-round reaction was not performed, and all reactions were incubated for 30 min at 37°C except where shorter incubation periods are noted. Heparin was omitted to permit multiple initiations of transcription.
Estimation of probabilities of blockage
The probability of blockage p was estimated as
where Iz and Ir are intensities of the truncated transcription product within the Z-DNA forming sequence and the ribozyme cleavage product, respectively; nz and nr are numbers of radioactive nucleotides within corresponding products.
For continuous (multiple-round) transcription we could assume that the transcription products are uniformly labeled, and thus n would be proportional to the length of the corresponding product. Thus the length of the products could be substituted in the above equation instead of n.
For single-round transcription n could be estimated as
where n0 is the number of radioactive nucleotides in the initially labeled starting sequence (in our case n0 = 3), L is the length of the product and m is the ratio of concentrations of nonradioactive nucleotide to the radioactive nucleotide in the final elongation reaction (i.e. after addition of heparin and extra nonradioactive nucleotides). We estimated m as ∼100.
RESULTS
Partial transcription blockage within the Z-DNA forming sequence
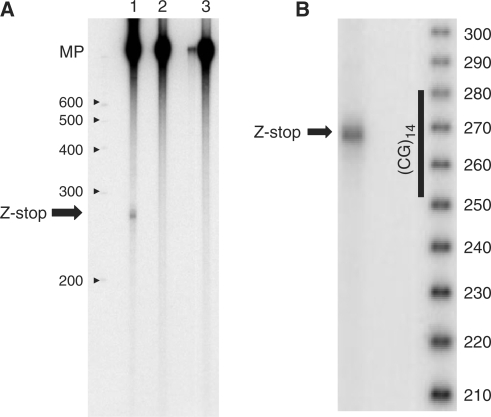
Some truncated transcription products were observed during in vitro transcription of the supercoiled plasmid pCG14, which contains the Z-DNA forming insert (CG)14 (Figure 2A, lane 1), in addition to the major transcription product (MP), indicating partial transcription blockage. For the control plasmids pCON and pTRANS, which have the same flanking regions as pCG14, but lack the Z-DNA forming insert (Figure 1), no truncation product was detected (Figure 2A, lanes 2 and 3, respectively), showing that the effect is sequence specific. By comparison with the denatured DNA size standards, we estimate that this blockage occurs roughly in the middle of the Z-DNA forming sequence (Figure 2B).
Figure 2.
Partial transcription blockage within a Z-DNA forming sequence. The arrow ‘Z-stop’ indicates the position of the RNA truncation product. MP (major product) is the ‘complete’ transcript from a circular DNA template. (A) The blockage is observed in the presence of the (CG)14 insert, but not in the control sequences. Lane 1: plasmid with the Z-DNA forming sequence pCG14. Lanes 2 and 3: control plasmids pCON and pTRANS, respectively. Transcription was single round in this experiment. The positions of the denatured DNA size markers are shown; the numbers indicate the lengths of the marker fragments (nucleotides). (B) More precise mapping of the blockage site position. Numbers indicate the lengths of denatured DNA marker fragments (nucleotides).
The intensity of the transcription blockage increases with negative supercoiling
Z-DNA is stabilized by negative supercoiling; if the transcription blockage were due to Z-DNA formation, its intensity should increase with increased negative supercoiling. To test this, we prepared samples of the pCG14 plasmid with increasing superhelical densities (Figure 3) and transcription from these samples was analyzed (Figure 4A and B). The results show that the intensity of blockage increases with increased negative supercoiling, supporting the hypothesis of Z-DNA involvement in the transcription blockage.
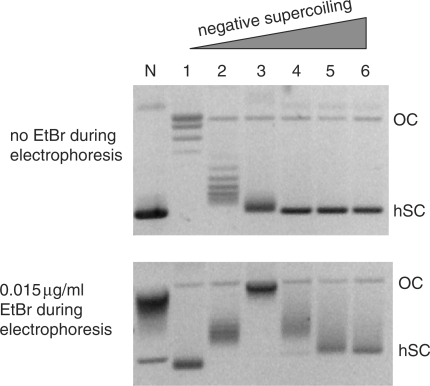
Figure 3.
Characterization of pCG14 plasmids with various levels of negative supercoiling. Lane N corresponds to plasmid with native supercoiling. Lanes 1–6 show plasmid samples treated with calf thymus Topo I in the presence of increasing amounts of EtBr and consequently having increased negative supercoiling. The concentrations of EtBr during incubation were 0 μg/ml, 1.25 μg/ml, 2.5 μg/ml, 5 μg/ml, 7.5 μg/ml and 10 μg/ml for lanes 1, 2, 3, 4, 5 and 6, respectively. To resolve the samples at lower superhelical densities, gel electrophoresis was performed in the absence of EtBr (upper panel). To resolve the samples at higher superhelical densities, gel electrophoresis was performed in the presence of 0.015 μg/ml EtBr (lower panel). OC (open circles) designates nicked plasmids which migrate close to relaxed circular closed plasmid, and hSC designates highly supercoiled plasmids. In the absence of EtBr (upper panel), the higher the negative superhelical density of the plasmid, the faster it migrates in the gel. The minimum mobility corresponds to relaxed plasmid (lane 1). Starting from some superhelical density, the mobility reaches saturation, and the plasmid samples are not resolved (lanes N, 4, 5 and 6). Binding of EtBr to the circular closed plasmid introduces positive supercoiling into the plasmid, the magnitude of which depends on EtBr concentration. In the presence of EtBr (lower panel), plasmid in lane 3 has the lowest mobility, close to the mobility of open circles (OC), which means that the negative supercoiling in this plasmid is almost exactly compensated by positive supercoiling introduced by EtBr. Plasmids with less negative supercoiling than those in lane 3 (lanes 1 and 2) in the presence of EtBr become positively supercoiled, and plasmids with higher negative supercoiling than for lane 3 (lanes N, 4, 5 and 6) in the presence of EtBr remain negatively supercoiled, but with somewhat lower superhelical density, which allows better resolution. It can be seen that the native superhelical density (lane N) is close to the superhelical density of the plasmid in lane 3.
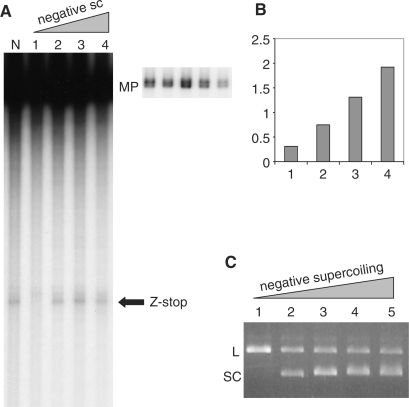
Figure 4.
Transcription blockage within the CG14 sequence increases with increased negative supercoiling. (A) Gel electrophoresis of the transcription products. The arrow designated ‘Z-stop’ shows the position of the blockage within the Z-DNA forming sequence. MP designates ‘major’ transcription products from the circular plasmid. On the right panel a shorter exposure of the region of the gel containing the MP band is shown. Lane N corresponds to the plasmid with native supercoiling. Lanes 1, 2, 3 and 4 correspond to the same plasmid samples with increasing negative superhelical densities, as shown in Figure 3, lanes 1, 3, 4 and 6, respectively. (B) Relative intensities of truncation products from A. The intensities of blockage bands (Z-stop) for lanes N, 1, 2, 3 and 4 were normalized to the intensities of corresponding MP bands. The results are presented as the ratios of normalized intensities for lanes 1, 2, 3 and 4 to the normalized intensity for the lane N. Transcription was single round in this experiment. (C) BssHII digest of the plasmid samples with increasing negative supercoiling. Lanes 1, 2, 3, 4 and 5 correspond to the samples shown in Figure 3, lanes 1, 3, 4, 5 and 6, respectively. L designates linear plasmid, SC designates supercoiled plasmids.
To demonstrate that Z-DNA really forms within the range of supercoiling at which we observe partial transcription blockage, we treated samples of the pCG14 plasmid at various superhelical densities with the restriction enzyme BssHII, which cleaves the sequence GCGCGC when it is in the B-conformation, but not when in the Z-conformation (32). Figure 4C shows that the relaxed DNA (lane 1) is completely converted to the linear form (L) due to BssHII cleavage, while for the supercoiled samples (lanes 2–5) a significant amount (more than 50%) of the plasmid remains in the circular closed supercoiled (SC) state, indicating inhibition of BssHII cleavage and thus Z-DNA formation within the same diapason of supercoiling for which transcription blockage occurs. Note that this analysis gives an underestimate of the portion of Z-DNA in the sample, because even under the conditions for which the B–Z equilibrium within the (CG)14 sequence is strongly shifted toward Z-DNA formation, the sequence spends a nonnegligible amount of time in the B-form where it could be cleaved by the enzyme.
Using a ribozyme sequence to estimate the amount of blockage
Since Z-DNA formation requires negative supercoiling, our transcription assay had to be performed on a circular DNA substrate. The major transcription product (MP) for these substrates could be used as a normalization factor to compare the relative probabilities of transcription blockage under various conditions, but it could not be used to estimate absolute probabilities of the blockage (i.e. the molar percentage of the truncated product; see the last subsection of Materials and Methods section), because its length is unknown, and in our experiments the radioactive signal in the transcription product is proportional to the length of the product RNA.
To overcome this problem, we cloned a self-cleaving ribozyme sequence (31,33) about 300-bp downstream from the Z-DNA forming sequence. This sequence cleaves itself with 80–90% efficiency (28,31,33), providing a well-defined band of known length (Figure 5). Using this approach, we estimated that the probability of blockage at Z-DNA in natively supercoiled substrates was ∼4%. The probabilities were similar for single-round and multiple-round transcription reactions, suggesting that interactions between multiple RNAPs do not play a major role in the observed effects. Note that the small apparent probability of the blockage is not due to the presence of Z-DNA in only a small fraction of the plasmid sample, because the analysis in Figure 4C shows that at negative supercoiling levels close to native, more than 50% of the plasmids contain Z-DNA. Thus, in most of the transcription events, T7 RNAP passes the sequence in Z-conformation without formation of the truncated products (see Discussion section).
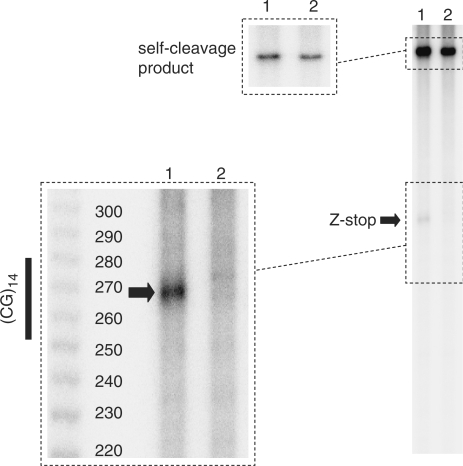
Figure 5.
Use of a ribozyme-containing plasmid to determine the probability of transcription blockage. The plasmid map on the left side of the figure shows the positions of the T7 promoter, the Z-DNA forming sequence and the ribozyme cleavage site. The numbers in parentheses show the distance from the T7 promoter. The gel image on the right side of the figure shows truncated transcripts due to blockage within the Z-DNA forming sequence, and those due to ribozyme self-cleavage. Lanes 1 and 2 correspond to pCG14 plasmids with (pCG14ribo) and without the ribozyme insert. Transcription was single round in this experiment.
A self-complementary (palindromic) sequence of the same length and base composition as (CG)14, but with disrupted purine–pyrimidine alternation, does not produce pronounced transcription blockage
Because CG-repeats are self-complementary, in addition to Z-DNA they could form cruciform structures in supercoiled DNA and hairpins both in DNA and in nascent RNA. To address the question of whether these other structures contribute to the effect on transcription, we designed a palindromic sequence PG (Figure 1). This sequence was obtained from (CG)14 by three C-G permutations that are symmetrical relative to the center of the sequence. These permutations preserve the self-complementarity required for cruciform and hairpin formations, but disrupt purine–pyrimidine alternation required for Z-DNA formation. We observed (Figure 6, lane 2) that this sequence does not have a pronounced effect on transcription, supporting the hypothesis that Z-DNA, rather than hairpin or cruciform structures are responsible for the observed effects.
Figure 6.
The transcription blockage band is observed in the Z-DNA forming sequence but not in the palindromic sequence with the same base composition but with disrupted purine–pyrimidine alternation. Lanes 1 and 2 correspond to the plasmids with sequences CG14 and PD, respectively. Both plasmids contain the ribozyme self-cleavage sequence, and the self-cleavage product is shown at the top of the gel. It can be seen that the blockage band (Z-stop) is pronounced only in the presence of the CG14 sequence. The panel at left shows the detail of the region in the vicinity of the Z-stop band, and the denatured 10-bp DNA ladder. The size of the truncation products suggests that the Z-stop is localized within the Z-DNA forming sequence. Transcription was multiple round in this experiment.
DISCUSSION
In the present work we observed partial transcription blockage induced by the (CG)14 sequence with T7 RNA polymerase. This blockage increased with increasing negative supercoiling in the template DNA. The dependence of the observed effect upon negative supercoiling implicates an unusual DNA structure, rather than some pausing signal, because sequence-specific pausing or termination signals do not require negative supercoiling, while non B-form DNA structures, like Z-DNA or cruciforms, are strongly dependent upon negative supercoiling. The blockage was not pronounced when the purine–pyrimidine alternation in this sequence, which is required for Z-DNA formation, was disrupted without changing self-complementarity, length or base composition (see Figure 1, the sequence PD), i.e. the factors which define the ability to form cruciforms. This suggests that the Z-DNA, rather than the cruciform is responsible for the observed effect. In addition, this control sequence can form an RNA hairpin of the same length and base composition as (CG)14. Thus, if the pausing or termination were due to the hairpin formation in the transcript, as is the case for many pausing and termination signals (34), it should be similar for (CG)14 and PD. Moreover, the PD sequence also has an undisrupted (CG)6 sequence in the middle (Figure 1). Hairpin-independent transcription termination signals are usually shorter than 10 bp [for example, see (35)]. Thus, if the (CG) motif is some unknown hairpin-independent pausing or transcriptional termination signal, then again the (CG)14 and PD would be expected to have similar effects on transcription. In contrast, the probability of Z-DNA formation is strongly related to the length of the undisrupted (CG)-motif (7,9), and consequently Z-DNA formation is much more favored in the sequence (CG)14 than in the sequence PD containing only (CG)6. Thus, the effect on transcription of the (CG)14 sequence, and the absence of a pronounced effect of the PD sequence, together with the dependence upon negative supercoiling strongly implicates the Z-DNA formation in the observed effect.
What is the mechanism of partial transcription blockage by Z-DNA?
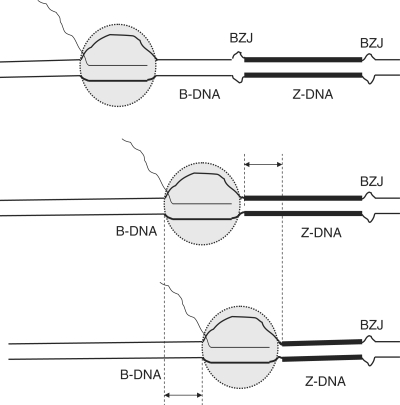
Consider RNAP moving toward a DNA sequence in the Z-conformation (Figure 7, from top to bottom). When RNAP reaches a DNA sequence in the Z-conformation, it partially ‘displaces’ the Z-DNA from part of the sequence (shown by a double-headed arrow), while the region of the same length that was unwound within the transcription complex is ‘restored’ to normal B-structure behind the RNAP. At sufficiently high negative superhelical stress, Z-DNA is more energetically favorable than B-DNA. Thus, the process of moving RNAP into the sequence in the Z-conformation is energetically unfavorable. This could increase the probability of transcription pausing or terminating upon RNAP movement into this sequence. The higher the negative supercoiling, the more energetically favorable would be Z-DNA in comparison with B-DNA, and, consequently, the larger the energetic barrier for RNAP movement into the sequence in Z-conformation. However, even at relatively high negative supercoiling, which is sufficient to convert the (CG)14 insert into a Z-conformation with more than 50% probability (see Figure 4 C), the absolute value of the probability of blockage is still low (around 4%; see Figure 5), thus most of the RNAPs pass through Z-DNA forming sequences.
Figure 7.
Possible mechanism for partial transcription blockage within the Z-DNA forming sequence. BZJ stands for B–Z junctions. B-DNA is shown by thin lines, and Z-DNA is shown by thick lines. When RNAP (depicted as a gray circle) moves into the sequence in the Z-conformation, it disrupts a region in that conformation (shown by a double-headed arrow), while behind the polymerase, a region of the same length is restored to the B-conformation.
The position of the blockage site inside the Z-DNA sequence is consistent with this model, although the model does not predict the exact position of the predominant blockage. We did not observe two pronounced blockage bands at B–Z junctions, as previously reported for a slightly longer sequence (CG)16 (19). Using multiple rounds of transcription as well as decreasing the time of transcription from 30 to 1 min (data not shown) had no effect on the position of the blockage and did not result in the production of additional truncation products. The position of the blockage site(s) might depend on the length of the Z-DNA forming insert and on the flanking sequences; further experiments will be required to investigate these issues. It is likely that in addition to the simple energetic factor considered above, other factors based on the structural difference between Z-DNA and B-DNA could be involved in the effect of Z-DNA on transcription (19). Another interesting possibility is the contribution from the short hairpin formed in the transcript from the first half of the CG-sequence. Our experiment with the control sequence PD (Figure 6) showed that the hairpin by itself does not cause a stop. However, it could contribute to the blockage, making the mechanism similar to the usual hairpin-dependent termination signal, where instead of the formation of an energetically unfavorable rU/dA duplex downstream from the hairpin-forming sequence there is an energetically unfavorable ‘invasion’ into the rest of the Z-DNA.
Other structures which could affect the transcription are R-loops. The R-loops which are formed by G-rich transcripts are unusually stable (36,37), and could strongly interfere with transcription elongation (38). According to recent data, stable R-loop formation depends upon the presence of G-stretches rather than GC-content (37); thus, if R-loop formation were solely responsible for the observed effect, then it would be stronger in the control sequence PD which has one G7 stretch. However, it cannot be excluded that the R-loop formation in part of the Z-DNA forming sequence might contribute to the effect.
Recently in was shown that the (CG)14 sequence studied here can induce double-strand breaks (resulting in large deletions) in mammalian cells, and the effect is increased by transcription through this sequence, while the control sequence (CON, Figure 1) with a random nucleotide composition did not show this effect (29). In our experiments, the (CG)14 sequence, but not the control sequence CON, partially inhibited T7 transcription. As already mentioned, a self-complementary sequence with the same length and base composition as (CG)14, but with base permutations which disrupt the CG alternation required for Z-DNA formation did not have a pronounced effect on transcription. It would be interesting to investigate the effect of similar permutations in Z-DNA forming sequences on genomic instability.
ACKNOWLEDGEMENTS
This work was supported by National Cancer Institute (CA77712 to P.C.H.; CA93729 to K.M.V.). We thank Ed Grabczyk for help with ribozyme constructions, Ann Ganesan, Allen Smith and Graciela Spivak for critical reading of the manuscript. Funding to pay the Open Access publication charges for this article was provided from the National Cancer Institute, of the National Institutes of Health in the Department of Health and Human Services, USA.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wang AH, Quigley GJ, Kolpak FJ, Crawford JL, van Boom JH, van der Marel G, Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979;282:680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- 2.Herbert A, Rich A. Left-handed Z-DNA: structure and function. Genetica. 1999;106:37–47. doi: 10.1023/a:1003768526018. [DOI] [PubMed] [Google Scholar]
- 3.Singleton CK, Klysik J, Stirdivant SM, Wells RD. Left-handed Z-DNA is induced by supercoiling in physiological ionic conditions. Nature. 1982;299:312–316. doi: 10.1038/299312a0. [DOI] [PubMed] [Google Scholar]
- 4.Nordheim A, Lafer EM, Peck LJ, Wang JC, Stollar BD, Rich A. Negatively supercoiled plasmids contain left-handed Z-DNA segments as detected by specific antibody binding. Cell. 1982;31:309–318. doi: 10.1016/0092-8674(82)90124-6. [DOI] [PubMed] [Google Scholar]
- 5.Haniford DB, Pulleyblank DE. Facile transition of poly[d(TG) x d(CA)] into a left-handed helix in physiological conditions. Nature. 1983;302:632–634. doi: 10.1038/302632a0. [DOI] [PubMed] [Google Scholar]
- 6.Frank-Kamenetskii MD, Vologodskii AV. Thermodynamics of the B-Z transition in superhelical DNA. Nature. 1984;307:481–482. doi: 10.1038/307481a0. [DOI] [PubMed] [Google Scholar]
- 7.Mirkin SM, Lyamichev VI, Kumarev VP, Kobzev VF, Nosikov VV, Vologodskii AV. The energetics of the B-Z transition in DNA. J. Biomol. Struct. Dyn. 1987;5:79–88. doi: 10.1080/07391102.1987.10506376. [DOI] [PubMed] [Google Scholar]
- 8.Johnston BH, Rich A. Chemical probes of DNA conformation: detection of Z-DNA at nucleotide resolution. Cell. 1985;42:713–724. doi: 10.1016/0092-8674(85)90268-5. [DOI] [PubMed] [Google Scholar]
- 9.Peck LJ, Wang JC. Energetics of B-to-Z transition in DNA. Proc. Natl Acad. Sci. USA. 1983;80:6206–6210. doi: 10.1073/pnas.80.20.6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brahms S, Vergne J, Brahms JG, Di Capua E, Bucher P, Koller T. Natural DNA sequences can form left-handed helices in low salt solution under conditions of topological constraint. J. Mol. Biol. 1982;162:473–493. doi: 10.1016/0022-2836(82)90539-3. [DOI] [PubMed] [Google Scholar]
- 11.Pohl FM, Thomae R, DiCapua E. Antibodies to Z-DNA interact with form V DNA. Nature. 1982;300:545–546. doi: 10.1038/300545a0. [DOI] [PubMed] [Google Scholar]
- 12.Johnston BH. Chemical probing of the B-Z transition in negatively supercoiled DNA. J. Biomol. Struct. Dyn. 1988;6:153–166. doi: 10.1080/07391102.1988.10506488. [DOI] [PubMed] [Google Scholar]
- 13.Singleton CK, Klysik J, Wells RD. Conformational flexibility of junctions between contiguous B- and Z-DNAs in supercoiled plasmids. Proc. Natl Acad. Sci. USA. 1983;80:2447–2451. doi: 10.1073/pnas.80.9.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ha SC, Lowenhaupt K, Rich A, Kim YG, Kim KK. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature. 2005;437:1183–1186. doi: 10.1038/nature04088. [DOI] [PubMed] [Google Scholar]
- 15.Wang G, Vasquez KM. Z-DNA, an active element in the genome. Front. Biosci. 2007;12:4424–4438. doi: 10.2741/2399. [DOI] [PubMed] [Google Scholar]
- 16.Wells RD. Non-B DNA conformations, mutagenesis and disease. Trends Biochem. Sci. 2007;32:271–278. doi: 10.1016/j.tibs.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 17.Rich A, Zhang S. Timeline: Z-DNA: the long road to biological function. Nat. Rev. Genet. 2003;4:566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- 18.Peck LJ, Wang JC. Transcriptional block caused by a negative supercoiling induced structural change in an alternating CG sequence. Cell. 1985;40:129–137. doi: 10.1016/0092-8674(85)90316-2. [DOI] [PubMed] [Google Scholar]
- 19.Droge P, Pohl FM. The influence of an alternate template conformation on elongating phage T7 RNA polymerase. Nucleic Acids Res. 1991;19:5301–5306. doi: 10.1093/nar/19.19.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durand R, Job C, Zarling DA, Teissere M, Jovin TM, Job D. Comparative transcription of right- and left-handed poly[d(G-C)] by wheat germ RNA polymerase II. EMBO J. 1983;2:1707–1714. doi: 10.1002/j.1460-2075.1983.tb01646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anshelevich VV, Vologodskii AV, Frank-Kamenetskii MD. A theoretical study of formation of DNA noncanonical structures under negative superhelical stress. J. Biomol. Struct. Dyn. 1988;6:247–259. doi: 10.1080/07391102.1988.10507711. [DOI] [PubMed] [Google Scholar]
- 22.Hanawalt PC. Transcription-coupled repair and human disease. Science. 1994;266:1957–1958. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- 23.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 24.Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 25.Hanawalt PC. Subpathways of nucleotide excision repair and their regulation. Oncogene. 2002;21:8949–8956. doi: 10.1038/sj.onc.1206096. [DOI] [PubMed] [Google Scholar]
- 26.Wang G, Seidman MM, Glazer PM. Mutagenesis in mammalian cells induced by triple helix formation and transcription-coupled repair. Science. 1996;271:802–805. doi: 10.1126/science.271.5250.802. [DOI] [PubMed] [Google Scholar]
- 27.Wang G, Vasquez KM. Naturally occurring H-DNA-forming sequences are mutagenic in mammalian cells. Proc. Natl Acad. Sci. USA. 2004;101:13448–13453. doi: 10.1073/pnas.0405116101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belotserkovskii BP, De Silva E, Tornaletti S, Wang G, Vasquez KM, Hanawalt PC. A triplex-forming sequence from the human c-MYC promoter interferes with DNA transcription. J. Biol. Chem. 2007;282:32433–32441. doi: 10.1074/jbc.M704618200. [DOI] [PubMed] [Google Scholar]
- 29.Wang G, Christensen LA, Vasquez KM. Z-DNA-forming sequences generate large-scale deletions in mammalian cells. Proc. Natl Acad. Sci. USA. 2006;103:2677–2682. doi: 10.1073/pnas.0511084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tornaletti S, Patrick SM, Turchi JJ, Hanawalt PC. Behavior of T7 RNA polymerase and mammalian RNA polymerase II at site-specific cisplatin adducts in the template DNA. J. Biol. Chem. 2003;278:35791–35797. doi: 10.1074/jbc.M305394200. [DOI] [PubMed] [Google Scholar]
- 31.Grabczyk E, Usdin K. The GAA*TTC triplet repeat expanded in Friedreich's ataxia impedes transcription elongation by T7 RNA polymerase in a length and supercoil dependent manner. Nucleic Acids Res. 2000;28:2815–2822. doi: 10.1093/nar/28.14.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Azorin F, Hahn R, Rich A. Restriction endonucleases can be used to study B-Z junctions in supercoiled DNA. Proc. Natl Acad. Sci. USA. 1984;81:5714–5718. doi: 10.1073/pnas.81.18.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grabczyk E, Usdin K. Alleviating transcript insufficiency caused by Friedreich's ataxia triplet repeats. Nucleic Acids Res. 2000;28:4930–4937. doi: 10.1093/nar/28.24.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landick R. RNA polymerase slides home: pause and termination site recognition. Cell. 1997;88:741–744. doi: 10.1016/s0092-8674(00)81919-4. [DOI] [PubMed] [Google Scholar]
- 35.He B, Kukarin A, Temiakov D, Chin-Bow ST, Lyakhov DL, Rong M, Durbin RK, McAllister WT. Characterization of an unusual, sequence-specific termination signal for T7 RNA polymerase. J. Biol. Chem. 1998;273:18802–18811. doi: 10.1074/jbc.273.30.18802. [DOI] [PubMed] [Google Scholar]
- 36.Reaban ME, Lebowitz J, Griffin JA. Transcription induces the formation of a stable RNA. DNA hybrid in the immunoglobulin alpha switch region. J. Biol. Chem. 1994;269:21850–21857. [PubMed] [Google Scholar]
- 37.Roy D, Yu K, Lieber MR. Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol. Cell. Biol. 2008;28:50–60. doi: 10.1128/MCB.01251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krasilnikova MM, Samadashwily GM, Krasilnikov AS, Mirkin SM. Transcription through a simple DNA repeat blocks replication elongation. EMBO J. 1998;17:5095–5102. doi: 10.1093/emboj/17.17.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]