Abstract
Expansions of trinucleotide repeats cause at least 15 heritable human diseases. Single-stranded triplet repeat DNA in vitro forms stable hairpins in a sequence-dependent manner that correlates with expansion risk in vivo. Hairpins are therefore considered likely intermediates during the expansion process. Unwinding of a hairpin by a DNA helicase would help protect against expansions. Yeast Srs2, but not the RecQ homolog Sgs1, blocks expansions in vivo in a manner largely dependent on its helicase function. The current study tested the idea that Srs2 would be faster at unwinding DNA substrates with an extrahelical triplet repeat hairpin embedded in a duplex context. These substrates should mimic the relevant intermediate structure thought to occur in vivo. Srs2 was faster than Sgs1 at unwinding several substrates containing triplet repeat hairpins or another structured loop. In contrast, control substrates with an unstructured loop or a Watson–Crick duplex were unwound equally well by both enzymes. Results with a fluorescently labeled, three-way junction showed that Srs2 unwinding proceeds unabated through extrahelical triplet repeats. In summary, Srs2 maintains its facile unwinding of triplet repeat hairpins embedded within duplex DNA, supporting the genetic evidence that Srs2 is a key helicase in Saccharomyces cerevisiae for preventing expansions.
INTRODUCTION
Expansions of certain crucial DNA trinucleotide repeats (TNRs) in humans are the mutagenic cause of some 15 hereditary neurological diseases (1,2). The risk of an expansion is closely tied to cis-acting features such as the sequence of the TNR. In 14 of the 15 diseases, the repeat is CNG where N represents any of the four nucleotides. In vitro studies showed a close correlation between the ability of single-stranded TNR DNA to form aberrant structures, usually a hairpin, and the likelihood of that sequence undergoing an expansion in vivo (3). Numerous biological studies support the central role of a TNR hairpin in the mutagenic process (4–9). Models of the expansion process suggest that TNR hairpins are embedded within duplex DNA (1,2), possibly making a hairpin more difficult to access and/or remove by cellular proteins. These considerations indicate that a hairpin within a duplex context is likely the relevant intermediate structure on the way to an expansion.
The hairpin model predicts that cellular proteins could help avoid expansions by either preventing hairpin formation or accelerating its removal. One such protein is the yeast DNA helicase Srs2, which was identified by a mutant screen to identify proteins that reduce expansion risk (10). Genetic studies indicate that Srs2 selectively blocks TNR expansions in yeast, largely through its DNA helicase function, based on the finding that a helicase-deficient point mutant of Srs2 was nearly as defective as a null allele at blocking expansions (10). In contrast, mutation of the related helicase Sgs1 was without phenotype in CTG·CAG repeat expansion assays, and a high-copy plasmid with wild-type SGS1 was unable to rescue the elevated expansion rates in an srs2 mutant (10). Another study found that loss of Sgs1 actually stabilized CCG·CGG tracts (11). Despite other genetic and biochemical similarities (12–14), Srs2 and Sgs1 are clearly distinguishable by their triplet repeat phenotypes.
The srs2 strains exhibit an unusually specific mutator phenotype. Expansions of hairpin-forming TNRs occur at higher rates in the absence of Srs2. In contrast, there was no change in the rates of CTG repeat contractions of dinucleotide repeats or of forward mutations in the CAN1 reporter gene (10). Thus, Srs2 in vivo selectively blocks expansions. Biochemical experiments showed that TNR sequences in duplex substrates or in an intramolecular hairpin were unwound faster by Srs2 than by Sgs1 or by Escherichia coli DNA helicase II, whereas all three enzymes were equally active on control substrates (15). Together, this information suggests that Srs2 is inherently fast at unwinding TNR-mediated structure, and that this property may underlie Srs2's selectivity for preventing TNR expansions. However, the previous biochemical assays used simple substrates where the helicase encountered triplet repeat sequences immediately upon entering the duplex (15). Essentially, this tested the ‘standing start’ ability of the helicases to unwind TNR sequences as the first duplex DNA they encountered. The previous work also found that when triplet repeats were distal to the helicase entry point, the kinetic advantage of Srs2 over Sgs1 was diminished (15). In vivo, a helicase loading site might be positioned at some distance from the TNR hairpin, requiring the enzyme to proceed through Watson–Crick duplexes first before encountering the aberrant, extrahelical structure. Thus, a test of ‘running start’ helicase activity is informative to see if Srs2 retains its kinetic advantage under these conditions.
The purpose of this study was to provide information about the DNA helicase activity of Srs2 on more biologically relevant model substrates containing a triplet repeat hairpin embedded in a duplex region (1,2). There is only limited information available for unwinding of such substrates (16,17), and the availability of purified Srs2 and Sgs1 provides the opportunity for comparative helicase studies under ‘running start’ conditions. Evidence is presented that Srs2 retains its kinetic advantage over Sgs1 in unwinding embedded triplet repeat hairpins, primarily by proceeding unabated through the structured extrahelical segment.
MATERIALS AND METHODS
Helicase substrates
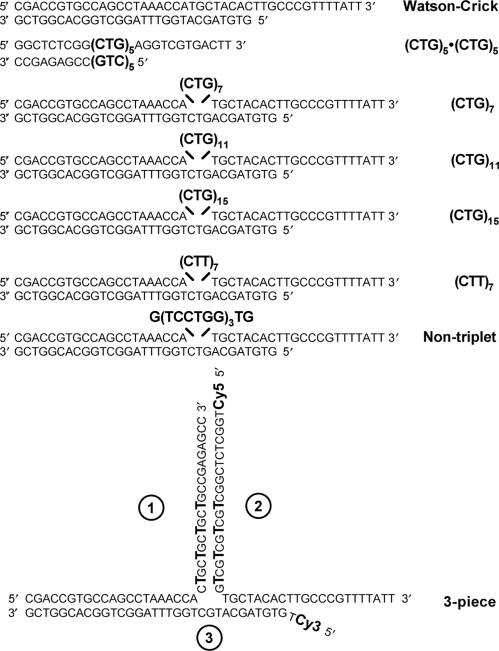
The substrates used for helicase assays are listed in Figure 1. Single-stranded oligonucleotides were purified on denaturing polyacrylamide gels prior to use. For duplex substrates, the shorter strand was end-labeled with 32P using T4 polynucleotide kinase (New England Biolabs, Beverly, MA, USA) according to manufacturer's protocol. The labeled strand was mixed at equimolar concentrations with the unlabeled longer strand, heated at 95°C for 5 min, annealed at 70°C for 10 min and slowly cooled to room temperature to generate double-stranded DNA. To confirm that the strands annealed properly and formed the predicted hairpin structure, the (CTG)7, (CTT)7 and nontriplet duplexes were subjected to digestion with mung bean nuclease (New England Biolabs). The digestion was done according to manufacturer's protocol and the products were analyzed on high resolution denaturing polyacrylamide gels as described previously (10). For the three-piece substrate (Figure 1), the three oligonucleotides (1, 2 and 3) were mixed at equimolar concentrations, heated at 95°C for 5 min, followed by initial annealing at 78°C for 20 min and a second annealing at 67°C for 10 min, and slow cooling to room temperature.
Figure 1.
Substrates for helicase assays in this study. All duplexes except the three-piece were radiolabeled on the 5′-end of the shorter strand. Equimolar amounts of two strands were mixed and annealed. Trinucleotide repeats and inserted sequences are indicated by the larger, bold font. In all cases, the DNA duplexes ran as discrete single bands on nondenaturing polyacrylamide gels, consistent with a well-defined structure. The three-piece substrate is obtained by annealing equimolar amounts of oligonucleotides 1, 2 and 3. Base-base mismatches are indicated by the larger, bold font. Oligonucleotides 2 and 3 are labeled with Cy5 or Cy3, respectively, at the 5′end of the terminal thymidine residue.
Protein expression and purification
The expression plasmid p11c::His9-Srs2 was a generous gift from Patrick Sung (Yale University). The plasmid encodes Srs2 with an N-terminal 9-histidine tag. His-tagged Srs2 was purified to homogeneity using a slightly modified five-step procedure. The crude extract was prepared using a French press and subsequently processed by ammonium sulfate precipitation and Q-sepharose and SP-sepharose chromatography as described (18). As identified by immunoblotting and silver staining of SDS–polyacrylamide gels, Srs2 eluted from the SP-sepharose column in about 360 mM KCl. The Srs2 pool was purified on Ni-NTA agarose column previously equilibrated with K buffer (20 mM KH2Po4, pH 7.5, 10% glycerol, 0.5 mM EDTA, 0.01% Igepal, 1 mM 2-mercaptoethanol) with 0.5 M KCl and 10 mM imidazole. Srs2 was eluted with 16 ml of 300 mM imidazole in K buffer containing 0.5 M KCl. The fractions containing Srs2 protein were diluted in K buffer to yield a protein solution that had a conductivity equivalent to 150 mM KCl. The protein solution is applied onto a 1 ml Mono S Column (GE Healthcare, Buckinghamshire, UK) and fractionated with a 20 ml gradient of 195 to 575 KCl in K buffer. Srs2 eluted from the Mono S column in 375 mM KCl. The final Srs2 pool is concentrated to about 0.5 mg/ml and stored in 2 µl aliquots at −80°C. Expression of 6-His-tagged Sgs1 (amino acids 400–1268 of 1447 full-length protein) was induced by galactose and the protein was purified using nickel chelate chromatography (14). Purified fractions were eluted from Ni-NTA resin column with a buffer (20 mM Tris–HCl, pH 7.4, 200 mM NaCl) including 80 mM imidazole.
Helicase assays
Most helicase assays measure the unwinding of 32P-labeled DNA from duplex substrates and the products were separated by 12% nondenaturing polyacrylamide gels, followed by phosphorimaging. For the three-piece substrate, visualization utilized fluorescence imaging. The radiolabeled or fluorescently labeled DNA substrates (0.5 nM) were incubated with enzyme (10 nM unless otherwise specified) for a given period of time, as indicated in the figure legends, under standard helicase assay conditions. Srs2 helicase assays were performed at 30°C in the presence of 25 mM Tris–HCl (pH 7.5), 1 mM dithiothreitol (DTT), 100 µg/ml bovine serum albumin and 2.5 mM ATP. The reactions were terminated with 0.3% SDS, 10mM EDTA, 5% glycerol and 0.1% bromophenol blue. Sgs1 helicase assay were performed at 30°C in the presence of 20 mM Tris–HCl (pH 7.5), 2 mM MgCl2, 2 mM ATP, 2 mM DTT and 100 µg/ml bovine serum albumin. After the incubation at 30°C, the reactions were terminated with 0.5% (w/v) Proteinase K, 100 mM Tris–HCl (pH 7.5), 200 mM EDTA and 2.5% (w/v) SDS, and the mixtures were incubated at 37°C for an additional 10 min. To help prevent reannealing of the products, a 10-fold molar excess of unlabeled oligonucleotide was added as described previously (15). All assays were performed two to three times with similar results.
RESULTS
Rationale and substrate design
In the current study, we tested the idea that Srs2 would be faster at unwinding complex DNA substrates with a TNR hairpin embedded in a duplex region. ‘Running start’ substrates used in the current study more closely mimic the hairpin intermediate thought to occur in vivo (1,2), and which Srs2 would therefore need to resolve to block expansions. (‘Standing start’ and ‘running start’ substrates are used in the DNA polymerase field to distinguish the activities of translesion polymerases when they encounter template lesions. These substrates are useful tools to help dissect enzymatic mechanisms, so there is precedent for both the concept and the nomenclature.) Sgs1 provides a useful comparison because both enzymes unwind in the 3′ to 5′ direction (13,14) and because both proteins modulate genetic recombination (12). However, the two enzymes are clearly not interchangeable for TNR stabilization. Defects in Srs2 increase expansion rates for several structure-forming TNRs (10,19), whereas loss of Sgs1 does not affect (CTG)n expansion rates (10), and actually increases stability of (CGG)n arrays (11).
The helicase substrates used in this study are listed in Figure 1. The presence of the 14 nt 3′ tail in all substrates, plus the known polarity of Srs2 and Sgs1 from 3′ to 5′, indicate that unwinding will occur from right to left (Figure 1). The Watson–Crick substrate and the (CTG)5·(CTG)5 partial duplex were included as controls to validate the current enzyme preparations and to compare the activity of purified Srs2 and Sgs1 with previous results (15). The upper strands, containing the inserted sequences from (CTG)7 onward, are predicted to form secondary structure with differing abilities depending on the insertion. For example, CTG sequences are known to adopt a hairpin conformation, whereas CTT repeats have little to no structure-forming propensity (3). The nontriplet insert has similar potential C-G base pairing and T-T mismatches as the (CTG)n substrates, but without any repeating triplets. The three-piece substrate (Figure 1) is comprised of three separate oligonucleotides. The stem of the arm formed between oligonucleotides 1 and 2 contains CTG sequences on both strands, followed by nine base pairs of Watson–Crick DNA for thermodynamic stabilization. Oligonucleotides 2 and 3 were 5′ labeled with Cy5 and Cy3 thymidine residues, respectively, to allow independent monitoring of unwinding of these two strands.
The kinetic advantage of Srs2 on TNR-containing partial duplex substrates is retained in the his-tagged enzyme
The purified Srs2 used for the current study contains an N-terminal 9-histidine tag, which differs from our previous preparation (15). The Watson–Crick duplex and the (CTG)5·(CTG)5 substrate were assayed to confirm earlier findings and identify any change in activity due to the presence of the histidine tag. Supplementary Figure 1 shows the profile of unwinding by Srs2 and Sgs1 at 10 nM concentration. For the Watson–Crick duplex, both enzymes were equally active, reaching ∼70% unwinding within 5 min (Supplementary Figure 1A and C). The reaction curves for the two enzymes are nearly superimposable over the full-time range, consistent with equivalent helicase activity at equal enzyme concentrations (Supplementary Figure 1C). In contrast, unwinding of the (CTG)5·(CTG)5 partial duplex substrate was 3- to 4-fold faster for Srs2 over the linear portion of the time curves (Supplementary Figure 1B and D). These findings are consistent both qualitatively and quantitatively with our previous work (15), and they indicate that the his-tagged Srs2 preparation used here retains the unusual property of unwinding a TNR-containing partial duplex substrate significantly faster than Sgs1. Furthermore, the assay times and enzyme concentrations used here are suitable for further investigating potential differences between the two helicases.
CTG hairpins embedded within a duplex region are unwound faster by Srs2 than Sgs1
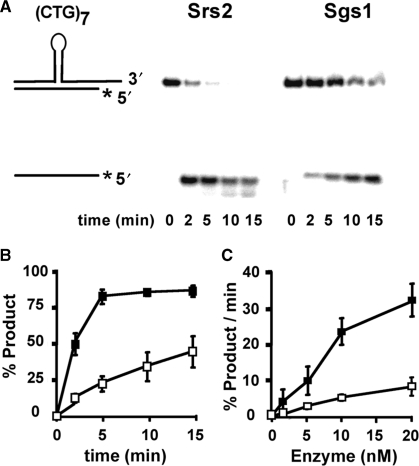
Current models for TNR expansions envision an intermediate in which the TNR hairpin is embedded within normal B-form duplex DNA (1,2). Thus, a more stringent test of Srs2 is to examine its unwinding activity on substrates that better approximate the proposed intermediate. The expected structure of the (CTG)7 hairpin is shown schematically in Figure 2A. Both Srs2 and Sgs1 at 10 nM unwind the (CTG)7 substrate; however, Srs2 retains a ∼4-fold kinetic advantage (Figure 2A and B). Faster unwinding by Srs2 was observed over protein concentrations from 2.5 to 20 nM (Figure 2C). Thus, more rapid unwinding by Srs2 occurs over a range of moderate enzyme concentrations. When the enzyme concentration was increased to a high level, 200 nM, both helicases completely unwound the molecule within 2 min (data not shown). Thus, high enzyme concentrations are required to overcome the slower unwinding of the (CTG)7 hairpin by Sgs1.
Figure 2.
Time- and enzyme concentration-dependent unwinding of (CTG)7 hairpin-containing duplexes. The schematic diagram shows the structured hairpin embedded within normal duplex regions. The shorter strand is radiolabeled on the 5′ end, as denoted by the asterisk. Purified Srs2 and Sgs1 were incubated with the DNA substrates (0.5 nM) for the indicated times. For 0 min, the substrate was incubated without protein. The reactions were quenched and the products were resolved on 12% nondenaturing polyacrylamide gel and visualized by phosphorimaging. (A) Srs2 or Sgs1 (10 nM each) were incubated with the substrate for 0–15 min. (B) Time dependence of unwinding from two–three repetitions (including that shown in panel A). (C) Enzyme concentration effects on unwinding rates in the linear time range, calculated as percentage of product unwound per minute of reaction. For (B) and (C), error bars are the range of values observed; filled squares, Srs2; unfilled squares, Sgs1.
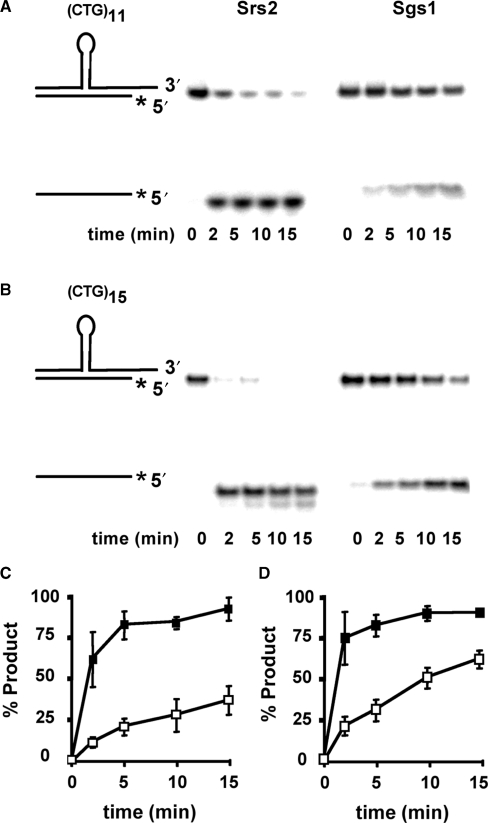
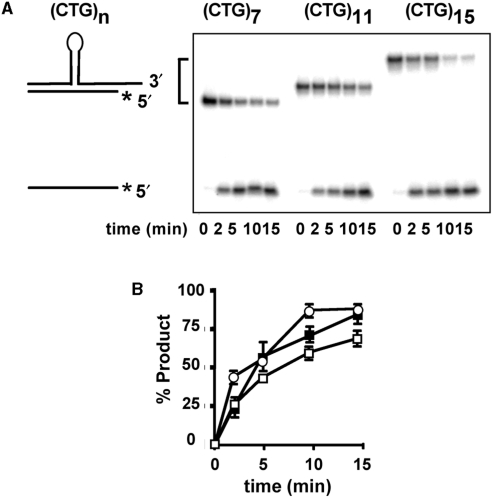
The kinetic advantage of Srs2 was also observed for longer CTG repeat-containing substrates, which are intended to mimic in vivo intermediates of larger expansion mutations. Figure 3 shows that substrates containing (CTG)11 or (CTG)15 inserts were also subject to ∼3- to 4-fold faster unwinding by Srs2, compared to Sgs1. Thus, Srs2 continues to proceed more quickly through longer CTG hairpins than does Sgs1. Interestingly, comparison of the Srs2 results for (CTG)7, (CTG)11 and (CTG)15 substrates show approximately equal rates of unwinding, despite the longer insertion lengths (compare Figure 2B with Figure 3C and D). The same is true for Sgs1, albeit at a slower pace. When the helicase assays were repeated at a reduced Srs2 concentration of 5 nM, to slow the reaction and reveal possible differences in rate, the unwinding profile was still very similar for (CTG)7, (CTG)11 and (CTG)15 substrates (Figure 4). These results imply that unwinding of the hairpin is not rate limiting for these substrates, and that Srs2 unwinding through the hairpin may occur at rates similar to that in the duplex region. This possibility is addressed later with the three-piece substrate.
Figure 3.
Effect of longer CTG repeat tracks on helicase activity. Helicase assays were performed as described earlier. (A) (CTG)11 hairpin-containing duplex DNA. (B) (CTG)15 hairpin-containing duplex DNA. (C and D) Quantitation of helicase activity on (CTG)11 (C) and (CTG)15 substrates (D). The results were averaged from three repetitions (including that shown in panels A and B). Error bars are ± 1 SD; filled squares, Srs2; unfilled squares, Sgs1. The shorter strand is radiolabeled on the 5′ end, as denoted by the asterisk.
Figure 4.
Unwinding of (CTG)7, (CTG)11 and (CTG)15 hairpin substrates by 5 nM Srs2. Purified Srs2 was incubated with DNA substrates (0.5 nM) for the indicated times. For 0 min, the substrate was incubated without protein. The reactions were quenched and the products were resolved on 12% nondenaturing polyacrylamide gel and visualized by phosphorimaging. (A) Representative unwinding results for (CTG)7, (CTG)11 and (CTG)15. (B) Quantitation of helicase activity on each substrate. The results were averaged from two repetitions (including that shown in panel A). Error bars are the range of values. Filled squares, (CTG)7; unfilled squares, (CTG)11; unfilled circles, (CTG)15. The shorter strand is radiolabeled on the 5′ end, as denoted by the asterisk.
Unwinding of alternative sequences
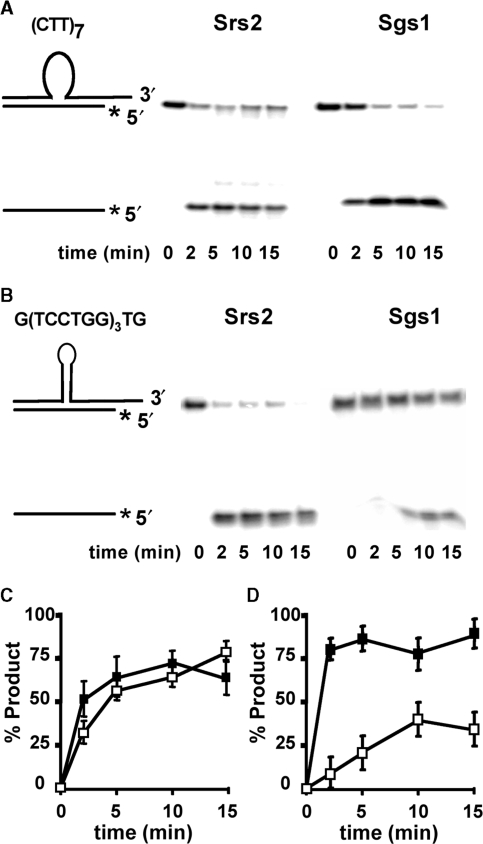
The faster unwinding by Srs2 of CTG repeats could be due to the repeating DNA sequence, its secondary structure or to both factors. To address these possibilities, assays were performed with two additional substrates to help distinguish between the influence of sequence and structure. The first contains a (CTT)7 insert, which retains a triplet repeat sequence but which has a low likelihood of secondary structure formation (3). Figure 5A shows that Srs2 and Sgs1 rapidly unwind the substrate with (CTT)7 insert, and the time course curves are essentially superimposable (Figure 5C). Thus, the kinetic advantage of Srs2 was overcome in a substrate expected to be devoid of secondary structure. The unwinding of (CTT)7 by Srs2 and Sgs1 is similarly rapid as the Watson–Crick control duplex (Supplementary Figure 1), consistent with previous results for unstructured sequences (15). The (CTT)7 results indicate that repeats of three nucleotides are not the major factor distinguishing unwinding by Srs2 and Sgs1.
Figure 5.
Effect of alternative sequences on unwinding by Srs2 and Sgs1. (A) The unwinding of duplex containing (CTT)7, a nonstructure forming loop. (B) The unwinding of duplexes with (TCCTGG)3 structured loop substrate. (C and D) Quantification of helicase activity on (CTT)7 (C) and the structured loop substrate (D). Filled squares denote Srs2 and unfilled squares represent Sgs1. The shorter strand is radiolabeled on the 5′ end, as denoted by the asterisk.
To address structural influence, we created a hexameric sequence TCCTGG, repeated thrice (Figure 1). The presence of potential G-C base pairs and T-T mismatches are similar to those of the (CTG)7 repeat, but without a triplet repeat sequence. This nontriplet sequence was unwound rapidly by Srs2 (Figure 5B and D). In contrast, Sgs1 was significantly slower at unwinding the nontriplet substrate. Both the qualitative pattern and the quantitative rate measurements on the nontriplet molecule closely resemble what was seen in the (CTG)n substrates in Figures 2 and 3. Thus, facile unwinding of the hexameric sequence is specific to Srs2. Together with the (CTT)7 results, we conclude that structure, not sequence, is the key determinant for the kinetic advantage of Srs2 over Sgs1.
Unwinding of a three-piece substrate suggests a model for faster activity by Srs2
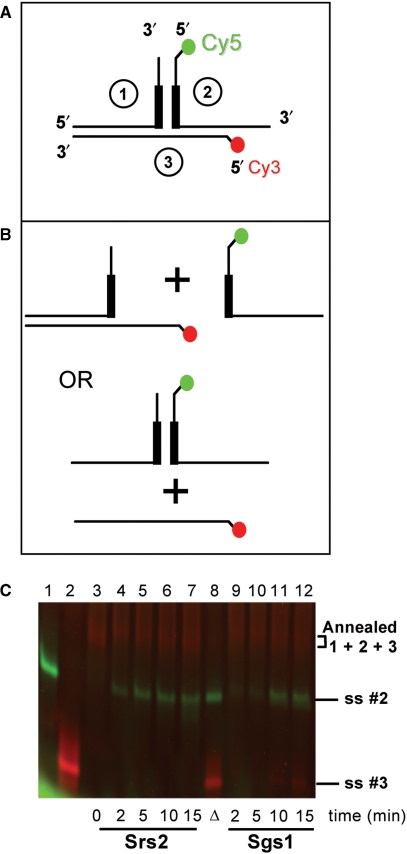
The hairpin substrates tested so far were all radiolabeled on the bottom, nonhairpin strand. The fate of the upper, hairpin-containing strand following helicase action was of interest to determine whether the hairpin was unwound. The three-piece substrate (Figure 6A) was used to address this question. Strands 2 and 3 were 5′ labeled with Cy5 and Cy3, respectively, to allow visualization of release of individual strands. In effect, we asked whether Srs2 and Sgs1 translocate along the upper strand (which contains the 3′ tail for initial binding), enter the hairpin at the three-way junction and unwind it to release oligonucleotide 2 (Figure 6B, top). Alternatively, a helicase might dissociate from its bound strand when it encounters the base of the hairpin and then rebind on the distal side before recommencing displacement to release oligonucleotide 3. If so, the hairpin would not be unwound (Figure 6B, bottom).
Figure 6.
Unwinding of a three-piece model substrate by Srs2 and Sgs1. (A) Structure of the three-piece model substrate. The substrate was created as described in Materials and methods section. The 5′end residue of oligonucleotide 2 was coupled to Cy5, shown in green. The 5′ terminal residue of oligonucleotide 3 was coupled to Cy3, shown in red. Thick bars denote location of the CTG repeats. (B) Potential helicase products. Srs2 or Sgs1 is presumed to bind initially to the 3′ tail of oligonucleotide 2, then unwind from right to left. Top, displacement of oligonucleotide 2 is expected if the helicase unwinds, while remaining bound to the initial strand. Bottom, displacement of oligonucleotide 3 is predicted from the dissociation–reassociation model described in the text. (C) Release of Cy5- and Cy3-labeled products by Srs2 or Sgs1. Each enzyme (10 nM) was incubated with the annealed three-piece substrate for the indicated times. For 0 min, the substrate was incubated without protein. The reactions were quenched, the products were resolved on 12% nondenaturing polyacrylamide gel and visualized by fluorescence imaging. Lane 1, single-stranded oligonucleotide 2; lane 2, single-stranded oligonucleotide 3; lane 3, annealed three-piece substrate; lanes 4–7, unwinding by Srs2 for the times indicated; lane 8, heat-denatured control; lanes 9–12, unwinding by Sgs1.
These models were evaluated for Srs2 and Sgs1 at 10 nM concentration. The time course of Srs2 unwinding shows release of the Cy5-labeled strand 2 as early as 2 min into the reaction, and additional product release from 5 to 15 min (Figure 6C, lanes 4–7). There was no apparent difference in unwinding rate for the three-piece substrate compared to any of the other TNR-containing substrates tested here. Furthermore, there was no detectable release of the Cy3-labeled strand 3, suggesting that Srs2 binds initially to the 3′ tail of strand 2 and remains bound to that strand until unwinding is complete, including displacement of the triplet repeat region in the stem-loop. At very high enzyme concentrations (200 nM; not shown), both labeled strands were released, probably due to initial displacement of strand 2, rebinding of Srs2 to the newly exposed 3′ end of strand 1, then unwinding of strands 1 and 3. Thus, Cy3-labeled strand 3 can be released in a sequential mechanism by high levels of Srs2.
With 10 nM Sgs1, there was primarily release of Cy5-labeled strand 2 but at later times than seen for Srs2 (Figure 6C, compare lanes 9–12 with lanes 4–7). This slower unwinding is consistent with results from other structured inserts seen in this study and from previous work (15). Unlike Srs2, the Sgs1 data in Figure 6C show a small amount of unwinding of Cy3-labeled strand 3. This is perhaps due to a minority of events, where Sgs1 leads to some sequential unwinding of strand 2 followed by strand 3. At 200 nM Sgs1, both fluorescently labeled strands are released (data not shown), consistent with sequential unwinding. Overall, the results with the three-piece substrate support the idea that both Srs2 and Sgs1 remain bound to the upper strand at the three-way junction, but that Srs2 unwinds the triplet repeat junction faster due to its unabated progress into and through the extrahelical structure.
DISCUSSION
These biochemical results with ‘running start’ hairpin substrates support the genetic findings that Srs2 selectively inhibits triplet repeat expansions in vivo. This study shows that purified Srs2 is ∼3- to 4-fold faster than Sgs1 at unwinding CTG repeat-containing hairpins embedded within duplex DNA. Srs2 maintains rapid unwinding even when initially loaded into the duplex region. Further analysis showed that a structured but nontriplet repeat substrate was also preferentially unwound by Srs2, suggesting that structure, not sequence, is the most important specificity determinant for Srs2. The second major finding was that Srs2 and Sgs1 were equally active on Watson–Crick duplexes and on unstructured loop substrates. Thus, the difference in activity cannot be ascribed to trivial reasons. The helicase specificity of Srs2 in vitro is consistent with its selective inhibition of expansions in vivo (10). These findings are similar both qualitatively and quantitatively to previous work with partial duplex TNR substrates (15). The third major finding of the work utilized a fluorescently labeled three-piece substrate to show that Srs2 unwinding proceeds through the triplet repeat region without apparent slowing or dissociation from the strand to which it originally binds. The enzyme can therefore unwind both the duplex region and the hairpin, and hairpin unwinding does not appear to be rate limiting for CTG tracts of 7 to 15 repeats. Together, the genetic and biochemical studies of Srs2 provide a paradigm for one way that triplet repeat expansions can be inhibited.
Srs2 likely acts to prevent expansions from the 3′ side of the hairpin, based both on its biochemical polarity (13,14) and also from recent genetic findings that Srs2 acts in concert with postreplication repair (PRR) to inhibit expansions (19). PRR is predicted to be recruited to an intermediate containing a 3′ end when replication stalls (20). Loss of PRR factors, including a pol30-K164R mutant that blocks ubiquitylation of PCNA, showed similar defects as a srs2 mutant on expansions CAG·CTG tracts (19). Furthermore, there was a synergistic effect on expansions in pol30-K164R rad27 double mutants. RAD27 encodes flap endonuclease 1, which is well characterized to inhibit expansions arising from 5′ flaps (6,21–25). By inference, expansions in yeast occur by both 5′ flap and 3′ slippage mechanisms, with Srs2 helping provide protection from the 3′ end.
While the ∼3- to 4-fold kinetic advantage is notable for Srs2 unwinding of TNR hairpins in vitro, the magnitude of expansion inhibition in vivo is significantly greater, up to 40-fold (10). It seems likely that additional reasons must exist to explain the full extent of protection against expansions in a cell. Perhaps Srs2 is preferentially recruited to a hairpin in vivo through protein–protein interactions, thus positioning Srs2 for unwinding. Previous work from other labs demonstrated interactions between Srs2 and the Pol32 subunit of DNA polymerase δ (26), and also for Srs2 with sumoylated PCNA (27). Subsequently it was shown genetically that Srs2 helps prevent expansions in concert with DNA polymerase δ and ubiquitylated PCNA (10,19). Further experimentation is required to test this recruitment model more directly.
The human protein hFBH1 has been cited as a possible functional orthologue of budding yeast Srs2, based on suppression of recombination defects and DNA damage sensitivity of an srs2 mutant by the human gene (28). It remains to be seen whether hFBH1 also suppresses triplet repeat expansion defects in a srs2 background. In vitro processing of slipped CTG·CTG repeats by human cell extracts leads to either correct repair, escape from repair or error-prone repair (29). The repair outcomes did not require the DNA repair proteins hMSH2, hMSH3, hMLH1, XPF, XPG or DNA polymerase β, but no helicases were examined. It will be interesting to see which, if any, of the human DNA helicases provides a function similar to Srs2 in preventing triplet repeat expansions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This study was funded by National Institutes of Health (GM61961) and Science Foundation of Ireland (06/IN.1/B73) to R.S.L. National Cancer Institute Cancer Center Support Grant (P30 CA36727) to the Eppley Institute. Funding to pay the Open Access publication charges for this article was provided by SFI 06/IN.1/B73.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pearson CE, Edamura KN, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 2.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 3.Gacy AM, Goellner G, Juranic N, Macura S, McMurray CT. Trinucleotide repeats that expand in human disease form hairpin structure in vitro. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 4.Kang S, Jaworski A, Ohshima K, Wells RD. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E. coli. Nat. Genet. 1995;10:213–218. doi: 10.1038/ng0695-213. [DOI] [PubMed] [Google Scholar]
- 5.Samadashwily GM, Raca G, Mirkin SM. Trinucleotide repeats affect DNA replication in vivo. Nat. Genet. 1997;17:298–304. doi: 10.1038/ng1197-298. [DOI] [PubMed] [Google Scholar]
- 6.Freudenreich CH, Kantrow SM, Zakian VA. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 7.Miret JJ, Pessoa-Brandao L, Lahue RS. Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1998;95:12438–12443. doi: 10.1073/pnas.95.21.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kovtun IV, Liu Y, Bjoras M, Klungland A, Wilson SH, McMurray CT. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–452. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claassen DA, Lahue RS. Expansions of CAG·CTG repeats in immortalized human astrocytes. Hum. Mol. Genet. 2007;16:3088–3096. doi: 10.1093/hmg/ddm270. [DOI] [PubMed] [Google Scholar]
- 10.Bhattacharyya S, Lahue RS. Yeast Srs2 DNA helicase selectively blocks expansions of trinucleotide repeats. Mol. Cell. Biol. 2004;24:7324–7330. doi: 10.1128/MCB.24.17.7324-7330.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White PJ, Borts RH, Hirst MC. Stability of the human Fragile X (CGG)n triplet repeat array in Saccharomyces cerevisiae deficient in aspects of DNA metabolism. Mol. Cell. Biol. 1999;19:5675–5684. doi: 10.1128/mcb.19.8.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein H. A radical solution to death. Nat. Genet. 2000;25:132–134. doi: 10.1038/75957. [DOI] [PubMed] [Google Scholar]
- 13.Rong J, Klein H. Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1993;268:1252–1259. [PubMed] [Google Scholar]
- 14.Bennett RJ, Sharp JA, Wang JC. Purification and characterization of the Sgs1 DNA helicase activity of Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:9644–9650. doi: 10.1074/jbc.273.16.9644. [DOI] [PubMed] [Google Scholar]
- 15.Bhattacharyya S, Lahue RS. Srs2 helicase of Saccharomyces cerevisiae selectively unwinds triplet repeat DNA. J. Biol. Chem. 2005;280:33311–33317. doi: 10.1074/jbc.M503325200. [DOI] [PubMed] [Google Scholar]
- 16.Fry M, Loeb LA. Human Werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 17.Kamath-Loeb AS, Loeb LA, Johansson E, Burgers P.MJ, Fry M. Interactions between the Werner syndrome helicase and DNA polymerase delta specifically facilitate copying of tetraplex and hairpin structures of the d(CGG)n trinucleotide repeat sequence. J. Biol. Chem. 2001;276:16439–16446. doi: 10.1074/jbc.M100253200. [DOI] [PubMed] [Google Scholar]
- 18.Van Komen S, Reddy MS, Krejci L, Klein H, Sung P. ATPase and DNA helicase activities of the Saccharomyces cerevisiae anti-recombinase Srs2. J. Biol. Chem. 2003;278:44331–44337. doi: 10.1074/jbc.M307256200. [DOI] [PubMed] [Google Scholar]
- 19.Daee DL, Mertz T, Lahue RS. Postreplication repair inhibits CAG·CTG repeat expansions in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:102–110. doi: 10.1128/MCB.01167-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ulrich HD. The RAD6 pathway: control of DNA damage bypass and mutagenesis by ubiquitin and SUMO. Chembiochem. 2005;6:1735–1743. doi: 10.1002/cbic.200500139. [DOI] [PubMed] [Google Scholar]
- 21.Schweitzer JK, Livingston DM. Expansions of CAG repeat tracts are frequent in a yeast mutant defective in Okazaki fragment maturation. Hum. Mol. Genet. 1998;7:69–74. doi: 10.1093/hmg/7.1.69. [DOI] [PubMed] [Google Scholar]
- 22.Spiro C, Pelletier R, Rolfsmeier ML, Dixon MJ, Lahue RS, Gupta G, Park MS, Chen X, Mariappan S.VS, McMurray CT. Inhibition of FEN-1 processing by DNA secondary structure at trinucleotide repeats. Mol. Cell. 1999;4:1079–1085. doi: 10.1016/s1097-2765(00)80236-1. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Bambara RA. Analysis of human flap endonuclease 1 mutants reveals a mechanism to prevent triplet repeat expansion. J. Biol. Chem. 2003;278:13728–13739. doi: 10.1074/jbc.M212061200. [DOI] [PubMed] [Google Scholar]
- 24.Spiro C, McMurray CT. Nuclease-deficient FEN-1 blocks Rad51/BRCA1-mediated repair and causes trinucleotide repeat instability. Mol. Cell. Biol. 2003;23:6063–6074. doi: 10.1128/MCB.23.17.6063-6074.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Zhang H, Veeraraghavan J, Bambara RA, Freudenreich CH. Saccharomyces cerevisiae flap endonuclease I uses flap equilibration to maintain triplet repeat stability. Mol. Cell. Biol. 2004;24:4049–4064. doi: 10.1128/MCB.24.9.4049-4064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang M-E, de Calignon A, Nicolas A, Galibert F. POL32, a subunit of the Saccharomyces cerevisiae DNA polymerase delta, defines a link between DNA replication and the mutagenic bypass repair pathway. Curr. Genet. 2000;38:178–187. doi: 10.1007/s002940000149. [DOI] [PubMed] [Google Scholar]
- 27.Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 28.Chiolo I, Saponaro M, Baryshnikova A, Kim J.-H, Seo Y.-S, Liberi G. The human F-box DNA helicase FBH1 faces S. cerevisiae Srs2 and post-replication repair pathway roles. Mol. Cell. Biol. 2007;27:7439–7450. doi: 10.1128/MCB.00963-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panigrahi GB, Lau R, Montgomery SE, Leonard MR, Pearson CE. Slipped (CAG)·(CTG) repeats can be correctly repaired, escape repair or undergo error-prone repair. Nat. Struct. Mol. Biol. 2005;12:654–662. doi: 10.1038/nsmb959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.