Abstract
I-SceI is a homing endonuclease that specifically cleaves an 18-bp double-stranded DNA. I-SceI exhibits a strong preference for cleaving the bottom strand DNA. The published structure of I-SceI bound to an uncleaved DNA substrate provided a mechanism for bottom strand cleavage but not for top strand cleavage. To more fully elucidate the I-SceI catalytic mechanism, we determined the X-ray structures of I-SceI in complex with DNA substrates that are nicked in either the top or bottom strands. The structures resemble intermediates along the DNA cleavage reaction. In a structure containing a nick in the top strand, the spatial arrangement of metal ions is similar to that observed in the structure that contains uncleaved DNA, suggesting that cleavage of the bottom strand occurs by a common mechanism regardless of whether this strand is cleaved first or second. In the structure containing a nick in the bottom strand, a new metal binding site is present in the active site that cleaves the top strand. This new metal and a candidate nucleophilic water molecule are correctly positioned to cleave the top strand following bottom strand cleavage, providing a plausible mechanism for top strand cleavage.
INTRODUCTION
I-SceI is a homing endonuclease that specifically cleaves its recognition sequence within the mitochondrial genome of Saccharomyces cerevisiae (1,2). I-SceI-mediated DNA cleavage initiates homing, a homologous recombination event that results in copying of the I-SceI homing endonuclease gene to loci that lack the element (3). The insertion of I-SceI recognition sites in the genomes of other organisms results in selective double-strand DNA cleavage at the inserted site, leaving the remainder of the genome intact (4). Moreover, I-SceI stimulates the frequency of recombination between a chromosomal locus containing the cleavage site and homologs located on exogenous DNA more than 1000-fold (5,6), making I-SceI an ideal choice for gene targeting studies (7–10) and for the study of DNA break repair mechanisms (11,12). The enzyme has also been successfully employed to study nonhomologous end joining (13–15).
I-SceI belongs to the LAGLIDADG homing endonuclease family, whose members cleave long DNA substrates (>14 bp) to generate four base 3′-overhangs (16). LAGLIDADG endonucleases are either monomers, such as I-SceI, that are composed of two pseudosymmetric subdomains that recognize asymmetric substrates (2) (Figure 1) or homodimers, such as I-CreI, that recognize palindromic or pseudopalindromic substrates (17). In both cases, the LAGLIDADG endonucleases fold into a β-saddle architecture, a common motif for nucleic acid binding proteins. The catalytic center is located at the interface of the monomer subdomains or dimer subunits and is defined by two juxtaposed signature LAGLIDADG helices (18,19).
Figure 1.
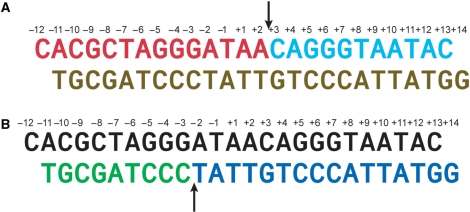
Nicked DNA duplexes used in crystallization studies. (A) A DNA duplex containing a nicked top strand assembled from two oligonucleotides (colored red and cyan) and an intact bottom strand oligonucleotide (brown). (B) A DNA duplex containing a nicked bottom strand assembled from two oligonucleotides (colored green and dark blue) and an intact top strand (colored black). The arrows indicate the cleavage sites on the top and bottom strands.
A two-metal-ion mechanism, similar to that of phosphoryl transferases (20) and restriction endonucleases (21,22) has been proposed for the phosphodiester cleavage reaction by LAGLIDADG endonucleases. The mechanism is based on structural work of I-CreI complexed with its DNA target sequence in the presence of Ca2+, Mg2+ or Mn2+ (23,24). Two overlapping actives sites that each cleave one DNA strand are defined by three metal ions and two conserved aspartates located at the carboxyl-termini of the LAGLIDADG helices. Each of the aspartates coordinates two of the three metals, one that is specific for only one of the two active sites and is termed the ‘unshared’ metal and one that is ‘shared’ by both sites. The aspartates are essential for catalysis since their substitution with alanine abolishes activity (24,25). In each active site, the unshared metals coordinate one in-line attacking nucleophilic water molecule that cleaves one of the DNA strands while the shared metal stabilizes the developing phosphoanion transition state and protonates the 3′ hydroxyl leaving group. The general bases that are responsible for the formation of the attacking nucleophilic water have not been identified. Instead, the solvent water network present at the active sites is proposed to act as nucleophile by a proton relay mechanism (24).
Previously, we reported the X-ray structure of I-SceI in a complex with its 18-bp DNA substrate (26). The two DNA strands of the asymmetric I-SceI recognition site have different sequences and are arbitrarily designated as the top and bottom strands (Figure 1). The crystal structure showed the bottom strand scissile phosphate buried in the active sites region and readily accessible for cleavage while the top strand was distant from any protein residues. This observation corroborated kinetic evidence that showed that the bottom strand in I-SceI is cleaved at a faster rate than the top strand (27). The crystal structure was obtained in the presence of Ca2+, which can substitute for Mg2+ in binding to I-SceI, but does not permit catalysis, and showed an unusual distribution of metals. As in the case of the I-CreI structure, three metals were found at the I-SceI catalytic center (Figure 2A), but the bound metals were not symmetrically distributed and reflected the asymmetry of the DNA substrate and the catalytic center. The arrangement of two of the metals and a putative nucleophilic water molecule resembled that observed at the I-CreI active sites and was consistent with a two-metal-ion mechanism for cleavage of the bottom strand. One metal, Ca2+ 1, was not shared between the active sites and was coordinated to the scissile phosphate of the bottom strand at −3 while a second, Ca2+ 3, was believed to be shared between the active sites and was coordinated to the conserved Asp 44 and Asp 145 and the scissile phosphates at −3 and +3. We suggested that a water molecule ligated to metal 1 (water 25) was the nucleophile that mediated bottom strand cleavage. This water was, in turn, hydrogen-bonded to Lys 223 which was initially thought to be a general base in the bottom strand phosphodiester cleavage reaction. However, Lys 223 is unlikely to play a catalytic role in the reaction since a Lys 223 alanine mutant is partially active (Yan et al., in preparation). In contrast to metal 1, metal 2, which coordinated the top strand scissile bond at +3, was buried deep within the DNA minor groove and was not coordinated to any protein residues. Therefore, unlike metal 1, which was ideally positioned to coordinate a water molecule to effect bottom strand cleavage, metal 2 was unlikely to play a similar role in catalyzing top strand cleavage. Thus, there was substantial uncertainty about the role of this metal and the definition of the second active site. We speculated that a conformational change may bring the top strand cleavage site in close proximity to active sites residues.
Figure 2.
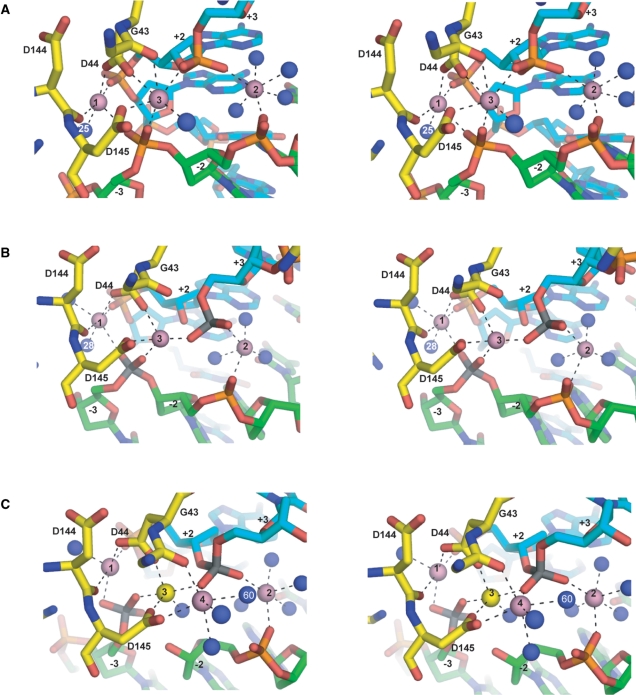
Stereo views of metal coordinations at the I-SceI/DNA active sites. (A) I-SceI uncleaved DNA complex. (B) I-SceI nicked top strand complex. (C) I-SceI nicked bottom strand complex. Ca2+ are depicted as pink spheres and the Na+ as a yellow sphere. Ordered water molecules are depicted as blue spheres. The scissile phosphoruses are colored gray. Metal coordination spheres are indicated by black dotted lines. The protein backbone is colored yellow, and the top and bottom DNA backbones are colored cyan and green, respectively.
In order to gain further insight into the cleavage mechanism of the bottom strand and to elucidate the cleavage mechanism of the top strand by I-SceI, we determined the crystal structures of complexes of I-SceI with duplex DNAs containing nicks in either the bottom or the top strands (Figure 1). The structures resemble intermediates along the DNA cleavage pathway and constitute the first report of a LAGLIDADG homing endonuclease in complex with nicked DNAs. The nicked bottom strand and nicked top strand structures reveal a different number and arrangement of metals coordinated to active site residues. In the nicked bottom strand structure, a new metal site that is absent in the uncleaved DNA structure, coordinates to the top strand scissile phosphate and to a water molecule that is a putative nucleophile for top strand cleavage. Since the bottom strand is cleaved at a much faster rate than the top strand, the nicked bottom strand structure may resemble the predominant intermediate along the DNA cleavage reaction pathway.
MATERIALS AND METHODS
Protein purification
A gene encoding I-SceI was cloned in pET15 (Novagen). Escherichia coli cells containing this plasmid were grown to OD600 = 0.5 and induced with 0.2 mM isopropyl-β-d-galactopyranoside (IPTG) overnight at 18°C. Cells were lyzed in 200 mM KCl, 1 mM EDTA, 1.4 mM β-mercaptoethanol (BME), 5% glycerol and 100 mM Tris–HCl, pH 8.0, by sonication and the lysate was clarified by centrifugation. The lysate was treated with 0.06% polyethylenimine to remove DNA and the protein was precipitated using ammonium sulfate. The precipitate was resuspended in 0.1 mM EDTA, 1.4 mM BME, 5% glycerol and 10 mM Tris–HCl, pH 8.0 and dialyzed overnight against resuspension buffer plus 50 mM KCl. The lysate was applied to a Q-Sepharose column (GE HealthCare) and the flowthrough fraction was dialyzed against 1 mM EDTA, 5% glycerol, 1.4 mM BME, 50 mM KCl and 10 mM HEPES, pH 8.0 and applied to a Mono S (GE Healthcare) column. I-SceI was eluted using a KCl gradient. Fractions containing I-SceI were pooled and dialyzed against 5% glycerol, 1.4 mM BME, 80 mM KCl, 0.1 mM EDTA 20 mM potassium phosphate, pH 7.6, and applied to a HiTrap heparin-Sepharose column (GE Healthcare). Fractions containing I-SceI were dialyzed against 100 mM KCl and 10 mM Tris–HCl, pH 7.5, for crystallization studies.
Crystallization and data collection
Synthetic DNA oligonucleotides were purchased from Integrated DNA Technologies (IDT). A 5′-terminal phosphate was introduced at the site of the DNA nick by phosphorylation of the appropriate oligonucleotide using T4 polynucleotide kinase according to the manufacturer's protocols. To prepare nicked DNA substrates three oligonucleotides were combined in equimolar amounts and annealed by a slow cooling protocol from 95° to 4°C over 3 h. I-SceI and the nicked DNA were combined in a 1:1 molar ratio and incubated on ice for 1 h. The protein–nicked DNA complexes were then concentrated to ∼6 mg/ml. Crystals were obtained by mixing 1.5 μl of the protein–DNA complex with 1.5 μl of a solution containing 22–26% 2-methyl-2,4-pentanediol (MPD) (v/v), 20 mM CaCl2, 1 mM dithiothreitol and 100 mM sodium acetate, pH 4.6, at 4°C in hanging drops. Crystals that grew within a week have dimensions 0.2 × 0.2 × 0.2 mm and properties shown in Table 1. Crystals were transferred to a cryoprotectant solution containing 30% MPD and 5% glycerol and flash-frozen in liquid nitrogen. Data sets for I-SceI/nicked bottom strand and I-SceI/nicked top strand crystals were collected on a ADSC Quantum CCD detector at the SBC-19ID beamline at the Advanced Photon Source. The I-SceI/nicked bottom strand data were processed with D*TREK version 9.7 (28) and the I-SceI/nicked top strand data were processed with HKL200 (29). Anomalous Ca2+ datasets were collected for both complexes at Rigaku (The Woodlands, Tx, USA) using a chromium rotating anode and a Rigaku Raxis IV imaging plate detector and processed with D*TREK version 9.7.
Table 1.
Diffraction data collection and refinement statistics
| Data set | Native nicked bottom strand | Native nicked top strand | Anomalous Ca2+ nicked bottom strand | Anomalous Ca2+nicked top strand | |
|---|---|---|---|---|---|
| Space group | P41212 | P41212 | P41212 | P41212 | |
| Wavelength (Å) | 0.979 | 1.03 | 2.29 | 2.29 | |
| Unit cell dimensions (Å) | a = b = 80.61 c = 127.09 | a = b = 80.68 c = 128.32 | a = b = 80.59 c = 127.93 | a = b = 80.60 c = 128.68 | |
| Asymmetric unit content | 1 complex | 1 complex | 1 complex | 1 complex | |
| D-min (Å) | 2.1 | 2.3 | 2.7 | 2.9 | |
| Completeness (%) | 99.7 | 99.7 | 100 | 100 | |
| R-syma (%, outer shell) | 6.1 (51.6) | 4.4 (43.4) | 7.5 (43.0) | 6.6 (38.7) | |
| Redundancy (outer shell) | 9.2 (9.2) | 5.3 (5.1) | 11.1 (10.8) | 11.0 (9.8) | |
| Refinement | |||||
| Resolution (Å) | 50–2.2 | 50–2.3 | |||
| R-factorb (%) | 26.8 | 25.5 | |||
| R-freeb (%) | 28.7 | 28.2 | |||
| Number of atoms | |||||
| Protein | 1855 | 1855 | |||
| Nucleic acid | 978 | 978 | |||
| Solvent | 98 | 58 | |||
| Metal ions | 4 | 3 | |||
| Rmsd bond lengths (Å) | 0.008 | 0.007 | |||
| Rmsd bond angles (°) | 1.53 | 1.33 | |||
| Ramachandran (%) | |||||
| Allowed | 98.5 | 99.5 | |||
| Generously allowed | 1.5 | 0.5 |
aRsym = ΣhΣi|Ii(h) − 〈I(h)〉|/ΣhΣiI(h).
bR-factor = Σ(|Fobs| − k|Fcal|)/Σ|Fobs. R-free was calculated using a random 5% of the reflection data that was omitted in the refinement.
Phasing and refinement
The structures of the I-SceI/nicked bottom strand and I-SceI/nicked bottom strand were determined by molecular replacement using MOLREP (30) and the I-SceI/DNA structure (pdb entry 1R7M) with waters and metals removed as the search model. The protein coordinates were refined with CNS (31) by rigid body refinement followed by conjugate gradient minimization and isotropic B-factor refinement. Examination of a Fo−Fc anomalous map followed by a Ca2+ anomalous difference map revealed the presence of four metal peaks in the vicinity of the active sites in the nicked bottom strand structure and three metal peaks in the nicked top strand structure. Waters were added to the coordinate list for peaks with I/σ >3.5 and deleted if the temperature factor was >60 Å2 following refinement. Alternate conformation for residues Thr 98 and Arg 180 were built into the nicked top strand structure and for Arg 180 in the nicked bottom strand structure.
RESULTS
Structure determination
The I-SceI/DNA complexes were crystallized in the presence of CaCl2 to prevent cleavage of the intact DNA strand. The structures of the nicked top strand and nicked bottom strand complexes were determined by molecular replacement using the I-SceI/uncleaved DNA structure (26) and refined to 2.3 Å and 2.2 Å, respectively. Difference Fourier maps revealed the presence of metal sites at the active sites region. To confirm the identity of the metal sites as Ca2+ ions, X-ray datasets for crystals of the complexes with the nicked top strand and the nicked bottom strand were collected at the Ca2+ edge. Data collection and refinement statistics are summarized in Table 1.
Absence of global conformational changes
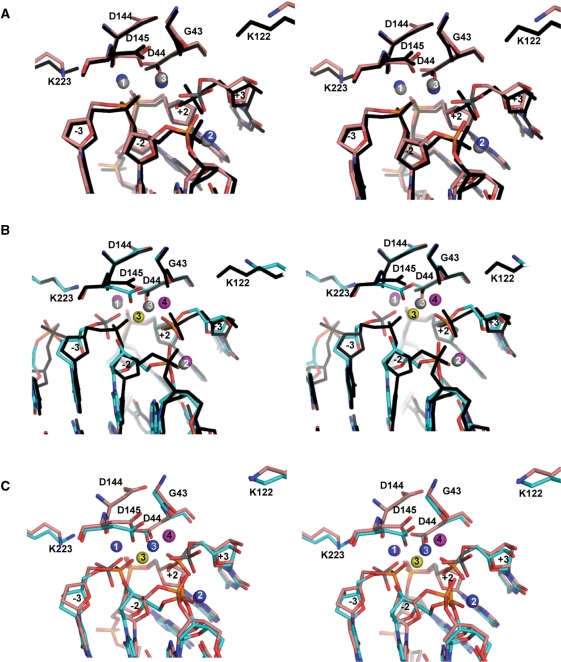
Previously, we raised the possibility that in order for I-SceI to cleave the top DNA strand, it was necessary for the protein and/or DNA to undergo conformational changes following cleavage of the bottom DNA strand (26). This is not the case as no major conformational differences exist between the nicked bottom strand structure and the uncleaved DNA structure (Figure 3B). Nor does the nicked top strand structure reveal any major conformational changes with respect to the uncleaved DNA (Figure 3A) and the nicked bottom strand structures (Figure 3C). The Cα r.m.s.d. of the nicked bottom strand and nicked top strand structures with respect to the uncleaved DNA structure are 0.91 Å and 1.0 Å, respectively and 0.30 Å with respect to each other. Protein conformational changes are restricted mainly to the side-chains of residues at the catalytic center region, which are discussed in a later section, and to the 116–123 loop which moves away from the active sites in both the nicked top strand and nicked bottom strand structures. This loop is close to the catalytic center and contains Lys 122, a residue whose substitution decreases the catalytic rate of the cleavage reaction of the top strand (Yan et al., in preparation). We believe that the different conformation of the loop may be due to lattice contacts that it makes with neighboring molecules that are not present in the structure of the uncleaved DNA complex, which was crystallized in a different space group.
Figure 3.
Stereo views of superpositions of I-SceI/DNA active sites. (A) Nicked top strand and uncleaved DNA complexes. (B) Nicked bottom strand and uncleaved DNA complexes. (C) Nicked bottom strand and nicked top strand complexes. The nicked top strand complex is shown with the backbone in salmon, the uncleaved DNA complex in black and the nicked bottom strand complex is shown with the backbone colored in cyan. Ca2+ are depicted as magenta, gray and blue spheres in the nicked bottom strand, uncleaved DNA and nicked top strand structures, respectively. The Na+ in the nicked bottom strand complex is depicted as a yellow sphere. The scissile phosphoruses at −3 in the nicked bottom strand complex and at +3 in the nicked top strand complex are colored gray.
Metal binding in the nicked top strand structure
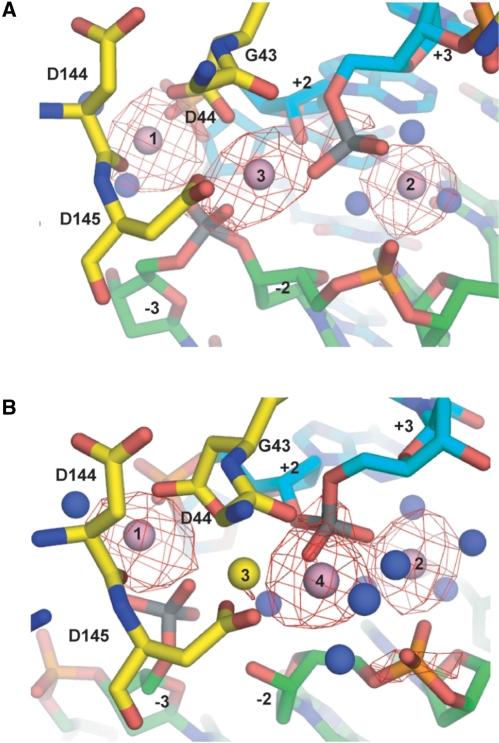
Difference Fourier maps of the refined nicked top strand structure revealed three metals (labeled 1–3 in Figure 2B), with peak heights of 14.8, 9.9 and 9.4 I/σ, respectively. These metals are arranged similarly as those bound to the active sites of the uncleaved DNA structure. Anomalous difference Fourier maps calculated with X-ray data collected at the Ca2+ edge showed electron density consistent with the presence of three Ca2+ ions (Figure 4A). The Ca2+ ions exhibit similar coordination spheres as those in the uncleaved DNA structure (Figure 2A), and the coordination distances are listed in Table 2. Metal 1 is coordinated to the carboxylate OD2 of Asp 44, the carbonyl oxygen of Asp 144, the O2P phosphate oxygen at +2, the O1P of the uncleaved scissile phosphate of the bottom strand at −3, and two water molecules. One of the waters (water 28) is the putative nucleophile for bottom strand cleavage and is in identical arc position as water 25 in the uncleaved DNA structure (Figure 2A and B). Metal 2 coordinates to the phosphate oxygen O2P at −2 and O3P at +3, and three water molecules. The shared metal 3 coordinates to the carboxylate OD1 of Asp 145 and Asp 44, and the O2P oxygens of the scissile phosphates at +3, and −3. It does not, however, coordinate to the backbone carbonyl of Gly 43 as in the uncleaved DNA structure.
Figure 4.
Anomalous Ca2+ difference Fourier electron density maps. (A) Nicked top strand complex (B) Nicked bottom strand complex. The maps, shown as red meshes, were contoured at 3.0 σ around the metal sites. The coloring scheme is the same as in Figure 2.
Table 2.
Metal coordination spheres
| Nicked bottom strand complex | Nicked top strand complex | Uncleaved DNA complexa | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Residue | Atom | Distance (Å) | Residue | Atom | Distance (Å) | Residue | Atom | Distance (Å) | |
| Metal 1 | D44 | OD2 | 2.2 | D44 | OD2 | 2.4 | D44 | OD2 | 2.3 |
| D144 | O | 2.4 | D144 | O | 2.3 | D144 | O | 2.4 | |
| A(+2) | O2P | 2.2 | A(+2) | O2P | 2.3 | A(+2) | O2P | 2.4 | |
| C(−3) | O1P | 2.4 | C(−3) | O1P | 2.5 | C(−3) | O2P | 2.4 | |
| C(−3) | O2P | 2.6 | W28 | O | 2.4 | W25 | O | 2.5 | |
| W77 | O | 2.3 | W35 | O | 2.2 | ||||
| Metal 2 | C(+3) | O2P | 2.2 | T(−2) | O2P | 2.4 | C(+3) | O2P | 2.3 |
| T(−2) | O2P | 2.3 | C(+3) | O3P | 2.3 | T(−2) | O2P | 2.3 | |
| W10 | O | 2.4 | W12 | O | 2.7 | W26 | O | 2.3 | |
| W17 | O | 2.3 | W32 | O | 2.0 | W89 | O | 2.3 | |
| W20 | O | 2.6 | W33 | O | 2.2 | W94 | O | 2.3 | |
| W36 | O | 2.3 | W160 | O | 2.4 | ||||
| Metal 3 | D44 | OD1 | 2.1 | D44 | OD1 | 2.4 | D44 | OD1 | 2.5 |
| D145 | OD1 | 2.3 | D145 | OD1 | 2.1 | D145 | OD1 | 2.2 | |
| C(+3) | O1P | 2.4 | C(+3) | O2P | 1.9 | G43 | O | 2.4 | |
| C(−3) | O2P | 2.0 | C(−3) | O1P | 2.2 | C(+3) | O1P | 2.3 | |
| W90 | O | 2.2 | C(−3) | O1P | 2.5 | ||||
| W78 | O | 2.2 | |||||||
| Metal 4 | D145 | OD2 | 2.4 | ||||||
| G43 | O | 2.3 | |||||||
| C(+3) | O1P | 2.4 | |||||||
| W18 | O | 2.5 | |||||||
| W37 | O | 2.2 | |||||||
| W60 | O | 2.4 | |||||||
aData from PDB 1R7M.
Metal binding in the nicked bottom strand structure
Difference Fourier maps of the refined nicked bottom strand structure revealed the presence of an additional metal site that was absent in the nicked top strand structure. The four observed metal sites, labeled 1 through 4, in the catalytic center of I-SceI (Figure 2C) have peak heights of 17, 13.4, 8.8 and 11.4 I/σ, respectively. The shared metal 3 shifts towards metal 1 and is now 3.7 Å distant from metal 1 as opposed to being 5.1 Å distant in the uncleaved DNA structure (compare Figure 2A and C), thereby allowing the binding of the new metal 4. Anomalous difference Fourier maps calculated with X-ray data collected at the Ca2+ edge revealed electron density consistent with the presence of three Ca2+ at metal sites 1, 2 and 4 (Figure 4B). No electron density is evident in the Ca2+ anomalous difference map for metal site 3, and it has been assigned as Na+.
In the nicked bottom strand structure, metals 1 and 2 lie at the same position and have similar spheres of coordination as in the uncleaved DNA structure. The main difference is the absence of the water 25 (Figure 2A), which was coordinated to metal 1 in the uncleaved DNA structure, and is believed to be the nucleophile in the cleavage of the bottom strand (26). Metal 1 is coordinated to the carboxylate OD2 of Asp 44, the carbonyl oxygen of Asp 144, the O1P and O2P of the free phosphate at −3, the phosphate oxygen O2P at +2 and a water molecule. Metal 2 is coordinated to the O2P of the top strand scissile phosphate at +3, the O2P phosphate oxygen at −2, and four water molecules. The shared metal 3 coordinates to the carboxylate OD1 oxygens of Asp 44 and Asp 145, the O1P of the top strand scissile phosphate at +3, the O2P of bottom strand scissile phosphate at −3 and a water molecule. It no longer coordinates to the carbonyl of Gly 43 as observed in the uncleaved DNA structure. The new metal 4 coordinates to the carboxylate OD2 of Asp 145, the carbonyl oxygen of Gly 43, the O1P of the top strand scissile phosphate at +3, and three water molecules. One of the waters ligated to metal 4, water 60, is well positioned to attack the top strand scissile phosphate.
Conformational changes at the catalytic center
Conformational changes in the catalytic center that result from cleavage of one of the DNA strands reflect the changes in metal positions that occur during catalysis. Overlaps of the catalytic centers of the uncleaved DNA, nicked bottom strand and nicked top strand structures are shown in Figure 3. In the comparison of the nicked top strand and uncleaved DNA structures (Figure 3A), changes in the conformations of the catalytic center residues are subtle because there are no rearrangements in the positions of the metals. The most significant changes are the movement of the side-chain of Asp 145 toward the DNA by 0.8 Å and the 2.8 Å retraction of the side-chain of Lys 122 from the catalytic center as a consequence of the conformational change of loop 116–123. The side-chain of Lys 223 undergoes a minor conformational change and is too far (3.6 Å) to form a hydrogen bond to the nucleophilic water 25 as it does in the uncleaved DNA structure.
In the nicked bottom strand structure (Figure 3B), the side-chain of Asp 145 undergoes a larger shift (1.5 Å) than in the nicked top strand structure and the side-chain of Asp 44 shifts 0.8 Å (Figure 3C). Both residues maintain their coordination to metal 3, which has also shifted 2.6 Å. In addition the side-chain of Asp 145 is also coordinated to the new metal 4. As in the case of the nicked top strand structure, Lys 122 moves 2.5 Å away from the active sites relative to its position in the uncleaved DNA structure, which may be the result of crystal packing forces.
The nicked strand in both the nicked top strand and nicked bottom strand structures does not undergo major conformational changes relative to the intact DNA and the narrowing of the minor groove that was observed in the uncleaved DNA is maintained. In the nicked bottom strand structure, the free phosphate of −3 moves about 2.0 Å toward the side-chain of Lys 223 occupying the position of the putative nucleophilic water 25 found in the uncleaved DNA structure. The distance between the carboxylate OD1 of Asp 145 and the phosphate O2P of –3 is only 3.0 Å. This repulsive interaction is weakened by coordination of metal 3 to both the OD1 of Asp 145 and the O2P phosphate oxygen at –3. In the nicked top strand structure the free phosphate of +3 moves 1.8 Å toward the position occupied by the putative nucleophilic water 60 and metal 4 of the nicked bottom strand structure.
DISCUSSION
Metal rearrangements
Rearrangements of metal binding sites that permit binding of additional metals have been observed in the structure of other DNA endonucleases. A comparison of the crystal structures of wild-type and mutant EcoRV–DNA complexes suggests that a catalytic metal shifts its position as the enzyme progresses from the initial state in which the DNA is uncleaved to the transition state (32). This shift permits the binding of a second metal that is essential for cleavage of the phosphodiester bond. Similarly, a shift of metal 3 in I-SceI that occurs once the bottom strand has been cleaved permits the binding of metal 4 that is thought to be involved in the cleavage of the top strand. One possibility is that this shift and the binding of metal 4 only occur following cleavage of the bottom strand. However, since metal 3 is apparently not a Ca2+ in the nicked bottom strand structure, it cannot be ruled out that the displacement of metal 3 is actually caused by the substitution of Ca2+ rather than Mg2+ at the active sites.
The structure of the nicked bottom strand structure illustrates the problem inherent in substituting Ca2+ for Mg2+ in structural studies which is usually explained in terms of steric crowding since Ca2+ has a much larger ionic radius (0.99 Å) than Mg2+ (0.65 Å). In I-SceI the binding of the additional Ca2+ at metal site 4 may prevent the binding of a Ca2+ to the shared metal site, which is only 3.9 Å distant. Consequently, binding of a smaller ion such as Na+ derived from the crystallization buffer fills the metal binding site instead. A similar arrangement was observed in the structure of the I-CreI/DNA complex crystals where the shared metal site was not occupied by Ca2+ but was instead filled by a mixture of bound solvent and/or sodium crystals (24).
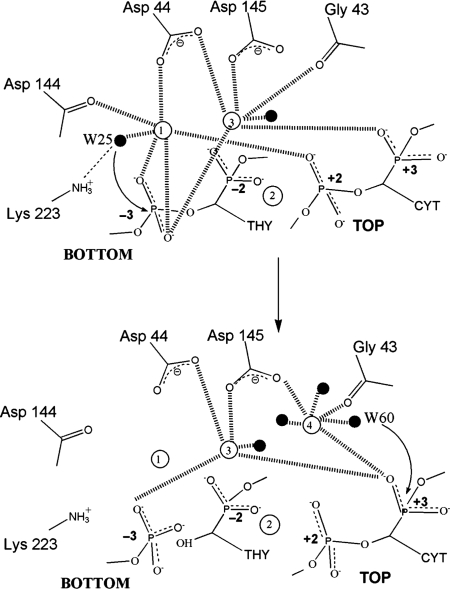
Mechanism of first- and second-strand cleavage of the bottom DNA strand
The uncleaved DNA structure provided a plausible mechanism for bottom strand cleavage that was directly based on the previous model proposed for I-CreI DNA cleavage (23) (Figure 5). Here, water 25 is activated for nucleophilic attack on the bottom strand phosphorus through coordination to metal 1. The phosphoanion transition state is stabilized by both metals 1 and 3. The fact that the active site responsible for bottom strand cleavage is poised for catalysis in the uncleaved DNA and nicked top strand structures is consistent with the bottom strand cleavage preference (26). Moreover, the important role of water 25 is reaffirmed by its presence in the uncleaved DNA and nicked top strand structures (water 28) and its absence in the nicked bottom strand structure (Figure 2).
Figure 5.
Proposed catalytic mechanism for bottom and top strand cleavage in the major catalytic pathway by I-SceI in which the bottom strand is cleaved first (based on the uncleaved DNA structure) followed by top strand cleavage (based on the nicked bottom strand structure). The metals are depicted by unfilled spheres and labeled 1–4. Ligated waters are depicted as black spheres. The putative nucleophilic water molecules are labeled as W25 (bottom strand cleavage) and W60 (top strand cleavage).
Mechanism of second-strand cleavage of the top DNA strand
The previously published structure of the uncleaved I-SceI/DNA complex did not shed light on the mechanism for top strand cleavage because there were no obvious metal ions correctly positioned to activate a nucleophile for attack on the scissile phosphate. In this report, the presence of an additional metal site that is evident in the nicked bottom strand structure provides a plausible mechanism for top strand cleavage following bottom strand cleavage (Figure 5). Metals 1, 3 and 4 in the I-SceI structure are arranged similarly as metals 1, 3 and 2, respectively, in the I-CreI structure (23) on which the two-ion mechanism for LAGLIDADG endonuclease cleavage was based. Metal 4, which is analogous to unshared metal 2 in the I-CreI structure, is thought to assist in cleavage of the top strand is conjunction with the shared metal 3. Metal 4 is coordinated to Asp 145 which is likely to be part of the second active site that is responsible for cleaving the top DNA strand. It also coordinates a water molecule (water 60) that is correctly positioned for in-line nucleophilic attack of the scissile phosphate of the top strand, just as water 25 is positioned in the uncleaved DNA and nicked top strand structures for nucleophilic attack of the bottom strand.
However, whereas water 25 in the bottom strand active site is hydrogen bonded to Lys 223, water 60 is not hydrogen bonded to Lys 122, which is the symmetric analog of Lys 223, in the top strand active site. One reason why Lys122 is not observed to hydrogen bond to water 60 may be due to crystallographic packing in the two nicked DNA structures that leads to the retraction of the 116–123 loop that harbors Lys122. Mutation of Lys 223 specifically reduces the extent of bottom strand cleavage and, in a similar manner, mutation of Lys 122 specifically reduces the degree of top strand cleavage (Yan et al., in preparation), hence neither lysine is essential for catalysis but both are important. In the nicked bottom strand structure the free phosphate of −3 moves toward the side-chain of Lys 223 indicating that this residue may be involved in the stabilization of the DNA product. Interestingly, mutation of two pseudosymmetric lysines in the intein-encoded LAGLIDADG homing enzyme PI-SceI that occupy similar positions as Lys 223 and Lys 122 also reduces the rates of the DNA cleavage reaction (33).
Mechanism of first-strand cleavage of the top DNA strand
Cleavage of the bottom DNA strand is not an obligatory first step in the reaction mechanism since kinetic experiments indicate that first-strand cleavage of the top strand can occur, albeit significantly slower than first-strand cleavage of the bottom strand (Yan et al., in preparation). The nicked top strand structure depicts a reaction intermediate in which the top strand has already been cleaved without providing information for how cleavage occurs. The absence of gross conformational changes between the uncleaved DNA and the nicked top strand intermediate structures indicates that cleavage of the top strand may take place in a protein–DNA conformation that favors bottom strand cleavage, which is in agreement with the low rate of top strand cleavage (27).
A novel feature of the three I-SceI structures (uncleaved, nicked top strand and nicked bottom strand) relative to those of other LAGLIDADG endonucleases is the presence of metal 2, which is completely buried in the DNA and is not coordinated to any protein residues. None of the other four crystal structures of LAGLIDADG proteins complexed to DNA (23,34–36) contain analogous metal ions. A plausible function for metal 2 would be that it plays a structural role by stabilizing the DNA bending at the catalytic center. However, the structures of the other LAGLIDADG enzymes contain similar DNA bends at the catalytic center without requiring an analog of metal 2 for DNA stabilization. In terms of catalysis, metal 2 is unlikely to be the equivalent of metal 1 in top strand cleavage. One of the waters ligated to metal 2 in both uncleaved DNA and nicked bottom strand structures is absent in the nicked top strand structure, but this water is not likely to act as a nucleophile since it is not correctly oriented to attack the top strand scissile phosphate. Moreover, if metal 2 were responsible for first-strand cleavage of the top strand (together with the shared metal 3), there would then be a different set of metals that catalyze top strand cleavage depending on whether it was cleaved first or second. However, a catalytic role of some kind for metal 2 in top strand cleavage cannot be entirely dismissed and metal 2 may be involved in the stabilization of the transition state.
We suggest that the new metal site 4 that we observe in the nicked bottom strand structure is also responsible for first-strand cleavage of the top strand. This site may be of low occupancy in the uncleaved DNA and nicked top strand structures since the top strand is only cleaved first 15% of the time (27) and the substitution of Ca2+ for Mg2+ may prevent its visualization. The higher rate of cleavage of the top strand after bottom strand cleavage (28) would correlate with a higher occupancy of metal site 4 in the nicked bottom strand structure, allowing its visualization. The absence of metal 4 from the nicked top strand structure may be also explained by the movement of the top strand free phosphate group at +3 toward the positions occupied by the nucleophilic water 60 and metal 4 in the nicked bottom strand structure, making metal site 4 unavailable.
CONCLUSION
The nicked bottom strand structure resembles an intermediate in which the enzyme is poised for cleavage of the top strand. The presence of a new metal site provides a mechanism for top strand cleavage in a more optimal chemical environment relative to the uncleaved DNA complex. Rearrangements of metal sites at catalytic centers require less energy than bringing about protein or DNA conformational changes so that enzymes such as I-SceI can catalyze reactions efficiently. Metals 1 and 4, which coordinate the nucleophilic waters, are responsible for the cleavage of the bottom and top DNA strands respectively in the major pathway of double-stranded DNA cleavage (Figure 5).
ACCESSION NUMBERS
Coordinates with accession numbers 3COW for the nicked bottom strand structure and 3COX for the nicked top strand structure have been deposited in the Protein Data Bank.
ACKNOWLEDGEMENTS
We thank Dr Meenakshi Vyas for collecting the nicked top strand structure data set and Dr James W. Pflugrath for collecting the anomalous datasets on both the nicked bottom strand and nicked top strand structures. We would also like to thank the personnel at the APS SBC-CAT for their assistance during data collection. The work was supported by a National Science Foundation grant MCB-0321550 and a National Institutes of Health grant (GM 070553) to FSG and the Welch Foundation to FAQ (Q-0581). Funding to pay the Open Access publication charges for this article was provided by National Institutes of Health grant (GM 070553).
Conflict of interest statement. None declared.
REFERENCES
- 1.Colleaux L, D’Auriol L, Betermier M, Cottarel G, Jacquier A, Galibert F, Dujon B. Universal code equivalent of a yeast mitochondrial intron reading frame is expressed into E. coli as a specific double strand endonuclease. Cell. 1986;44:521–533. doi: 10.1016/0092-8674(86)90262-x. [DOI] [PubMed] [Google Scholar]
- 2.Colleaux L, D’Auriol L, Galibert F, Dujon B. Recognition and cleavage site of the intron-encoded omega transposase. Proc. Natl Acad. Sci. USA. 1988;85:6022–6026. doi: 10.1073/pnas.85.16.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacquier A, Dujon B. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell. 1985;41:383–394. doi: 10.1016/s0092-8674(85)80011-8. [DOI] [PubMed] [Google Scholar]
- 4.Thierry A, Perrin A, Boyer J, Fairhead C, Dujon B, Frey B, Schmitz G. Cleavage of yeast and bacteriophage T7 genomes at a single site using the rare cutter endonuclease I-SceI. Nucleic Acids Res. 1991;19:189–190. doi: 10.1093/nar/19.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puchta H, Dujon B, Hohn B. Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res. 1993;21:5034–5040. doi: 10.1093/nar/21.22.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choulika A, Perrin A, Dujon B, Nicolas J.-F. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thermes V, Grabher C, Ristoratore F, Bourrat F, Choulika A, Wittbrodt J, Joly J. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech. Dev. 2002;118:91–98. doi: 10.1016/s0925-4773(02)00218-6. [DOI] [PubMed] [Google Scholar]
- 8.Grabher C, Joly JS, Wittbrodt J. Highly efficient zebrafish transgenesis mediated by the meganuclease I-SceI. Methods Cell. Biol. 2004;77:381–401. doi: 10.1016/s0091-679x(04)77021-1. [DOI] [PubMed] [Google Scholar]
- 9.Riu E, Grimm D, Huang Z, Kay MA. Increased maintenance and persistence of transgenes by excision of expression cassettes from plasmid sequences in vivo. Hum. Gene Ther. 2005;16:558–570. doi: 10.1089/hum.2005.16.558. [DOI] [PubMed] [Google Scholar]
- 10.Ogino H, McConnell WB, Grainger RM. High-throughput transgenesis in Xenopus using I-SceI meganuclease. Nat. Protoc. 2006;1:1703–1710. doi: 10.1038/nprot.2006.208. [DOI] [PubMed] [Google Scholar]
- 11.Sargent RG, Brenneman MA, Wilson JH. Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol. Cell. Biol. 1997;17:267–277. doi: 10.1128/mcb.17.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bellaiche Y, Mogila V, Perrimon N. I-SceI endonuclease, a new tool for studying DNA double-strand break repair mechanisms in Drosophila. Genetics. 1999;152:1037–1044. doi: 10.1093/genetics/152.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang F, Han M, Romanienko PJ, Jasin M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl Acad. Sci. USA. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nickoloff JA, Brenneman MA. Analysis of recombinational repair of DNA double-strand breaks in mammalian cells with I-SceI nuclease. Methods Mol. Biol. 2004;262:35–52. doi: 10.1385/1-59259-761-0:035. [DOI] [PubMed] [Google Scholar]
- 15.Rebuzzini P, Khoriauli L, Azzalin CM, Magnani E, Mondello C, Giulotto E. New mammalian cellular systems to study mutations introduced at the break site by non-homologous end-joining. DNA Repair. 2005;4:546–555. doi: 10.1016/j.dnarep.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 16.Stoddard BL. Homing endonuclease structure and function. Q. Rev. Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Kim HH, Yuan X, Herrin DL. Purification, biochemical characterization and protein-DNA interactions of the I-CreI endonuclease produced in Escherichia coli. Nucleic Acids Res. 1997;25:3767–3776. doi: 10.1093/nar/25.19.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duan X, Gimble FS, Quiocho FA. Crystal structure of PI-SceI, a homing endonuclease with protein splicing activity. Cell. 1997;89:555–564. doi: 10.1016/s0092-8674(00)80237-8. [DOI] [PubMed] [Google Scholar]
- 19.Heath PJ, Stephens KM, Monnat R.J., Jr., Stoddard BL. The structure of I-CreI, a group I intron-encoded homing endonuclease. Nat. Struct. Biol. 1997;4:468–476. doi: 10.1038/nsb0697-468. [DOI] [PubMed] [Google Scholar]
- 20.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl Acad. Sci. USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kostrewa D, Winkler FK. Mg2+binding to the active site of EcoRV endonuclease: a crystallographic study of complexes with substrate and product DNA at 2 Å resolution. Biochemistry. 1995;34:683–696. doi: 10.1021/bi00002a036. [DOI] [PubMed] [Google Scholar]
- 22.Viadiu H, Aggarwal AK. The role of metals in catalysis by the restriction endonuclease BamHI. Nat. Struct. Biol. 1998;5:910–916. doi: 10.1038/2352. [DOI] [PubMed] [Google Scholar]
- 23.Chevalier BS, Monnat R.J., Jr., Stoddard BL. The homing endonuclease I-CreI uses three metals, one of which is shared between the two active sites. Nat. Struct. Biol. 2001;8:312–316. doi: 10.1038/86181. [DOI] [PubMed] [Google Scholar]
- 24.Chevalier B, Sussman D, Otis C, Noel AJ, Turmel M, Lemieux C, Stephens K, Monnat R.J., Jr., Stoddard BL. Metal-dependent DNA cleavage mechanism of the I-CreI LAGLIDADG homing endonuclease. Biochemistry. 2004;43:14015–14026. doi: 10.1021/bi048970c. [DOI] [PubMed] [Google Scholar]
- 25.Gimble FS, Stephens BW. Substitutions in conserved dodecapeptide motifs that uncouple the DNA binding and DNA cleavage activities of PI-SceI endonuclease. J. Biol. Chem. 1995;270:5849–5856. doi: 10.1074/jbc.270.11.5849. [DOI] [PubMed] [Google Scholar]
- 26.Moure CM, Gimble FS, Quiocho FA. The crystal structure of the gene targeting homing endonuclease I-SceI reveals the origins of its target site specificity. J. Mol. Biol. 2003;334:685–695. doi: 10.1016/j.jmb.2003.09.068. [DOI] [PubMed] [Google Scholar]
- 27.Perrin A, Buckle M, Dujon B. Asymmetrical recognition and activity of the I-SceI endonuclease on its site and on intron-exon junctions. EMBO J. 1993;12:2939–2947. doi: 10.1002/j.1460-2075.1993.tb05956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr. D. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 30.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 31.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 32.Horton NC, Perona JJ. DNA cleavage by EcoRV endonuclease: two metal ions in three metal ion binding sites. Biochemistry. 2004;43:6841–6857. doi: 10.1021/bi0499056. [DOI] [PubMed] [Google Scholar]
- 33.He Z, Crist M, Yen H, Duan X, Quiocho FA, Gimble FS. Amino acid residues in both the protein splicing and endonuclease domains of the PI-SceI intein mediate DNA binding. J. Biol. Chem. 1998;273:4607–4615. doi: 10.1074/jbc.273.8.4607. [DOI] [PubMed] [Google Scholar]
- 34.Moure CM, Gimble FS, Quiocho FA. Crystal structure of the intein homing endonuclease PI-SceI bound to its recognition sequence. Nat. Struct. Biol. 2002;9:764–770. doi: 10.1038/nsb840. [DOI] [PubMed] [Google Scholar]
- 35.Chevalier B, Turmel M, Lemieux C, Monnat RJ, Stoddard BL. Flexible DNA target ite recognition by divergent homing endonuclease Isoschizomers I-CreI and I-MsoI. J. Mol. Biol. 2003;329:253–269. doi: 10.1016/s0022-2836(03)00447-9. [DOI] [PubMed] [Google Scholar]
- 36.Spiegel PC, Chevalier B, Sussman D, Turmel M, Lemieux C, Stoddard BL. The structure of I-CeuI homing endonuclease: Evolving asymmetric DNA recognition from a symmetric protein scaffold. Structure. 2006;14:869–880. doi: 10.1016/j.str.2006.03.009. [DOI] [PubMed] [Google Scholar]