Abstract
The Hoxa2 gene has a fundamental role in vertebrate craniofacial and hindbrain patterning. Segmental control of Hoxa2 expression is crucial to its function and several studies have highlighted transcriptional regulatory elements governing its activity in distinct rhombomeres. Here, we identify a putative Hox–Pbx responsive cis-regulatory sequence, which resides in the coding sequence of Hoxa2 and is an important component of Hoxa2 regulation in rhombomere (r) 4. By using cell transfection and chromatin immunoprecipitation (ChIP) assays, we show that this regulatory sequence is responsive to paralogue group 1 and 2 Hox proteins and to their Pbx co-factors. Importantly, we also show that the Hox–Pbx element cooperates with a previously reported Hoxa2 r4 intronic enhancer and that its integrity is required to drive specific reporter gene expression in r4 upon electroporation in the chick embryo hindbrain. Thus, both intronic as well as exonic regulatory sequences are involved in Hoxa2 segmental regulation in the developing r4. Finally, we found that the Hox–Pbx exonic element is embedded in a larger 205-bp long ultraconserved genomic element (UCE) shared by all vertebrate genomes. In this respect, our data further support the idea that extreme conservation of UCE sequences may be the result of multiple superposed functional and evolutionary constraints.
INTRODUCTION
Hom-C/Hox genes encode transcription factors involved in the patterning of the main body axis and limbs, as well as in multiple aspects of organogenesis (1–5). Further to the initial discovery of homeotic (Hom-C/Hox) genes in Drosophila, it appeared that these genes have been widely conserved through evolution and they were associated to the modelling of both invertebrate and vertebrate body plans. Moreover, although they have been duplicated up to four times in the vertebrate phyla, their arrangement in orderly chromosomal clusters has also been conserved (6). In the mouse genome, there are 39 Hox genes clustered on four chromosomal loci. Crucial for the fulfilment of their developmental roles is their proper regulation in space and time during embryogenesis (7,8). In particular, the accurate patterning of the rostro-caudal axis of the mouse embryo requires the different Hox genes to be activated in a nested fashion (9,10)
The Hoxa1, -a2, -b1 and -b2 genes interact to pattern rhombomeric territories in the hindbrain as well as the neural crest cells emanating from the hindbrain region (10–18). To establish and/or maintain their accurate expression patterns, these genes establish some stimulatory cross-regulatory loops involving the cooperation between Hox proteins and the three-amino acid loop extension (TALE) homoeodomain proteins Pbx and Prep/Meis (19–23).
By a reporter-based transgenic approach, Frasch et al. (24) analysed the activity of the genome fragment that extends from the beginning of the Hoxa2 coding region to the 5′ untranslated sequence of Hoxa1 and identified a 1.25-kb enhancer region, which mediates hindbrain-specific gene expression in rhombomere (r) 4. More recently, based on comparative sequence examination and functional assays in chicken and mouse embryos, Tümpel et al. (25) identified three Hox/Pbx and one Prep/Meis-binding sites defining a r4-specific enhancer residing in the intron of Hoxa2. These sites were shown to respond to Hoxb1, suggesting that the expression of Hoxa2 in r4 is switched on by a Hoxb1-mediated regulation. From these data, a model was proposed explaining the establishment of the r4 identity, involving an initiation phase relying on retinoids signalling and further cross-regulatory interactions between paralogue group 1 (i.e. Hoxa1 and Hoxb1) and group 2 (i.e. Hoxa2 and Hoxb2) Hox genes.
Here, we used comparative sequence examination, transfection assays, chromatin immunoprecipitation and chick electroporation to identify additional cis-regulatory elements involved in the control of segmental Hoxa2 regulation. We discovered a novel Hox/Pbx bipartite binding site active in r4 with the unusual feature to reside within the coding sequence of the first exon of Hoxa2. Furthermore, this regulatory site is also comprised in a 205-bp sequence interval recognized as an ultraconserved element (UCE) (26) largely overlapping with the Hoxa2 coding sequence.
UCEs consist in sequences of >200 bp that are perfectly conserved between orthologous genomic loci in man, mouse, rat and other mammals. Bejerano et al. (26) have identified 481 UCEs among which many reside nearby genes coding for transcription factors proposed to be involved in developmental tasks. In that context, long-range sequence comparisons over entire Hox complexes revealed some regions with very high sequence conservation (27–30), including a few UCEs (26).
The r4-specific regulatory sequence reported here responds to both paralogue group 1 and 2 Hox proteins. Considering that Hoxa2 intronic sequences were already shown to mediate cross-regulatory controls between Hox genes in r4, we further demonstrated that the Hox–Pbx element within the UCE critically contributes to the r4-specific activity of the 1.25-kb enhancer region of Frasch et al. (24) in the chick hindbrain. We thus found that both intronic and exonic regulatory elements cooperate to ensure r4-specific gene expression. In contrast, the 205-bp UCE was not able to confer enhancer activity when tested alone in chick hindbrain. Thus, while several UCE loci have been proposed to primarily act as transcriptional control elements, this does not appear to be the case for the Hoxa2 UCE when out of the context of the full 1.25-kb enhancer.
MATERIALS AND METHODS
Plasmids construction
The pAdML-Luc plasmid contains a luciferase reporter gene placed under the control of the TATA box and the transcription start site from the Adenovirus-2 Major Late promoter (AdML) (31). The 1.25-kb r4 enhancer region was isolated following restriction of the MZ20 plasmid (32), and cloned in the BamHI site of the pAdML-Luc plasmid (1.25-kb reporter vector). The mutation of the Hox responsive element (HRE) was created by site-directed mutagenesis using a PCR approach (mutagenic primer: 5′-GATACATTTC AAAGTAGCAG CATAAAGACC TCGACGCTT-3′) to give rise to the m1.25-kb reporter vector. The coding region from the first exon of Xenopus and zebrafish Hoxa2 genes were amplified from the vectors pCS2.xHoxa2 and pCS2.zHoxa2 (33), respectively, and cloned in the BamHI site of pAdML-Luc. The reporter constructs designed for chick electroporation were obtained by cloning the 1.25-kb or the m1.25-kb regions in pKS-β-globin-lacZ (BGZ40, 34) yielding the 1.25-kb-β-globin-lacZ and m1.25-kb-β-globin-lacZ constructs, respectively. The 1.25-kb-ΔPH1-3-β-globin-lacZ was obtained by a PCR-based deletion strategy (mutagenic primer used for deletion: 5′-TTTCCCTAAC TTGTGTAATG TAGGAGTGTT GTAGCTAATA TAAAGTTTGC-3′). The 1.25-kb-ΔUCE-β-globin-lacZ and the UCE-β-globin-lacZ were made by cloning PCR-amplified NotI–BamHI (5′-GAGGATCCCT CGCCACGGCG CTGGCGTTG-3′ and 5′-GAGGATCCCC CGCCGCTGCC ATCA-3′) and the EcoRI–NotI (5′-GAGGATCCCA ATAGTTTAAT AGTAGCG-3′ and 5′-GAGGATCCTC GACTTGGGGC GGCCGCCAA-3′) fragments of the 1.25-kb region into the pKS-β-globin-lacZ plasmid, respectively.
The expression vectors pCMVHoxa1, pCMVHoxa1(QN-AA), pCMVHoxa1(WM-AA), pCMVHoxa2, pCMVHoxa2(QN-AA) and pCMVPbx1a have been described elsewhere (35,36). The pCS2-Prep1 has been described by Goudet et al. (37). The HOXB1 and HOXB2 expression vectors have been described by Di Rocco et al. (20), and the Hoxd4 expression vector by Rambaldi et al. (38). The HOXB7 expression vector was obtained by cloning the PCR-amplified HOXB7 open reading frame in the pCR3.1 plasmid (Invitrogen, Carlsbad, CA, USA). The substitution of the WM amino acids of the hexapeptide motif of Hoxa2 was generated by mutagenic PCR (mutagenic primer: 5′-CCTGAGTATCCCGCGGCGAAGGAGAAGAAG-3′). Bacterial expression vectors for His-tagged Hoxa2, Hoxa1, Hoxa2(QN-AA) and Hoxa1(WM-AA) proteins were generated by cloning the corresponding coding sequences into pQE30 (Qiagen Benelux B.V., Venlo, NL). The lacZ reporter constructs pCMVlacZ and pSVK3lacZ used for co-transfection assays have been described previously (35).
Cell culture and transient transfections
P19 and COS7 cells were maintained and transfected as described by Remacle et al. (36). Cells were harvested for enzymatic assays 48 h after transfection. Lysis and enzymatic activity dosages were performed with the β-gal Reporter Gene Assay (Chemiluminescent) kit (Roche Diagnostics, Basel, CH) and the Luciferase Reporter Gene Assay (High sensitivity) kit (Roche). The luciferase activities were normalized to standard β-galactosidase activities resulting from the constitutive lacZ expression encoded by the pCMVlacZ (P19 cells) or pSVK3lacZ (COS7 cells) reporter plasmids.
Protein purification and electrophoresis mobility shift assay (EMSA)
His-tagged Hox proteins were purified on Sigma–Ni–Sepharose columns, as described by the manufacturer (Sigma, St Louis, MO, USA) and verified by poly-acrylamide gel electrophoresis and western blotting. Pbx1a proteins were produced using the in vitro transcription/translation TnT Coupled Reticulocyte Lysate Systems (Promega, Madison, WI, USA) as described by the manufacturer.
Oligonucleotides were labelled with γ-32P dATP (Amersham Biosciences, Piscataway, NJ, USA; AA0018 >5000 Ci/mmol) and purified on a ChromaSpin-10 column (BD Biosciences, Erembodegem, Belgium). EMSA were performed with various combinations of proteins or reticulocyte lysates, in a sample volume of 20 µl containing 10 mM Tris pH 7.5; 75 mM NaCl; 1 mM DTT; 1 mM EDTA; 540 ng/ml BSA; 12% glycerol; 8 µg/ml dI–dC; 60 000 c.p.m. paired oligonucleotides (1 ng) and in some cases, unlabelled competitor oligonucleotides. The samples were incubated at room temperature for 20 min and on ice for 20 min. Complexes were separated by electrophoresis in non-denaturating 6% Tris–borate–EDTA polyacrymlamide gel under 10 V/cm. Oligonucleotides used harboured the HRE site (top strand: 5′-TTTCAAAGTTCATCAATCAAGACCTCG-3′) or the mutated HRE site (top strand: 5′-TTTCAAAGTTGGTGGGGGAAGACCTCG-3′). For EMSA involving anti-Hoxa2 (Santa Cruz Biotechnology, Santa Cruz, CA, USA; sc-17149) or anti-Pbx1 (Santa Cruz sc-889) antibodies, 1 µl of antibody (at 200 µg/ml) was added to the proteins prior to incubation with paired oligonucleotides.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed according to the manufacturer instructions (EZ ChIP, Upstate, Charlottesville, VA, USA). Briefly, mouse embryonic carcinoma (EC) P19 cells were seeded on a 10 cm diameter tissue culture dish. After 4 h, all-trans retinoic acid was added to the medium (10–5 M). Cells were cultivated for 72 h then washed three times with ice-cold PBS containing protease inhibitor (Complete™, Roche). Cells were cross-linked with 1% formaldehyde for 10 min at room temperature. To quench excess formaldehyde, 500 µl of glycine 2.5 M was added for 5 min. A total of 4 × 106 treated cells were re-suspended in 100 µl SDS lysis buffer and sonicated to obtain chromatin fragments from 200 to 1000 bp. The sonicate was diluted 10 times with ChIP dilution buffer and cleared with protein-G-agarose beads and salmon sperm DNA (Upstate), for at least 2 h at 4°C. The supernatant was collected after centrifugation and 1% was collected as input chromatin and stored at 4°C until de-cross-linking. Immunoprecipitation was performed overnight at 4°C by the addition of 10 µg anti-Hoxa2 (Santa Cruz sc-17150), 10 µg of anti-Pbx1/2/3 antibody (Santa Cruz, sc-888x) or 10 µg anti-IgG antibody (anti-goat IgG, Santa Cruz sc-2028; anti-rabbit IgG, Santa Cruz sc-2027). The immune complexes were immobilized by protein-G-agarose beads conditioned with salmon sperm DNA, for 2 h at 4°C, and successively washed with low salt wash buffer (four times), higher salt buffer (once), LiCl buffer (once) and TE buffer (twice). After elution with 1 M NaHCO3, the protein–DNA complexes were de-cross-linked by incubation overnight at 65°C. Before DNA precipitation, all samples were treated with 1 µl of RNAseA, 20 mg/ml, at 37°C for 30 min, and then with proteinase K for 1 h at 45°C.
To evaluate the amount of precipitated DNA, quantitative real-time PCR was performed on a Roche lightcycler by using 2xQuantiTect SYBR Green PCR Master Mix (Qiagen), with primers flanking the HRE sequence (5′-CGCTGAGTGCCTGACATCT-3′ and 5′-GAGTGTGAAAGCGTCGAGGT-3′) or amplifying a region located 1883 bp 5′ to the Hoxa2 transcriptional start site (5′-GACCCTATTGCTGAAAGCCAC-3′ and 5′-GCAATCACCTCATTATTTGTATTCC-3′), and with primers specific of the Hprt gene used as ChIP negative control (5′-TTATCTGGGAATCCTCTGGG-3′ and 5′-AAAGGCAGTTCCGGAACTCT-3′). Standard curves quantification were used for each individual primers pair. Input DNA values were used to normalize the values from ChIP samples.
In Ovo electroporation
Chick eggs were incubated in a humidified chamber and embryos were staged according to Hamburger and Hamilton (HH) (39). DNA constructs were injected into lumens of HH stage 10–12 chick embryonic hindbrains. Electroporation was performed using a square wave electroporator (BTX) (40). Electroporated embryos were harvested 24 h after electroporation and stained for lacZ reporter expression as previously described (41). Vector DNA concentrations for injection were: 0.3 mg/ml of reporter construct; 0.7 mg/ml of Hox expression vectors and 0.5 mg/ml of co-injected pCMV/EGFP as a tracer of electroporated cells and as internal control.
RESULTS
A Hox–Pbx regulatory site lies within an UCE encompassing the coding sequence of Hoxa2
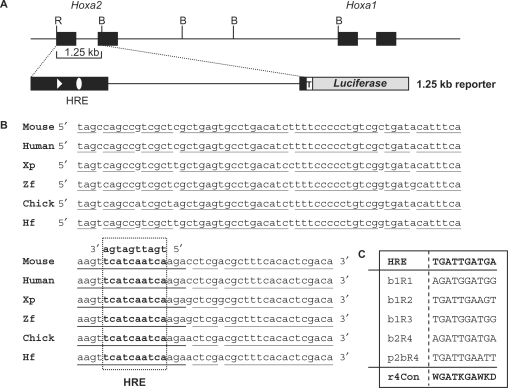
Recent genomic analyses of the vertebrate Hox complexes revealed several sequences sharing high similarity from sharks to mammals (28,29). Among those, the 5′end of Hoxa2 appeared among the best-conserved sequences in the HoxA cluster. In fact, we found that the first 182 bp of the Hoxa2 coding frame belongs to a 205-bp long UCE previously identified by Bejerano et al. (26) that, however, was not initially reported to lie within the Hoxa2 gene (see Supplementary Table 1 in ref. 26). The 205-bp sequence perfectly conserved among the seven mammalian genomes analysed extends from a few nucleotides upstream the Hoxa2 ATG codon to the middle of the coding sequence of the first exon (Supplementary Figure 1). The conserved sequence also shares strong similarities among the more distant vertebrate species. Multiple alignments involving 15 vertebrate Hoxa2 orthologues revealed an overall 83% sequence identity over 186 bp.
Notably, the nucleotide sequence conservation of this UCE goes beyond what might be expected solely from the functional conservation of the Hoxa2 protein, particularly when considering that nucleotide sequence variation is allowed by the degeneracy of the genetic code. We therefore hypothesized that other sequence-coded functions, such as cis-regulatory elements, may lie within the coding sequence of the first exon of Hoxa2. In this respect, the Hoxa2 UCE is included in a 1.25-kb genomic fragment previously identified by Frasch et al. (24) (Figure 1A) that specifically directs reporter gene expression in the r4 of the developing mouse hindbrain. Interestingly, we additionally found one decanucleotide element within the UCE sequence obeying the consensus sequence for binding of a Hox–Pbx heterodimer (41–43) (Figure 1B and C). This putative regulatory site, hereafter referred to as HRE, thus resides in the coding sequence of the first exon of the Hoxa2 gene.
Figure 1.
Schematic representation of the Hoxa2-Hoxa1 locus and of the 1.25-kb reporter vector (A) and sequence alignment of a 100-bp region encompassing the HRE Hox-Pbx responsive element from the mouse, man, Xenopus (Xp), zebrafish (Zf), chick and horn shark (Hf) (B). The HRE sequence (open box) is shown in bold cases. (C) Sequence alignment and consensus sequence (r4 con) of known Hox–Pbx-binding sites active in the r4 territory. Black rectangles represent exons; dark grey rectangle represents the luciferase reporter gene; the open triangle shows the AUG start codon of Hoxa2; the open ellipse represents the Hox–Pbx-binding site; the open rectangle with a T shows the minimal (TATA box) promoter of the reporter construct. B: BamHI; R: EcoRI.
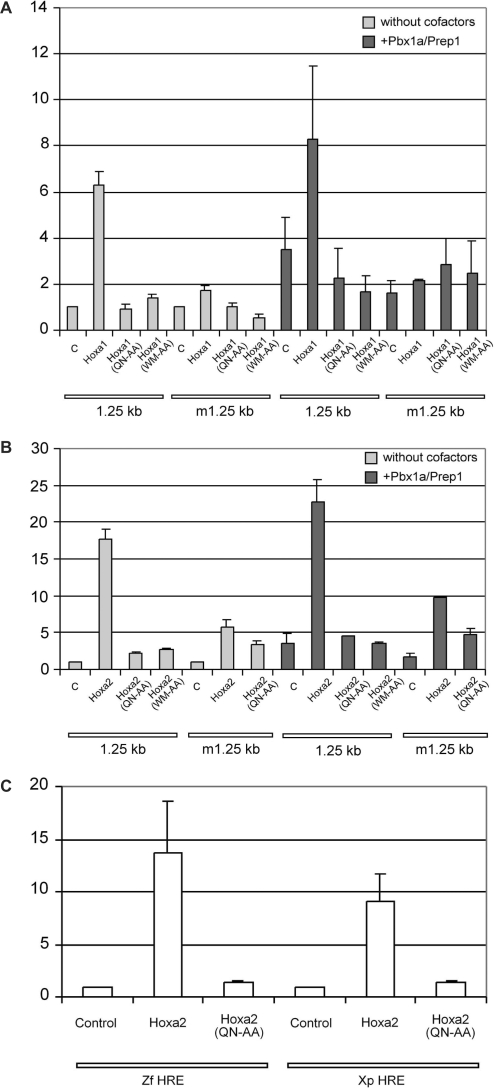
To determine whether this site is required for Hox-mediated regulatory activity, we first cloned the mouse 1.25-kb fragment upstream of a minimal promoter (AdML) and a luciferase cassette, and tested for its ability to activate reporter expression in mouse teratocarcinoma (EC) P19 cells in the presence of Hox proteins known to be involved in hindbrain development. Specifically, co-transfection with Hoxa1 or Hoxa2 expression vectors revealed 6- and 18-fold reporter activation, respectively (Figure 2A and B). A reporter construct devoid of the 1.25-kb fragment did not respond to the Hox proteins (data not shown). In addition, no reporter activation was observed upon co-transfection of DNA-binding defective Hoxa1(QN-AA) or Hoxa2(QN-AA) in which the critical residues glutamine 50 and asparagine 51 of the homoeodomain were replaced by alanines (Figure 2). Most importantly, the mutation (TGATTGATGA > TtATgctgct) of the putative Hox–Pbx HRE-binding sequence severely impaired the ability of Hoxa1 or Hoxa2 to drive reporter activation (Figure 2A and B). Finally, similar results were obtained when the 1.25-kb DNA fragment was placed upstream of an IL6 gene minimal promoter (data not shown). Therefore, a cis-regulatory element resides at the beginning of the Hoxa2 coding sequence and responds to both Hoxa1- and Hoxa2-mediated transactivation in cell transfection assays.
Figure 2.
The exonic HRE element of Hoxa2 is responsive to Hox, Pbx and Prep proteins in teratocarcinoma P19 cells. (A) Reporter constructs based on the wild-type 1.25-kb regulatory fragment (1.25 kb) or its mutant derivative with a modified HRE site (m1.25 kb) were transfected alone (control, c) or in combination of expression vectors for Hoxa1, Hoxa1(QN-AA) or Hoxa1(WM-AA), as well as for Pbx1a and Prep1 proteins. (B) Similar transfection experiments were performed involving Hoxa2, Hoxa2(QN-AA) or Hoxa2(WM-AA) expression vectors as well as, (C) with reporter constructs based on the first exon of the Xenopus (Xp) and zebrafish (Zf) Hoxa2 gene containing the conserved HRE sequence. Values are expressed as fold activation over transfection of the reporter plasmid alone. Bars indicate the standard deviation of at least four independent experiments, except for experiments involving the Hoxa2(WM-AA) expression vector, the Xp HRE reporter and the Zf HRE reporter that were reproduced twice.
We next addressed the sufficiency of the HRE-containing UCE to drive Hox-mediated regulation. As the UCE and the HRE sequence are conserved in a broad range of vertebrate species (Figure 1B), we designed 325 bp reporter constructs containing both the zebrafish and Xenopus UCE homologues. Upon co-transfection in cell culture, reporter expression was readily activated by Hoxa2 (Figure 2C) supporting the conservation of regulatory control at the HRE site among vertebrates.
Contribution of Pbx and Prep proteins to HRE activity
For efficient transcriptional regulation of Hox target genes, Hox proteins often need to form complexes with Pbx and Prep co-factors (19,22,23,41,44). In addition, Prep is required to control the nuclear versus cytoplasmic distribution of Pbx (45).
To address the influence of Pbx and Prep on Hox-mediated HRE activity, we tested Pbx1a and Prep1 vectors in co-transfection experiments in P19 cells. Transfected Pbx1a and Prep1 proteins were active on the 1.25-kb fragment harbouring the HRE site but not its mutated derivative (Figure 2), supporting that Pbx1a and Prep1 are active on the HRE element. However, the reporter activations observed in these experiments were rather low (3- to 4-fold). Similarly, Pbx1a and Prep1 only modestly increased the Hoxa1- and Hoxa2-mediated reporter activations (1.2-fold; Figure 2A and B). The absence of a significant synergistic effect is likely due to the considerable endogenous levels of Pbx and Prep proteins in P19 cells (36, S.R. and R.R., unpublished data). Nonetheless, a way to evaluate the partnership between Hox and Pbx is to disrupt their interaction by substituting the WM residues of a conserved hexapeptide motif by alanines (46). Indeed, transfected Hoxa1(WM-AA) and Hoxa2(WM-AA) hexapeptide mutants behaved like loss-of-function mutants on the 1.25-kb enhancer region (Figure 2A and B). This strongly indicated that the activity provided by Hoxa1 and Hoxa2 on HRE actually relied on their interactions with Pbx and Prep proteins.
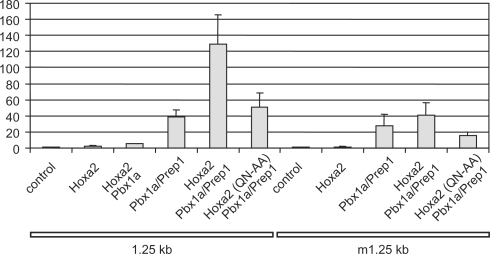
To further address this issue, we assayed the activity of Pbx1a and Prep1 in COS7 cells that did not show high constitutive Pbx expression but displayed intense nuclear accumulation of Pbx1a upon Pbx1a and Prep1 co-transfection (data not shown). In this cell line, expression of Pbx1a and Prep1 resulted in an important 40-fold activation of the reporter (Figure 3). The further addition of Hoxa2 resulted in a very strong synergistic response, up to 100- to 150-fold reporter activation, whereas Hoxa2 or Hoxa2–Pbx1a alone did not or hardly stimulated reporter expression (Figure 3). Finally, the mutation of the HRE site in the 1.25-kb fragment reduced the Hoxa2/Pbx1a/Prep1-mediated activation by >70% (Figure 3).
Figure 3.
Hoxa2, Pbx and Prep synergize onto HRE in COS7 cells. Reporter constructs based on the wild-type 1.25-kb regulatory fragment (1.25 kb) or its HRE mutant derivative (m1.25 kb) were transfected alone (control) or in combination with expression vectors for Pbx1a, Prep1, Hoxa2 or Hoxa2(QN-AA) proteins. Values are expressed as fold activation over transfection of the reporter plasmid alone. Bars indicate the standard deviation of at least three independent experiments, except for experiments involving Hoxa2 and Pbx1a alone that were reproduced twice.
Altogether, the co-transfection experiments indicated that the transcriptional activation provided from the HRE enhancer element requires Hox, Pbx1a and Prep1 partnership.
Binding of Hox and Pbx factors on the HRE sequence
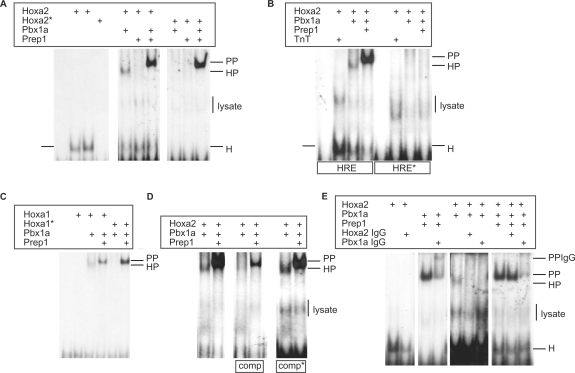
To test the physical interaction of Hox proteins and their Pbx cofactors with the HRE sequence, EMSA experiments were performed. The Hoxa2 protein alone was able to bind to oligonucleotides with the HRE sequence though not the DNA-binding defective Hoxa2(QN-AA) protein (Figure 4A). No detectable gel retarded complex was observed with the Hoxa1 protein alone (Figure 4C). Addition of in vitro synthesized Pbx1a proteins resulted in higher protein DNA complex formation confirming that Hox–Pbx interacted on the HRE sequence. Such Hox–Pbx complexes were observed both with Hoxa1 and Hoxa2, but not with proteins mutated in their homoeodomain (QN-AA substitution) or hexapeptide (WM-AA) (Figure 4A and C). Assays involving both Pbx1a and Prep1 proteins resulted in retarded complexes showing identical electrophoretic mobility with either wild-type or mutant Hoxa1 or Hoxa2 (Figure 4A and C). Since the mutant Hox proteins cannot bind either DNA or Pbx, the observed shifted complexes should contain only Pbx1a and Prep1, suggesting that no trimeric interaction can occur on the HRE sequence. This was confirmed by including anti-Hoxa2 or anti-Pbx1 antibodies in the assays. Although the anti-Hoxa2 antibody chased Hoxa2–DNA and Hoxa2–Pbx1–DNA complex formation, it did not chase complexes obtained in the presence of Pbx1a and Prep1 proteins (Figure 4E). Conversely, the anti-Pbx1 antibody chased and super-shifted the complexes obtained with Pbx1a and Prep1 proteins (Figure 4E).
Figure 4.
Hoxa2 and Pbx1a bind to the HRE sequence. (A) Gel retardation assays were performed with double-stranded oligonucleotides bearing the wild-type HRE sequence, in the presence of E. coli purified His-tagged Hoxa2 protein, its Hoxa2(QN-AA) mutant derivative (Hoxa2*) and in vitro translated Pbx1a and Prep1 proteins. Hoxa2 binds to the HRE sequence alone (H), or in combination with Pbx1a (HP). Addition of both Pbx1a and Prep1 generated a similar retarded complex (PP) whatever the wild-type or mutant Hoxa2 protein is involved (see text for comments). The binding assays involving the in vitro translated samples generated aspecific retarded complexes corresponding to the reticulocyte extracts (lysate). (B) Similar experiments were run with oligonucleotides containing a mutated HRE site. While the wild-type sequence (HRE) allowed complex formation with Hoxa2 (H), Hoxa2–Pbx1a (HP) or Pbx1a and Prep1 (PP), no binding was observed on the mutant sequence (HRE*). Assays involving the purified Hoxa2 protein and reticulocyte extracts devoid of expression vectors (TnT) reveal aspecific complex formation (lysate). (C) The HRE sequence is recognized by Hoxa1 and Pbx1a (HP) while not by Hoxa1. The hexapeptide mutant Hoxa1(WM-AA) does not bind the HRE sequence, neither alone nor in combination with Pbx1a. Again, addition of both Pbx1a and Prep1 generates a similarly retarded complex with either the wild-type or mutant Hoxa1 protein (see text for comments). (D) To address the specificity of Hoxa2 binding to the HRE sequence, competition experiments were performed with a 100-fold molar excess of unlabelled wild-type (comp) or mutant oligonucleotides (comp*) with respect to labelled probes. Only the wild-type competitor titrates out both Hoxa2–Pbx1a (HP) and Pbx1a-Prep1 (PP) complex formation. (E) Assays including anti-Hoxa2 or anti-Pbx1 antibodies were performed to confirm the identity of the proteins involved in the shifted complexes. Complex formation with Hoxa2 (H) and Hoxa2–Pbx1a (HP) was impaired by the anti-Hoxa2 antibody, whereas complexes obtained by involving Pbx1a and Prep1 or Pbx1a, Prep1 and Hoxa2 were not. This shows that only dimeric Pbx1a–Prep1 complexes (PP) were formed in the presence of Pbx1a and Prep1 proteins, and that no trimeric complexes including Hoxa2 were obtained. Conversely, the anti-Pbx1 antibody chased and super-shifted the Pbx1a–Prep1 containing complexes (PPIgG).
Nucleotide substitutions in the HRE sequence abolished the Hoxa2–DNA, Hoxa2–Pbx1a–DNA and Pbx1a–Prep1–DNA interactions (Figure 4B). Finally, competition experiments involving an excess of unlabelled mutant oligonucleotides with respect to the labelled wild-type confirmed that Hoxa2, Hoxa2–Pbx1a and Pbx1a–Prep1 specifically bound to the HRE sequence (Figure 4D).
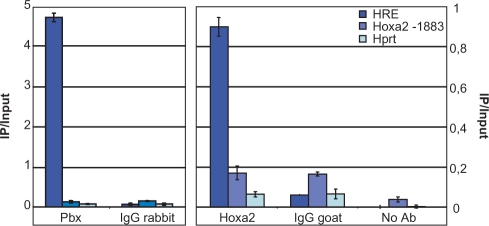
To provide evidence that Hoxa2 or Pbx-containing complexes were formed on the HRE in the context of chromatin, we performed ChIP assays with anti-Pbx and anti-Hoxa2 antibodies on EC P19 cells induced by retinoic acid (RA) for 72 h. Under these conditions, significant endogenous Pbx and Hoxa2 expression levels were present (data not shown). Following Pbx or Hoxa2 targeted immunoprecipitation and quantitative real-time PCR (qPCR), the HRE sequence was significantly amplified as compared to the Hprt locus used as negative control (Figure 5). As additional controls, immunoprecipitation involving unrelated, anti-IgG antibodies did not lead to significant HRE sequence recovery. Also, qPCR amplification directed to a 81-bp sequence located 1883-bp upstream of the Hoxa2 transcriptional start site revealed that this upstream sequence was not significantly immunoprecipitated. Altogether, these data demonstrate that the HRE was selectively retrieved by ChIP with both the anti-Pbx and the anti-Hoxa2 antibodies.
Figure 5.
ChIP experiments with anti-Hoxa2 and anti-Pbx antibodies resulted in the retrieval of the HRE-containing sequence. Quantitative real-time PCR reveals a significant enrichment of the HRE sequence (HRE) upon Hoxa2 (Hoxa2) and Pbx (Pbx) immunoprecipitations, by comparison with anti-IgG precipitation (IgG rabbit, IgG goat) or with the no antibody control (No Ab). A short sequence residing 1883 bp 5′ to the Hoxa2 transcription start site (Hoxa2-1883) was not specifically immunoprecipitated with the anti-Hoxa2 and anti-Pbx antibodies as compared to the anti-IgG IP. Similarly, no specific immunoprecipitation was observed for the unrelated Hprt locus (Hprt). ChIP values are expressed as percentage of input DNA (IP/Input, n = 2), for one representative experiment out of four independent ones.
Conservation of Hox-mediated HRE spatial activity in the hindbrain of chicken embryos
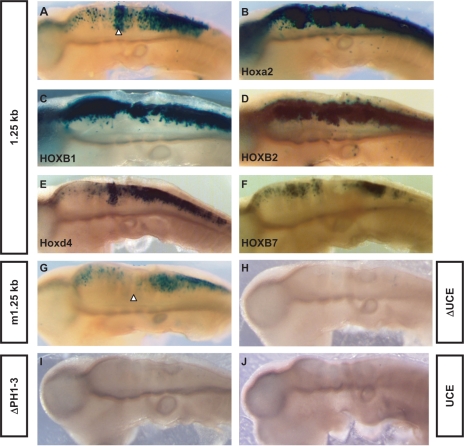
To evaluate the contribution of HRE to the regulation of spatial expression in vivo, we first inserted a reporter lacZ gene under the control of the 1.25-kb fragment (1.25kb-β-globin-lacZ plasmid, see ‘Materials and methods’ section for details) and electroporated the construct in the neural tube of chick embryos at HH stage 10–12. The 1.25-kb DNA fragment including the HRE element was previously shown to drive lacZ expression in the r4 of transgenic mouse embryos (24). A strong and reproducible lacZ activity was detected in chick r4, similar to the mouse (Figure 6A; see also in ref. 24). X-gal staining was also detected in the posterior hindbrain-rostral spinal cord, as well as weakly in rostral hindbrain (Figure 6A). In contrast, electroporation of a construct (m1.25kb-β-globin-lacZ) containing the same selective mutation in the HRE element as previously tested in cell transfection assays reproducibly resulted in loss or very severe reduction of r4 staining (Figure 6G). The remainder of the enhancer expression pattern in the neural tube was conserved, albeit detected at a lower level.
Figure 6.
The HRE element is active and responds to Hox-1 and -2 paralogues in the chick neural tube. X-gal staining reveals the expression pattern of the lacZ reporter gene under the control of the 1.25-kb region (A to F) or its mutant m1.25-kb derivative (G) in electroporated chick neural tube. Enhancement of reporter expression is observed upon co-electroporation of a Hox-1 and -2 expression vectors (B to D) while not upon Hoxd4 or HOXB7 (E, F). Deletion of either the UCE sequence (H, ΔUCE) or the intronic PH sites (I, ΔPH1-3) from the 1.25-kb region results in a loss of reporter activity. Consistently, the UCE sequence is not able to drive lacZ expression in electroporated chick neural tube on its own (J). White arrowheads show the localization of the fourth rhombomere.
The 1.25-kb genomic fragment used to drive reporter expression in our assay contained the three intronic Hox/Pbx-binding sites (PH1-3) previously shown to be active in r4 (25). Although these sites were sufficient to cooperatively drive lacZ expression in r4 from distinct reporter constructs electroporated in the chick neural tube, their presence was not sufficient to support gene stimulation when we deleted the HRE site from the 1.25-kb region (Figure 6H). Similarly, when deleting the intronic PH sites, the intact HRE site was not able to provide r4 expression on its own (Figure 6I). Finally, a reporter construct in which lacZ was inserted under the control of a 410-bp Hoxa2 fragment containing the UCE was not expressed in the chick neural tube showing that the UCE per se is not sufficient to provide gene expression (Figure 6J). These data clearly show that the activity of the 1.25-kb enhancer region in r4 relies on the molecular cooperation over a large distance of at least 4 PH cis-acting elements.
Co-electroporation of the wild-type reporter with expression vectors for group 1 or group 2 Hox proteins, i.e. Hoxa2, HOXB1 or HOXB2, all resulted in robust transactivation and consequent strong lacZ upregulation in the chick hindbrain and spinal cord in vivo (Figure 6B–D), whereas co-electroporation with Hoxd4 or HOXB7 expression vectors did not yield significant upregulation of the reporter construct (Figure 6E and F).
These data therefore demonstrate that the coding HRE element is necessary for selective control of enhancer activity in r4 and that the intronic PH sites and the exonic HRE are required together to provide r4-specific expression. Consistently, while the HRE is contained in a larger UCE, this UCE does not show enhancer activity per se. Finally, our data also demonstrate that the 1.25-kb enhancer is specifically responsive to paralogue group 1 and 2 Hox proteins in vivo.
DISCUSSION
Hox genes are involved in the patterning of the hindbrain and recent studies contributed to describe how cross talk among Hoxa1, Hoxb1, Hoxa2 and Hoxb2 confer and maintain specific segmental identities in the rostral hindbrain (16–19,22,23,25,47–50).
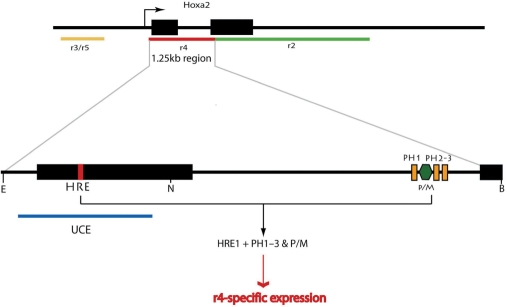
Frasch et al. (24) described 1.25- and 2.5-kb long DNA regions encompassing the Hoxa2 locus that were able to direct the expression of a reporter gene in the r4 and r2 territories, respectively. Recently, Tümpel et al. (25,51) identified evolutionary conserved r2- and r4-specific cis-regulatory sequences residing in the second exon and in the intron of the Hoxa2 gene, respectively. In particular, the r4 regulatory module consisted in three Hox/Pbx (PH) bipartite binding sites flanking a Pbx/Meis site (PM) and was shown to respond to Hoxb1 and Hoxa2. From their data, Tümpel et al. (25) proposed a model for the initiation, establishment and maintenance of r4 identity relying on Hox1 and Hox2 gene cross- and auto-regulations. Here, we identified a Hox–Pbx–Prep responsive element (HRE) residing in the first exon of Hoxa2, thus located within the 1.25-kb fragment driving expression in r4. We demonstrated that this element is bound by Hoxa2 and Pbx in the chromatin of differentiating EC cells, is required for gene expression in r4 in vivo and critically cooperates with the previously reported PH-PM sites (Figure 7).
Figure 7.
The r4-specific expression of Hoxa2 results from the cooperative activity of exonic and intronic Hox–Pbx regulatory elements. Three Hox–Pbx elements (PH1-3, orange rectangles) associated to a Prep/Meis-binding site (P/M, dark green hexagon) reside in the intron of Hoxa2 (25) and synergize with the HRE sequence (red box) embedded in the UCE (blue line) overlapping with the coding sequence of the first exon (black rectangles: Hoxa2 coding sequence). Our data demonstrated that both the intronic and exonic elements are required to provide a reproducible reporter expression in the r4 domain of the developing hindbrain. The r3/r5, r4 and r2-specific enhancer regions governing the spatial pattern of Hoxa2 expression in the developing hindbrain are represented as yellow, red and green lines underlying the Hoxa2 locus, respectively (arrow: transcription initiation site of Hoxa2).
The intronic PH sites identified by Tümpel et al. (25) were reported to be active in r4 when contained in reporter constructs which did not include the HRE. It is worth noticing that these authors injected 2.5 to 7 times more reporter DNA for the chick electroporation than in our experiments (25). In this work, we show that at lower DNA concentrations the HRE and PH sites were required together to provide detectable reporter activity in the chick neural tube. This finding highlights that those intragenic regulatory modules critically cooperate for the segmental expression of Hoxa2 in r4. Notably, the mutation of the HRE sequence selectively impaired the activity of the 1.25-kb region in r4, while the spatial distribution of the remainder of the enhancer activity was unchanged and showed persistent reporter expression, albeit at lower levels, in the hindbrain and spinal cord (Figure 6G).
On the basis of EMSA data and chick neural tube co-electroporation, it has been proposed that the intronic PH and PM regulatory sequences take part in Hoxb1-to-Hoxa2 cross-regulation as well as in Hoxa2 auto-regulation. Here we supported and further extended this proposal by providing evidence that the r4 enhancer activity of the 1.25-kb genomic fragment bearing both the intronic and HRE sequences was responsive to HOXB1, HOXB2 and Hoxa2, while not so to Hox proteins of other paralogue groups. We also showed that both Hoxa1 and Hoxa2 physically bound to the HRE together with Pbx proteins. Our data therefore bring further support to the idea that multiple cross- and auto-regulatory controls between group 1 and 2 Hox proteins contribute to establish and maintain the r4 identity.
Interestingly, the mouse 1.25-kb region containing the PH-PM sites and HRE was active in the developing hindbrain of the chick embryo, with a prominent expression in r4 (Figure 6A), similar to its pattern of activity in the mouse embryo (24), thus supporting conservation of Hox-dependent regulatory mechanisms in vertebrate hindbrain development (4,52,53). The PH–PM sites have been shown to be evolutionary conserved among vertebrates, although Xenopus genomes intriguingly lacked them. All vertebrate species share the exonic HRE. In particular, we showed that zebrafish and Xenopus Hoxa2 displayed a conserved HRE which was also responsive to Hox proteins in cell transfections. Therefore, one possibility is that in Xenopus the HRE might account for the r4-specific expression of Hoxa2 in the absence of the intronic module.
A striking feature of the HRE is that it is located in an UCE, which corresponds to the 5′ end of the Hoxa2 coding sequence. This UCE has been previously identified by Bejerano et al. (26) as ultraconserved region (uc.). However, it was not reported as lying in the Hoxa2 coding sequence. UCEs have been recently defined as genome segments of 200 bp or more that are absolutely conserved between orthologous regions of mammalian genomes (26). The extreme sequence conservation of UCEs suggests that these elements play vital roles for their host; however, deletion of UCEs was reported to yield viable mice (54). A total of 481 UCEs have been reported so far among which 12 reside within or in the vicinity of the Hox complexes (26). Five of these UCEs were described as partially overlapping with Hox coding sequences. The function of the UCEs is supposed to be diverse. Intergenic UCEs have been proposed to play a role in regulatory networks of transcriptional control (55–58). However, some in vivo enhancer analyses aiming to address UCEs functions involved reporter constructs based on large genomic fragments not strictly confined to the UCE sequences (56). It is of particular significance that although the HRE-bearing UCE contributes to Hoxa2 regulation, the UCE sequence on its own did not seem to provide r4-enhancer activity.
Exonic UCEs are known to play roles in RNA processing control (55,59,60). Others corresponding to non-coding RNAs (ncRNA) have been proposed to contribute to post-transcriptional gene regulation, in parallel to or together with miRNAs (61). Whether the latter possibilities also apply for the Hoxa2 UCE function remains to be addressed. However, our study reveals a more intricate situation because while being exonic and coding, Uc.212 also contains the HRE sequence and takes part in transcriptional control. Intricate and superposed regulatory functions for an UCE have been revealed for an element involved in the transcriptional regulation of Dlx5/6. This UCE contains an intergenic enhancer (ei) recognized by Dlx2 and is also transcribed as a part of a non-coding RNA molecule which acts as a transcriptional co-activator of Dlx2 and thereby contributes to the ei enhancer activity (57). Alone, the presence of HRE cannot account for the extreme sequence conservation observed over 205 bp. It is therefore reasonable to assume that other cis-(DNA) or trans-(RNA) regulatory elements reside in this sequence interval and take part in an integrative gene control. In this respect, it should be mentioned that phylogenetically conserved enhancers may not only share the spatial arrangement of conserved cis-regulatory sequences but also the sequence of the spacers between them, which may underscore as yet unknown functional and evolutionary constraints (62).
Together with a recently reported r2-specific cis-regulatory element residing in the second exon of Hoxa2 (51), it is the first time that a cis-acting element is identified in the coding sequence of a Hox gene, and to our knowledge, only very few exonic enhancers have been reported so far, all lying within untranslated gene regions (63–65). A trivial explanation might be that searches for enhancers/silencers of transcription are biased towards non-coding regions of the genome. Softwares and databases devoted to the identification of putative cis-regulatory elements often exclude exonic regions from their analyses (66–69). Alternatively, the paucity of regulatory sequences in gene coding regions may be explained in evolutionary terms, to uncouple the evolution of expression patterns from that of coding sequences. In the particular context of the homeotic complexes, the strong link between the integrity of Hox gene clusters, Hox gene expression patterns and Hox gene function may have led to unusual coupling between gene regulation and gene function determinants (70).
The possibility of such a coupling is reinforced by the observation that the role of Hoxa2 in craniofacial and neural development of the vertebrate embryo has been highly conserved throughout vertebrate evolution including in teleost fishes, amphibians, birds or mammals (33,71–74). Altogether, these data indicate that the identified Hox responsive element HRE, which resides in a coding UCE is a crucial element with a conserved role in Hox-dependent r4 regulation in the vertebrate embryo.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Dr Marc Geisen for help with in ovo electroporation experiments and fruitful discussions, Dr Damien Hermand for support in ChIP assays, Dr Bernard Peers for kindly providing the pCS2-Prep1 vector and Dr Mark S. Featherstone for the pAdMLluc reporter and Hoxd4 expression vector. This work was supported by the Belgian Fund for Scientific Research (Crédit aux chercheurs, FNRS) and the Fonds Spéciaux de Recherche (FSR) of the Université catholique de Louvain (UCL). X.L. held a FRIA fellowship from the FNRS, a FSR grant (UCL); and fellowships from the Association Française contre les Myopathies (AFM) and Fondation pour la Recherche Médicale (FRM). C.M. held a FRIA fellowship from the FNRS; S.R. held a FRIA fellowship from the FNRS, an FSR grant from the UCL and a Télévie grant (FNRS). O.A.S. was supported by La Ligue Nationale Contre Le Cancer and Association pour la Recherche sur le Cancer (ARC). Work in FMR′s laboratory was supported by grants from the FRM (‘Equipe labelisée’), AFM, Agence Nationale pour la Recherche (ANR), Association pour la Recherche contre le Cancer (ARC), and by institutional funds from CNRS and INSERM. Funding to pay the Open Access publication charges for this article was provided by UCL.
Conflict of interest statement. None declared.
REFERENCES
- 1.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 2.Lumsden A, Krumlauf R. Patterning the vertebrate neuraxis. Science. 1996;274:1109–1115. doi: 10.1126/science.274.5290.1109. [DOI] [PubMed] [Google Scholar]
- 3.Capecchi MR. Hox genes and mammalian development. Cold Spring Harb. Symp. Quant. Biol. 1997;62:273–281. [PubMed] [Google Scholar]
- 4.Rijli FM, Gavalas A, Chambon P. Segmentation and specification in the branchial region of the head: the role of the Hox selector genes. Int. J. Dev. Biol. 1998;42:393–401. [PubMed] [Google Scholar]
- 5.Kmita M, Duboule D. Organizing axes in time and space; 25 years of colinear tinkering. Science. 2003;301:331–333. doi: 10.1126/science.1085753. [DOI] [PubMed] [Google Scholar]
- 6.Hoegg S, Meyer A. Hox clusters as models for vertebrate genome evolution. Trends Genet. 2005;21:421–424. doi: 10.1016/j.tig.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Graham A, Papalopulu N, Krumlauf R. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell. 1989;57:367–378. doi: 10.1016/0092-8674(89)90912-4. [DOI] [PubMed] [Google Scholar]
- 8.Dolle P, Izpisua-Belmonte JC, Falkenstein H, Renucci A, Duboule D. Coordinate expression of the murine Hox-5 complex homoeobox-containing genes during limb pattern formation. Nature. 1989;342:767–772. doi: 10.1038/342767a0. [DOI] [PubMed] [Google Scholar]
- 9.Deschamps J, van Nes J. Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development. 2005;132:2931–2942. doi: 10.1242/dev.01897. [DOI] [PubMed] [Google Scholar]
- 10.Lufkin T, Dierich A, LeMeur M, Mark M, Chambon P. Disruption of the Hox-1.6 homeobox gene results in defects in a region corresponding to its rostral domain of expression. Cell. 1991;66:1105–1119. doi: 10.1016/0092-8674(91)90034-v. [DOI] [PubMed] [Google Scholar]
- 11.Gendron-Maguire M, Mallo M, Zhang M, Gridley T. Hoxa-2 mutant mice exhibit homeotic transformation of skeletal elements derived from cranial neural crest. Cell. 1993;75:1317–1331. doi: 10.1016/0092-8674(93)90619-2. [DOI] [PubMed] [Google Scholar]
- 12.Rijli FM, Mark M, Lakkaraju S, Dierich A, Dolle P, Chambon P. A homeotic transformation is generated in the rostral branchial region of the head by disruption of Hoxa-2, which acts as a selector gene. Cell. 1993;75:1333–1349. doi: 10.1016/0092-8674(93)90620-6. [DOI] [PubMed] [Google Scholar]
- 13.Barrow JR, Capecchi MR. Targeted disruption of the Hoxb-2 locus in mice interferes with expression of Hoxb-1 and Hoxb-4. Development. 1996;122:3817–3828. doi: 10.1242/dev.122.12.3817. [DOI] [PubMed] [Google Scholar]
- 14.Goddard JM, Rossel M, Manley NR, Capecchi MR. Mice with targeted disruption of Hoxb-1 fail to form the motor nucleus of the VIIth nerve. Development. 1996;122:3217–3228. doi: 10.1242/dev.122.10.3217. [DOI] [PubMed] [Google Scholar]
- 15.Gavalas A, Davenne M, Lumsden A, Chambon P, Rijli FM. Role of Hoxa-2 in axon pathfinding and rostral hindbrain patterning. Development. 1997;124:3693–3702. doi: 10.1242/dev.124.19.3693. [DOI] [PubMed] [Google Scholar]
- 16.Gavalas A, Trainor P, Ariza-McNaughton L, Krumlauf R. Synergy between Hoxa1 and Hoxb1: the relationship between arch patterning and the generation of cranial neural crest. Development. 2001;128:3017–3027. doi: 10.1242/dev.128.15.3017. [DOI] [PubMed] [Google Scholar]
- 17.Barrow JR, Stadler HS, Capecchi MR. Roles of Hoxa1 and Hoxa2 in patterning the early hindbrain of the mouse. Development. 2000;127:933–944. doi: 10.1242/dev.127.5.933. [DOI] [PubMed] [Google Scholar]
- 18.Davenne M, Maconochie MK, Neun R, Pattyn A, Chambon P, Krumlauf R, Rijli FM. Hoxa2 and Hoxb2 control dorsoventral patterns of neuronal development in the rostral hindbrain. Neuron. 1999;22:677–691. doi: 10.1016/s0896-6273(00)80728-x. [DOI] [PubMed] [Google Scholar]
- 19.Popperl H, Bienz M, Studer M, Chan SK, Aparicio S, Brenner S, Mann RS, Krumlauf R. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell. 1995;81:1031–1042. doi: 10.1016/s0092-8674(05)80008-x. [DOI] [PubMed] [Google Scholar]
- 20.Di Rocco G, Mavilio F, Zappavigna V. Functional dissection of a transcriptionally active, target-specific Hox-Pbx complex. EMBO J. 1997;16:3644–3654. doi: 10.1093/emboj/16.12.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berthelsen J, Zappavigna V, Mavilio F, Blasi F. Prep1, a novel functional partner of Pbx proteins. EMBO J. 1998;17:1423–1433. doi: 10.1093/emboj/17.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobs Y, Schnabel CA, Cleary ML. Trimeric association of Hox and TALE homeodomain proteins mediates Hoxb2 hindbrain enhancer activity. Mol. Cell Biol. 1999;19:5134–5142. doi: 10.1128/mcb.19.7.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferretti E, Marshall H, Popperl H, Maconochie M, Krumlauf R, Blasi F. Segmental expression of Hoxb2 in r4 requires two separate sites that integrate cooperative interactions between Prep1, Pbx and Hox proteins. Development. 2000;127:155–166. doi: 10.1242/dev.127.1.155. [DOI] [PubMed] [Google Scholar]
- 24.Frasch M, Chen X, Lufkin T. Evolutionary-conserved enhancers direct region-specific expression of the murine Hoxa-1 and Hoxa-2 loci in both mice and Drosophila. Development. 1995;121:957–974. doi: 10.1242/dev.121.4.957. [DOI] [PubMed] [Google Scholar]
- 25.Tumpel S, Cambronero F, Ferretti E, Blasi F, Wiedemann LM, Krumlauf R. Expression of Hoxa2 in rhombomere 4 is regulated by a conserved cross-regulatory mechanism dependent upon Hoxb1. Dev. Biol. 2007;302:646–660. doi: 10.1016/j.ydbio.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 26.Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 27.Chiu CH, Amemiya C, Dewar K, Kim CB, Ruddle FH, Wagner GP. Molecular evolution of the HoxA cluster in the three major gnathostome lineages. Proc. Natl Acad. Sci. USA. 2002;99:5492–5497. doi: 10.1073/pnas.052709899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim CB, Amemiya C, Bailey W, Kawasaki K, Mezey J, Miller W, Minoshima S, Shimizu N, Wagner G, Ruddle F. Hox cluster genomics in the horn shark, Heterodontus francisci. Proc. Natl Acad. Sci. USA. 2000;97:1655–1660. doi: 10.1073/pnas.030539697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez P, Amemiya CT. Genomics of the HOX gene cluster. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2002;133:571–580. doi: 10.1016/s1096-4959(02)00121-5. [DOI] [PubMed] [Google Scholar]
- 30.Santini S, Boore JL, Meyer A. Evolutionary conservation of regulatory elements in vertebrate Hox gene clusters. Genome Res. 2003;13:1111–1122. doi: 10.1101/gr.700503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Popperl H, Featherstone MS. An autoregulatory element of the murine Hox-4.2 gene. EMBO J. 1992;11:3673–3680. doi: 10.1002/j.1460-2075.1992.tb05452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang M, Kim HJ, Marshall H, Gendron-Maguire M, Lucas DA, Baron A, Gudas LJ, Gridley T, Krumlauf R, Grippo JF. Ectopic Hoxa-1 induces rhombomere transformation in mouse hindbrain. Development. 1994;120:2431–2442. doi: 10.1242/dev.120.9.2431. [DOI] [PubMed] [Google Scholar]
- 33.Pasqualetti M, Ori M, Nardi I, Rijli FM. Ectopic Hoxa2 induction after neural crest migration results in homeosis of jaw elements in Xenopus. Development. 2000;127:5367–5378. doi: 10.1242/dev.127.24.5367. [DOI] [PubMed] [Google Scholar]
- 34.Studer M, Lumsden A, Ariza-McNaughton L, Bradley A, Krumlauf R. Altered segmental identity and abnormal migration of motor neurons in mice lacking Hoxb-1. Nature. 1996;384:630–634. doi: 10.1038/384630a0. [DOI] [PubMed] [Google Scholar]
- 35.Matis C, Chomez P, Picard J, Rezsohazy R. Differential and opposed transcriptional effects of protein fusions containing the VP16 activation domain. FEBS Lett. 2001;499:92–96. doi: 10.1016/s0014-5793(01)02532-7. [DOI] [PubMed] [Google Scholar]
- 36.Remacle S, Shaw-Jackson C, Matis C, Lampe X, Picard J, Rezsohazy R. Changing homeodomain residues 2 and 3 of Hoxa1 alters its activity in a cell-type and enhancer dependent manner. Nucleic Acids Res. 2002;30:2663–2668. doi: 10.1093/nar/gkf372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goudet G, Delhalle S, Biemar F, Martial JA, Peers B. Functional and cooperative interactions between the homeodomain PDX1, Pbx, and Prep1 factors on the somatostatin promoter. J. Biol. Chem. 1999;274:4067–4073. doi: 10.1074/jbc.274.7.4067. [DOI] [PubMed] [Google Scholar]
- 38.Rambaldi I, Kovacs EN, Featherstone MS. A proline-rich transcriptional activation domain in murine HOXD-4 (HOX-4.2) Nucleic Acids Res. 1994;22:376–382. doi: 10.1093/nar/22.3.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. 1951. Dev. Dyn. 1992;195:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- 40.Nakamura H, Funahashi J. Introduction of DNA into chick embryos by in ovo electroporation. Methods. 2001;24:43–48. doi: 10.1006/meth.2001.1155. [DOI] [PubMed] [Google Scholar]
- 41.Samad OA, Geisen MJ, Caronia G, Varlet I, Zappavigna V, Ericson J, Goridis C, Rijli FM. Integration of anteroposterior and dorsoventral regulation of Phox2b transcription in cranial motoneuron progenitors by homeodomain proteins. Development. 2004;131:4071–4083. doi: 10.1242/dev.01282. [DOI] [PubMed] [Google Scholar]
- 42.Lampe X, Picard JJ, Rezsohazy R. The Hoxa2 enhancer 2 contains a critical Hoxa2 responsive regulatory element. Biochem. Biophys. Res. Commun. 2004;316:898–902. doi: 10.1016/j.bbrc.2004.02.138. [DOI] [PubMed] [Google Scholar]
- 43.Ferretti E, Cambronero F, Tumpel S, Longobardi E, Wiedemann LM, Blasi F, Krumlauf R. Hoxb1 enhancer and control of rhombomere 4 expression: complex interplay between PREP1-PBX1-HOXB1 binding sites. Mol. Cell Biol. 2005;25:8541–8552. doi: 10.1128/MCB.25.19.8541-8552.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mann RS, Affolter M. Hox proteins meet more partners. Curr. Opin. Genet. Dev. 1998;8:423–429. doi: 10.1016/s0959-437x(98)80113-5. [DOI] [PubMed] [Google Scholar]
- 45.Berthelsen J, Kilstrup-Nielsen C, Blasi F, Mavilio F, Zappavigna V. The subcellular localization of PBX1 and EXD proteins depends on nuclear import and export signals and is modulated by association with PREP1 and HTH. Genes Dev. 1999;13:946–953. doi: 10.1101/gad.13.8.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phelan ML, Rambaldi I, Featherstone MS. Cooperative interactions between HOX and PBX proteins mediated by a conserved peptide motif. Mol. Cell Biol. 1995;15:3989–3997. doi: 10.1128/mcb.15.8.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maconochie MK, Nonchev S, Studer M, Chan SK, Popperl H, Sham MH, Mann RS, Krumlauf R. Cross-regulation in the mouse HoxB complex: the expression of Hoxb2 in rhombomere 4 is regulated by Hoxb1. Genes Dev. 1997;11:1885–1895. doi: 10.1101/gad.11.14.1885. [DOI] [PubMed] [Google Scholar]
- 48.Gavalas A, Studer M, Lumsden A, Rijli FM, Krumlauf R, Chambon P. Hoxa1 and Hoxb1 synergize in patterning the hindbrain, cranial nerves and second pharyngeal arch. Development. 1998;125:1123–1136. doi: 10.1242/dev.125.6.1123. [DOI] [PubMed] [Google Scholar]
- 49.Nonchev S, Vesque C, Maconochie M, Seitanidou T, Ariza-McNaughton L, Frain M, Marshall H, Sham MH, Krumlauf R, Charnay P. Segmental expression of Hoxa-2 in the hindbrain is directly regulated by Krox-20. Development. 1996;122:543–554. doi: 10.1242/dev.122.2.543. [DOI] [PubMed] [Google Scholar]
- 50.Studer M, Gavalas A, Marshall H, Ariza-McNaughton L, Rijli FM, Chambon P, Krumlauf R. Genetic interactions between Hoxa1 and Hoxb1 reveal new roles in regulation of early hindbrain patterning. Development. 1998;125:1025–1036. doi: 10.1242/dev.125.6.1025. [DOI] [PubMed] [Google Scholar]
- 51.Tumpel S, Cambronero F, Wiedemann LM, Krumlauf R. Evolution of cis elements in the differential expression of two Hoxa2 coparalogous genes in pufferfish (Takifugu rubripes) Proc. Natl Acad. Sci. USA. 2006;103:5419–5424. doi: 10.1073/pnas.0600993103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trainor PA, Krumlauf R. Patterning the cranial neural crest: hindbrain segmentation and Hox gene plasticity. Nat. Rev. Neurosci. 2000;1:116–124. doi: 10.1038/35039056. [DOI] [PubMed] [Google Scholar]
- 53.Kiecker C, Lumsden A. Compartments and their boundaries in vertebrate brain development. Nat. Rev. Neurosci. 2005;6:553–564. doi: 10.1038/nrn1702. [DOI] [PubMed] [Google Scholar]
- 54.Ahituv N, Zhu Y, Visel A, Holt A, Afzal V, Pennacchio LA, Rubin EM. Deletion of ultraconserved elements yields viable mice. PLoS Biol. 2007;5:e234. doi: 10.1371/journal.pbio.0050234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bejerano G, Lowe CB, Ahituv N, King B, Siepel A, Salama SR, Rubin EM, Kent WJ, Haussler D. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- 56.Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 57.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paparidis Z, Abbasi AA, Malik S, Goode DK, Callaway H, Elgar G, deGraaff E, Lopez-Rios J, Zeller R, Grzeschik KH. Ultraconserved non-coding sequence element controls a subset of spatiotemporal GLI3 expression. Dev. Growth Differ. 2007;49:543–553. doi: 10.1111/j.1440-169X.2007.00954.x. [DOI] [PubMed] [Google Scholar]
- 59.Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 60.Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O’Brien G, Shiue L, Clark TA, Blume JE, Ares M., Jr. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 62.Boffelli D, Nobrega MA, Rubin EM. Comparative genomics at the vertebrate extremes. Nat. Rev. Genet. 2004;5:456–465. doi: 10.1038/nrg1350. [DOI] [PubMed] [Google Scholar]
- 63.Frenkel B, Montecino M, Stein JL, Lian JB, Stein GS. A composite intragenic silencer domain exhibits negative and positive transcriptional control of the bone-specific osteocalcin gene: promoter and cell type requirements. Proc. Natl Acad. Sci. USA. 1994;91:10923–10927. doi: 10.1073/pnas.91.23.10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grad JM, Lyons LS, Robins DM, Burnstein KL. The androgen receptor (AR) amino-terminus imposes androgen-specific regulation of AR gene expression via an exonic enhancer. Endocrinology. 2001;142:1107–1116. doi: 10.1210/endo.142.3.8049. [DOI] [PubMed] [Google Scholar]
- 65.Jiang F, Li P, Fornace A.J., Jr, Nicosia SV, Bai W. G2/M arrest by 1,25-dihydroxyvitamin D3 in ovarian cancer cells mediated through the induction of GADD45 via an exonic enhancer. J. Biol. Chem. 2003;278:48030–48040. doi: 10.1074/jbc.M308430200. [DOI] [PubMed] [Google Scholar]
- 66.Rebeiz M, Reeves NL, Posakony JW. SCORE: a computational approach to the identification of cis-regulatory modules and target genes in whole-genome sequence data. Site clustering over random expectation. Proc. Natl Acad. Sci. USA. 2002;99:9888–9893. doi: 10.1073/pnas.152320899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sandelin A, Wasserman WW, Lenhard B. ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 2004;32:W249–W252. doi: 10.1093/nar/gkh372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marinescu VD, Kohane IS, Riva A. The MAPPER database: a multi-genome catalog of putative transcription factor binding sites. Nucleic Acids Res. 2005;33:D91–D97. doi: 10.1093/nar/gki103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun H, Palaniswamy SK, Pohar TT, Jin VX, Huang TH, Davuluri RV. MPromDb: an integrated resource for annotation and visualization of mammalian gene promoters and ChIP-chip experimental data. Nucleic Acids Res. 2006;34:D98–D103. doi: 10.1093/nar/gkj096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duboule D. Vertebrate hox gene regulation: clustering and/or colinearity? Curr. Opin. Genet. Dev. 1998;8:514–518. doi: 10.1016/s0959-437x(98)80004-x. [DOI] [PubMed] [Google Scholar]
- 71.Baltzinger M, Ori M, Pasqualetti M, Nardi I, Rijli FM. Hoxa2 knockdown in Xenopus results in hyoid to mandibular homeosis. Dev. Dyn. 2005;234:858–867. doi: 10.1002/dvdy.20567. [DOI] [PubMed] [Google Scholar]
- 72.Hunter MP, Prince VE. Zebrafish hox paralogue group 2 genes function redundantly as selector genes to pattern the second pharyngeal arch. Dev. Biol. 2002;247:367–389. doi: 10.1006/dbio.2002.0701. [DOI] [PubMed] [Google Scholar]
- 73.Santagati F, Rijli FM. Cranial neural crest and the building of the vertebrate head. Nat. Rev. Neurosci. 2003;4:806–818. doi: 10.1038/nrn1221. [DOI] [PubMed] [Google Scholar]
- 74.Creuzet S, Couly G, Le Douarin NM. Patterning the neural crest derivatives during development of the vertebrate head: insights from avian studies. J. Anat. 2005;207:447–459. doi: 10.1111/j.1469-7580.2005.00485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.