Abstract
The anti-termination protein, HutP, regulates the gene expression of the hut (histidine utilization) operon of Bacillus subtilis, by destabilizing the hut terminator RNA located upstream of the coding region encoding l-histidine degradation enzymes. On the basis of biochemical, in vivo and X-ray structural analyses, we now report that HutP uses its dual RNA-binding surfaces to access two XAG-rich regions (sites I and II) within the terminator RNA to mediate the destabilization process. In this process, HutP initiates destabilization at the 5′-end of its mRNA by binding to the first XAG-rich region (site I) and then accesses the second XAG-rich region (site II), located downstream of the stable G-C-rich segment of the terminator stem. By this action, HutP appears to disrupt the G-C-rich terminator stem, and thus prevents premature termination of transcription in the RNA segment preceding the regions encoding for the histidine degradation enzymes.
INTRODUCTION
Bacteria exploit a variety of mechanisms to regulate transcription elongation, to control gene expression in response to change in their environment. Among these, a common path is the modulation of mRNA secondary structures by RNA-binding proteins, either to pause transcription near the terminator region or to allow synthesis of the full-length transcript. The stability of these structures depends on the intrinsic stability of alternative secondary structures and also on the stabilization of one of the secondary structures by regulatory RNA-binding proteins. The binding of a regulatory protein to the nascent mRNA may result in transcription elongation; otherwise, the mRNA forms the default termination structures, as in the mRNA of cold-shock proteins and those from catabolic operons. In contrast, the transcription terminators are stabilized by the regulatory RNA-binding proteins; otherwise, the default mRNA structure is set for transcription elongation, as observed in the tryptophan biosynthetic operons in Bacillus subtilis (1). The former and latter mechanisms of transcription regulation are referred as anti-termination and termination, respectively. Several of these bacterial termination and anti-termination proteins that regulate terminator/anti-terminator structures have been described in Escherichia coli and B. subtilis, including TRAP, PyrR, LacT, BglG, SacT/SacY and GlpP (1–7). One such anti-terminator protein is HutP. It regulates hut (histidine utilization) operon transcription by an anti-termination mechanism in B. subtilis (8). The hut operon consists of six open reading frames. The HutP gene is located near the promoter, and five additional genes, hutH, hutU, hutI, hutG and hutM, are located downstream from hutP, which encode histidase, urocanase, imidazolone propionate amino hydrolase, formiminoglutamate hydrolase and histdine permease, respectively. Previous biochemical and genetic studies suggested that the hut mRNA forms a terminator structure (+515 to +553 nt) in between the hutP and hutH coding regions, and that this terminator is used to regulate transcription of the distal 5 genes of the hut operon by an anti-termination mechanism (8–10). HutP requires a ligand, l-histidine, but for complete activation, HutP also requires divalent metal ions (10–14). On the basis of in vitro selections and site-specific mutational analyses, we have identified UAGNNNNUAGNNNNUAG as the recognition motif (N indicates any base), with the identified residues constituting the important RNA chemical groups required for HutP recognition (12,13).
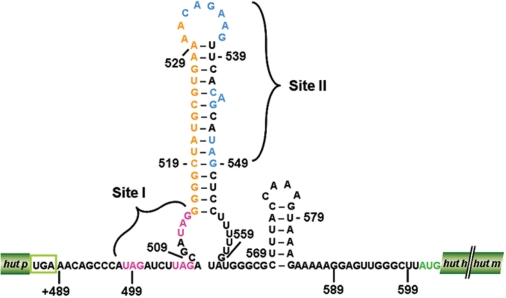
In order to obtain insights into the interactions of these ligands with HutP, and to clarify the mechanisms of HutP-mediated anti-termination of transcription, we previously solved the crystal structures of HutP (ligand free), and the complexes of HutP-l-histidine, HutP-l-histidine-Mg2+ and HutP-l-histidine-Mg2+-21-mer RNA (12,14). A comparison of these structures revealed snap-shots of the conformational changes of HutP in the presence of l-histidine and divalent metal ions needed for binding to specific regions within the hut terminator RNA (15) Although the above study revealed how HutP activation proceeds before HutP binding to the target sequence within the terminator RNA, it was important to establish how HutP initiates the destabilization process that alters the RNA terminator structure, in order to allow synthesis of the full-length transcript. The hut terminator RNA possesses two XAG-rich regions (X indicates any base), which potentially interact with HutP (Figure 1). In the present study, based on biochemical, in vivo and structural studies, we show that hexameric activated HutP accesses to two XAG-rich sites (I and II) within the terminator RNA on its two RNA-binding surfaces. Substitutions of bases within the XAG motifs in either site I or II impair the overall anti-termination process, suggesting that HutP uses these two binding sites to force the terminator RNA to undergo conformational changes, especially near the XAG motifs and also within the G-C-rich region located between sites I and II. Thus, it appears that HutP destabilizes the hut terminator RNA in a novel fashion.
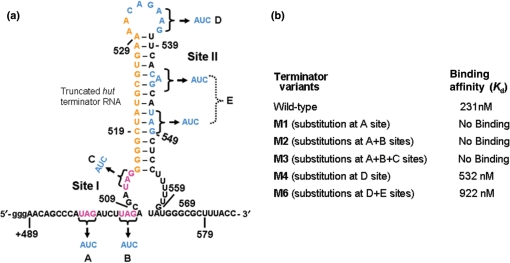
Figure 1.
The proposed hut mRNA-terminator structure. Potential HutP binding sites, site I and site II, are indicated by magenta and blue letters, respectively. The stop codon of the hutP gene is shown in a green box, and the start codon of the hutH gene is shown in green letters. The linker region (17-nt) between the two HutP binding sites is shown in orange letters.
MATERIALS AND METHODS
Expression and purification of the HutP protein
The nucleotide sequence encoding HutP, containing the Val 51 Ile mutation, was amplified and cloned into the pET5a vector (Promega). The resultant plasmid was transformed into E. coli strain BL21 (DE3). The HutP protein was overexpressed at mid-log phase by the addition of isopropyl-β-d-thiogalactopyranoside (1 mM), and was purified as described previously (12). The single-point mutant of HutP (Val 51 Ile) was found to bind with higher affinity to hut mRNA in the presence of l-histidine, compared to the wild-type HutP (12).
RNA synthesis, labeling and in-line mapping
The 55-mer RNA (Figure 2a) was synthesized chemically, deprotected, purified by PAGE and recovered as reported previously (12). Wild-type RNA was synthesized enzymatically using T7 polymerase. RNAs were labeled at the 3′-end with pCp and T4 RNA ligase (16) or at the 5′-end with (γ-32P) ATP and polynucleotide kinase (17). The in-line mapping assay was carried out by incubating labeled RNAs for 6 h at 25°C, in a buffer containing 5 mM MgCl2, 50 mM Tris–HCl (pH 8.3 at 25°C) and 100 mM KCl, in the absence or presence of HutP concentrations ranging from 1 to 10 µM, along with 10 mM l-histidine. After each incubation, the spontaneously cleaved products were resolved by 12% denaturing (8 M urea) PAGE and were visualized by autoradiography.
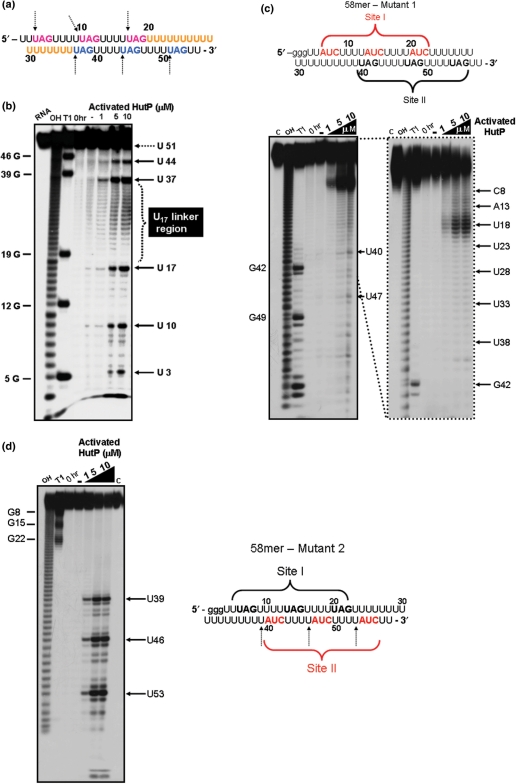
Figure 2.
The in-line mapping assay on the model 55-mer RNA and its mutants. (a) Sequence of the model 55-mer RNA. Linker bases are shown in orange. UAG residues are highlighted in magenta and blue as potential site I and site II residues, respectively. (b) A representative autoradiogram showing an in-line mapping assay of the interactions between the 55-mer model RNA and HutP. (c) A representative autoradiogram showing an in-line mapping assay of the interactions between mutant-1 RNA and HutP. (d) A representative autoradiogram showing an in-line mapping assay of the interactions between mutant-2 RNA and HutP. Red letters indicate the base substitution positions. OH-alkali digestion: T1–(specific for G bases). The labeled RNA was incubated for 6 h at 25°C, in a buffer containing 5 mM MgCl2, 50 mM Tris–HCl (pH 8.3 at 25°C) and 100 mM KCl, with HutP concentrations ranging from 1 to 10 µM, along with 10 mM l-histidine. After each incubation, spontaneously cleaved products were resolved by 12% denaturing (8 M urea) PAGE, followed by autoradiography. Cleavage sites are indicated by dotted arrows.
Enzymatic synthesis of wild-type terminator RNA and model RNA mutants
The PCR product was prepared from the templates (5′-AACAGCCCATAGATCTTAGACGATAGGGGGCTATGCGTGAAAACAGAAGTTCACAGCATAGC-3′ and 5′-AAGCCCAACTCCTTTTTCTTTACTTTGGTAAAGCGCCCATACAAAAAGGAGCTATGCTGTGAACTTCTG-3′) by the primers 5′-AGTAATACGACTCACTATAGGG AACAGCCCATAGATCT TA GAC-3′ and 5′-GGTAAA GCGCCCATACAAAAAGG-3′. Using the template DNA and the primers, we prepared the dsDNA template, containing a T7 promoter, using a commercial PCR kit (ExTaq kit, Takara, Japan). The reaction mixture was cycled at 94°C for 1.15 min, 50°C for 1.0 min and 72°C for 1.15 min for 10 cycles. The resulting PCR product was precipitated, and about 2 μg of template were used for in vitro T7 transcription, followed by isolation of RNAs as described (15).
Mutants of this model RNA with modified UAG motifs were prepared enzymatically, by using the following sets of oligos. Mutations at the first three UAG repeats were prepared from the template 5′ GGGTTATCTTTTATC TTTTATCTTTTTTTTTTTTTTTTTTAGTTTTTAGTTTTTAGTT 3′, and were amplified with the primers 5′ AGTAATACGACTCACTATAGGGTTATCTTTTATCTTTTATC 3′ and 5′ AACTAAAAACTAAA AACTAAA 3′. The second three UAG repeats were mutated with the template 5′ GGGTTTAGTTTTTAG TTTTTAGTTTTTTTTTTTTTTTTTATCTTTTATCTTT TATCTT 3′, and were amplified by using the primers 5′ AGTAATACGACTCACTATAGGG TTTAGTTTTTAGTTTTTAG 3′ and 5′ AAGATAAAAG ATAAAAGATAA 3′. In both cases, the forward primer included a T7 promoter region for in vitro transcription, as highlighted with italics.
Filter-binding assay
Shorter hut-derived terminator RNAs (89-mer), from the vector ST4 and its variants, were amplified by PCR, using the conditions described above, in the presence of primers (forward primer, 5′-AGTAATACGACTCACTATAGGGAACAGCCCATAGATC TTAGAC-3′ and reverse primer, 5′-GGTAAAGCGCCC ATACAAAAAGG-3′). To prepare internally labeled RNAs, the PCR products were transcribed in vitro, using T7 RNA polymerase in the presence of α-32P-CTP (GE Healthcare Biosciences), as previously reported (13). Transcription reactions were carried out at 37°C for 3 h. The resulting transcribed products were separated by denaturing PAGE (7 M urea) and the RNA was recovered from the gels. The resulting labeled RNAs were used for filter binding assays, as reported previously (15).
Construction of hut-LacZ fusion vectors and analysis of β-galactosidase activity
To analyze the importance of the UAG repeats within the hut mRNA for anti-termination of transcription mediated by the HutP protein in B. subtilis, we constructed hut-lacZ translation fusion vectors with substitutions in both the first and second binding sites. To prepare these vectors, we specifically mutated the UAG sites within the terminator region of the hut mRNA, using site-directed mutagenesis (QuikChange kit, Stratagene, USA). For this mutagenesis, we used primers (Supplementary Figure 1) and the previously constructed ST4 vector as the template (10) (kindly provided by Dr M. Oda, AIST). We prepared a total of four ST4-derived variants, ST4-1-1, ST4-1-3, ST4-2-1 and ST4-2-3, with substitutions in either the first or second site, by mutating either a single UAG motif or all three XAG motifs. All substitutions were confirmed at their respective positions by DNA sequencing, using the conditions recommended for the ABI PRISM 3100 sequencer (Applied Biosystems, Foster City, CA, USA). All of these hut-lacZ fusion vectors (ST4-derived) were then introduced into the amyE locus of the B. subtilis 1A1 and HUTPN1 (hutPΔN1) strains by transformation (18). All of these strains containing the fusions were grown at 37°C in Hut assay medium (8,19) containing 0.01% yeast extract, with or without histidine (0.1%), and the β-galactosidase activity was monitored as described previously (10). Induction ratios were calculated by dividing the β-galactosidase activity in the cells grown without l-histidine by that in the cells grown with l-histidine.
Crystallization of the RNA (55-mer)-HutP-L-his–Mg2+ ion complex
Crystals of the HutP-l-his–Mg2+-55mer RNA complex were grown at 20°C by the hanging drop vapor-diffusion method. Drops consisted of 2 μl of protein (10 mg/ml, in 50 mM HEPES, pH 7.4 and 10 mM of MgCl2), 1 μl of 0.1 M l-histidine, 2 μl of 55-mer RNA (300 μM) and 2 μl of well solution (30% MPD, 0.1 M HEPES, pH 7.4 and 0.1 M MgCl2). Diffraction data were collected with a Jupiter210 CCD detector (RIGAKU MSC Co., Tokyo, JAPAN) at the RIKEN Structural Genomics Beamline I, BL26B1, Spring8, Hyogo, Japan. Data sets for the quaternary complex structures were processed at 1.7 Å with the HKL2000 suite of programs.
Structure determination
The HutP-l-his–Mg2+-55mer RNA complex structure was determined by molecular replacement, using the HutP-l-histidine–Mg2+-21-mer RNA complex as a search model (PDB ID 1WMQ). Both the molecular replacement and structure refinement were carried out using CNS (20). The model was built using the program Quanta (21). The structure was refined to 1.7 Å resolution. Data collection and refinement statistics are provided in Supplementary Table 1. The final model contains three monomers (A, B and C) of HutP, one continuous 21-nt fragment from the 55-mer RNA, three Mg2+ ions, three l-histidines and 406 water molecules. A continuous 21-mer RNA was built from the electron density map. Figures were prepared using the program Pymol (22).
RESULTS
Mapping HutP binding sites on a 55-mer model RNA
To evaluate whether HutP binds to RNA with two XAG-rich sites (I and II), we chemically synthesized a model RNA (55-mer; Figure 2a), in which repeat XAG regions are joined by a linker region of 17 Us (U17; between the third and fourth XAG motifs: Figure 2a), similar to the length of the linker region present between sites I and II in the hut terminator RNA (Figure 1). We analyzed the HutP binding sites within the 55-mer RNA using an in-line mapping assay, which relies on the structure-dependent spontaneous cleavage of RNA in the presence of divalent metal ions. In this assay, the internucleotide linkages between the bases, which assume an in-line conformation defined by the pseudo-bond angle (τ) [O2′ (U2)-P(U3)–O5′ (U3)], are sensitive to spontaneous cleavage, predominantly in the nonstructured region of the RNA (23). This sensitive assay was previously used by our lab and others to map ligand-binding sites in RNA (17,24).
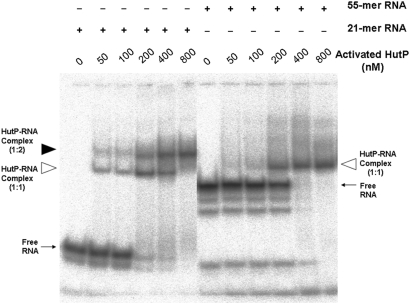
In-line mapping experiments were performed by incubating the 55-mer RNA with the HutP-l-histdine complex in the presence or absence of Mg2+ ions, to initiate spontaneous RNA cleavage. The cleavage sites can be identified by separating the cleaved RNA products by gel electrophoresis. Our in-line mapping revealed that effective cleavage was observed in the XAG regions at both sites within the 55-mer RNA, but only in the presence of activated HutP (HutP-l-histidine-Mg2+) (Figure 2b). These sites are analogous to the UpU sites before the AG residues of three repeats of a 7-mer RNA (5′-(UpUpUpApGpUpU)3 -3′), which we used previously in our structural analysis (15). Our quaternary complex structure with this RNA revealed that this region undergoes a conformational change (τ = 111.4°), reflecting its sensitivity to spontaneous cleavage (Supplementary Figure 2). In addition to the above sites, the linker region U17 between the two UAG-rich regions was also cleaved efficiently, in the presence of activated HutP. These results suggest that the two XAG-rich sites (I and II) in the model 55-mer RNA are recognized specifically by the hexameric HutP, probably on its two (top and bottom) surfaces, and that the binding causes conformational changes near the UAG sites and the 17-nt linker region. Next, we carried out similar mapping studies using two kinds of RNA, having substitutions of the residues in the UAG motifs, either at 5′-end (site I) or 3′-ends (site II). The cleavage patterns observed in these RNAs were significantly different from 55mer RNA having two XAG-rich regions (Figure 2c and d). The absence of cleavages in these two RNAs reflects the inability of HutP modulation, suggesting that until the two UAG motifs are fully bound by HutP, the linker region may not undergo a significant conformational change (Figure 2c and d). Moreover, these studies suggest that hexameric HutP can access two sites within the 55-mer RNA. To confirm this notion, we carried out a gel-shift analysis and found that hexameric HutP binds to model RNA in a cooperative manner, forming a 1:1 ratio of RNA to protein complex, as compared to the 21-mer RNA (Figure 3). Taken together, these studies revealed that both the upstream and downstream UAG motifs are important, not only for binding to the specific residues within the hexameric HutP but also for causing significant conformational changes in the linker region.
Figure 3.
Gel-shift assay showing HutP–RNA complexes. All reactions were carried out in binding buffer (15 mM HEPES pH 7.5, 30 mM NaCl) with 20 nM of RNA (labeled RNA 104 c.p.m.) containing 10 mM of l-his and Mg2+ ion. To this reaction mixture, various amount of purified HutP protein (to a final concentration of 50–800 nM) were added, and after an incubated for 15 min, the reactions were fractionated by 8% native PAGE. The positions of the free and complex RNA (protein to RNA, 1:1 ratio) are indicated by an arrow and an open arrowhead, respectively. The 21-mer RNA complex (protein to RNA, 1:2 ratio) is indicated by a filled arrowhead.
Mapping HutP-binding sites on a truncated hut terminator
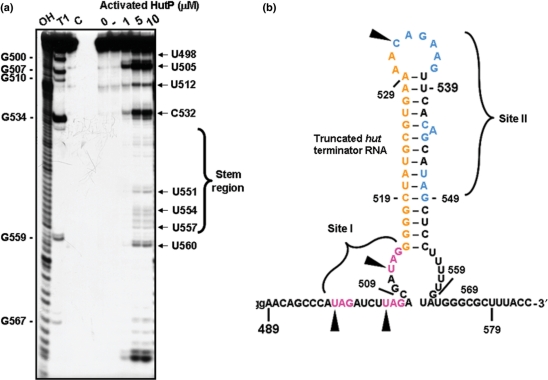
The above in-line mapping studies were then extended to the wild-type hut terminator RNA, containing two sites for HutP binding. For this, we initially prepared a truncated terminator RNA spanning the region from +489 to +574 nts (89-mer containing three non-terminator residues, ‘G’, which were added during T7 transcription). The secondary structure of the RNA in both the full-length and truncated versions was similar, in terms of the formation of a hairpin structure. The prepared RNA was initially tested for its binding to the activated HutP, and was found to bind as efficiently as the full-length terminator RNA (+489 to +600 nt, 112-mer). The in-line mapping assay revealed four efficient cleavage sites, identified as the U498, U505, U512 and C532 residues (Figure 4a and b). These sites are equivalent to the first four UAG sites in the model 55-mer RNA. In addition to these cleavages, we also observed weak cleavage within the G-C/U rich region located between sites I and II (Figure 4a). However, we did not observe cleavages at other XAG motifs in site II. This could be due to the effect of the surrounding residues on the cleavage efficiency. Considering the sensitivity of the spacer region for the in-line attack in the presence of activated HutP, the XAG motifs located at sites I and II within the terminator region probably interact with the cognate residues of HutP. In-line mapping studies on both the model 55-mer RNA and the truncated wild-type hut terminator RNA revealed that the two XAG-rich sites in the hut terminator RNA are recognized by hexameric HutP, and for this, a 17-nt linker length is sufficient for the placement of the next potential binding site.
Figure 4.
The in-line mapping assay on truncated wild-type hut terminator RNA. (a) Representative autoradiogram showing an in-line mapping assay of the interactions between HutP and truncated wild-type hut RNA. (b) Spontaneous cleavage sites at the XAG regions in the truncated wild-type hut RNA are indicated by filled arrows.
In vitro binding analysis of mutant hut terminator RNAs
Next, to understand whether the recognition of two XAG-rich sites by HutP is a common feature for the HutP-mediated anti-termination process, we compared the primary sequences of the hut terminator RNAs among Bacillus species. Interestingly, many of these hut terminator RNAs in Bacillus species have two conserved XAG-rich regions flanking the linker region of 12–17 nt (Supplementary Figure 3). To examine the importance of the XAG motifs at two sites for HutP binding, we prepared five different variants, with substitutions of their complementary residues in their XAG motifs, in either the first alone (M1) or first two (M2) or first three motifs (M3) in site I (truncated terminator RNA 1) and in site II (truncated terminator RNA 2) [spacer region (M4) and the last XAG motif (M6); Figure 5a]. Upon transcription of these RNAs, three additional ‘G’ residues were added to their 5′ ends, as extra nucleotides, and thus the RNAs were 58-nt long. As seen in Figure 5b, the activated HutP lost its ability to bind to the hut terminator RNA with a base substitution at either the first XAG motif or the first two or three (XAG) motifs (M1, M2 and M3). The loss of HutP binding could also be due to the perturbation of the secondary structures of these RNAs. However, a secondary structure prediction program suggested that these mutant terminator RNAs have similar secondary structures and are also thermodynamically stable (Supplementary Figure 4). On the other hand, in site II, the substitution of the bases in the spacer region (M4) and the last potential XAG residue along with other residues in site II (M6) marginally affected (2- to 5-fold) the binding to the activated HutP (Figure 5b). In the case of the M6 variant RNA, the mutation was also predicted to alter the secondary structure marginally, and thus, it was difficult to judge whether the reduced affinity to the HutP was due to the alteration of the secondary structure or the loss of important residues. These binding analyses suggested that the XAG-rich region at the 5′ end (site I) is very important, as compared to the second XAG-rich region (site II). Nevertheless, these variants should be analyzed directly for anti-termination of the hut terminator in B. subtilis, because binding alone may not reflect the overall anti-termination process.
Figure 5.
HutP-binding affinity of the site I and site II mutants of the truncated terminator RNA and the hut wild-type RNA, determined using a filter binding assay. (a) Base substitutions at site I and site II are indicated on the hut terminator RNA. (b) The equilibrium dissociation constant (Kd) for each mutant was determined. Binding reactions were performed with 10 nM of RNA and protein concentrations ranging from 250 to 2250 nM.
In vivo hut anti-termination assay
To evaluate directly the importance of the two XAG-rich regions within the hut terminator RNA in the overall anti-termination process, we constructed a set of hut-lacZ translational fusion vectors containing base substitutions in the XAG motifs located at either site I or site II (Figure 6a and Supplementary Figure 5). The resulting fusion vectors with different substitutions were each introduced into the amyE locus of the B. subtilis 1A1 and HUTPN1 strains by transformation. The 1A1 strain has the ability to produce HutP from the chromosomal hut operon, whereas the HUTPN1 strain has a four-base deletion in the coding region of the HutP gene (25). We used these two strains to examine whether the observed inducible expression is dependent on the HutP expression. The β-galactosidase activity in these cells was measured, and the induction ratios were calculated by dividing the β-galactosidase activity in the cells grown without l-histidine by the β-galactosidase activity in the cells grown with l-histidine.
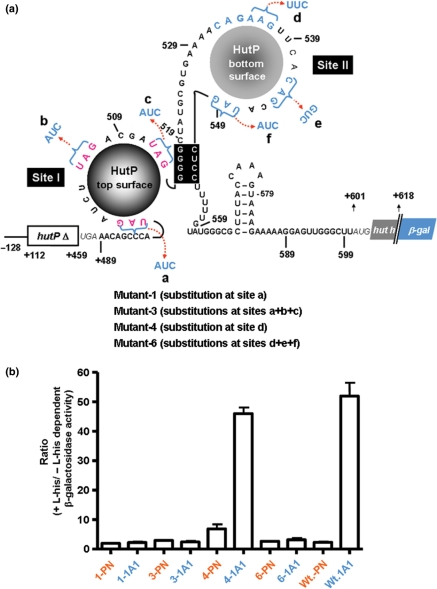
Figure 6.
In vivo analysis to identify the importance of XAG residues at sites (I and II) located within the hut terminator RNA. (a) Different variants for the in vivo anti-termination assay are shown on the possible secondary structure of the hut terminator, as a HutP-bound form. Deleted regions of the hut operon on the β-gal fusions are indicated by boxes with the open triangle symbol. (b) The ratio of the β-galactosidase activity was measured in the PN1 and 1A1 strains of Bacillus subtilis. The average from two experiments is indicated with error bars.
As seen in Figure 6b, the ratio of β-galactosidase activity was enhanced by about 55-fold when the hut terminator was transformed into the host 1A1, expressing HutP, but not significantly in the HUTPN1 strain. This suggests that the hut terminator was destabilized specifically in the presence of HutP, and a similar result was previously reported for the same construct (10). However, with the substitution of the first XAG motif at the 5′end (site I) of the terminator RNA, we observed that the ratio of induction was affected significantly in the 1A1 host, suggesting the loss of HutP-dependent anti-termination. Substitution of all three XAG bases from site I also affected the anti-termination process (M3; Figure 6b). This suggests that HutP can access only one site within the hut terminator, and thus only one surface of HutP binds the terminator RNA, as also observed with the 58-mer-RNA mutant 1 and mutant 2 (Figures 2c and d). On the other hand, substitutions in the spacer region (within site II), the hut-4-variant, marginally affected the ratio of induction in the HutP-expressing host. These results suggest that the HutP-binding site was not affected, which is consistent with the filter binding assay results showing that the spacer AAG (535–537 nt) is not essential for HutP binding. However, when the last XAG motif alone in site II was substituted along with other residues in site II (M6), the induction ratio for the β-galactosidase activity was significantly affected. Although in this variant, the other XAG sites within site II were not affected, the anti-termination ability mediated by HutP was abolished, similar to that observed with the substitution of all three XAG motifs within the site I (M3). These studies underscore the importance of two XAG-rich sites (I and II) for the anti-termination process mediated by HutP, probably to facilitate the opening of the stable G-C rich region of the terminator from both ends.
Structure of the quaternary complex with 55-mer RNA
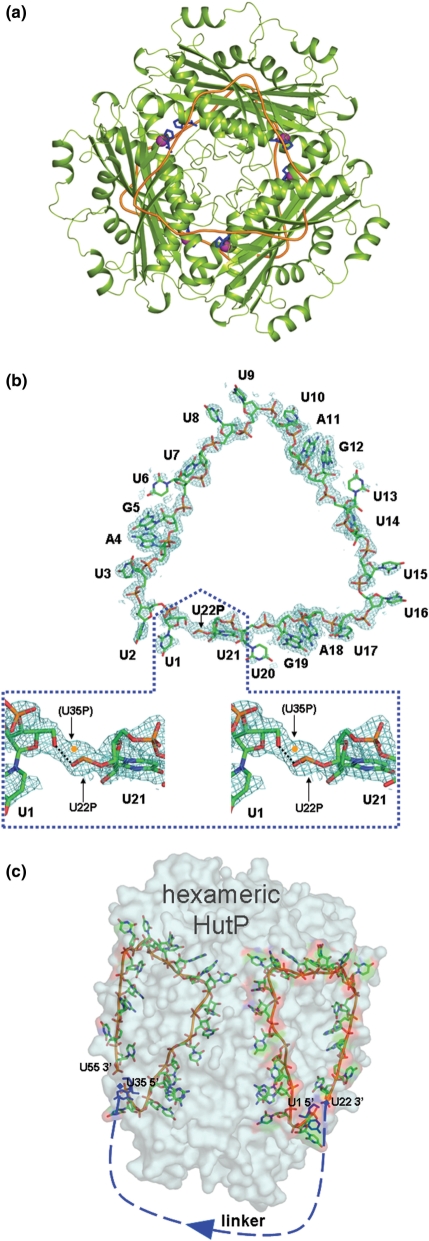
In the in-line mapping studies described above, we suggested that each of the two XAG-rich domains flanking the 17-nt linker region interacts with each surface of the HutP hexamer. In order to clarify this, we solved the crystal structure of the complex (HutP-l-his–Mg2+-55-mer RNA; Protein data bank accession code 2GZT). The important XAG sequence is repeated three times, with the spacer sequence U4 at both the 5′ and 3′ sides (Figure 1). The crystal of the present complex belongs to the monoclinic space group C2, in contrast to the crystal of the previous complex (HutP-21-mer RNA-l-histidine–Mg2+), with the rhombohedral space group R3 (15). The HutP protein forms a hexameric structure with D3 molecular symmetry (Figure 7a). The overall hexameric structure of HutP and the binding mode with l-histidine and Mg2+ are essentially the same as those described for the previous complex, with an r.m.s.d. of 0.17 Å between the Cα pairs. However, it should be noted that the hexamer is created by different crystallographic symmetries. In the present HutP hexamer, the three monomers within the trimer are related by noncrystallographic 3-fold symmetry, and the two trimers are related by crystallographic 2-fold symmetry. In contrast, in the previous complex, the three monomers are related by crystallographic 3-fold symmetry, and the two trimers are related by non-crystallographic 2-fold symmetry. The bound RNA is also related by 2-fold and 3-fold symmetries. The noncrystallographic 3-fold symmetry in the present structure allowed us to determine the entire structure of the 21-mer nucleotide fragment of the 55-mer RNA, thus providing more information than the previous complex, in which only a 7-mer was determined, and the bases of the spacer nucleotides were somewhat disordered (15). The r.m.s.d. among U1-U7, U8-U14 and U15-U21 in the present complex is within 0.44 Å. A continuous electron density along the fragment U1-U21 is clearly visible (Figure 7b), but that of the linker fragment U22–U34 is absent, except for the phosphate of U22. The phosphate oxygen atoms of U22 probably hydrogen bond to the O5′ atom of U1, as shown in Figure 7b. The electron density in this region demonstrates the clear connectivity between the U22 phosphate and the U1 ribose, which can be explained as follows. Figure 7b shows a close-up view of the electron density around U1 and U22P, where U1 is equivalent to U35 and U21 is equivalent to U55 according to crystallographic 2-fold symmetry. In this case the U35 phosphate is not superposed on the U22 phosphate but has been modeled closer to U1 (symmetry related U35). The modeled U35P is shifted by about 1 Å on the electron density, such that it can be covalently bonded to the U35 ribose. Thus, the electron densities for U22P and U35P are averaged by crystallographic 2-fold symmetry. The electron densities of the 21-mer fragments of U1–U21 and U35–U55 are visible, because they are superposed well, but in the linker region those of U22 and the following nucleotides, and those of U34 and the preceding nucleotides are not visible, suggesting they are not superposed. Therefore, the electron densities of the linker region disappear by symmetry averaging. Alternatively, the absence of electron density in this region is probably due to the large disorder of the linker, reflecting the high mobility of the linker region. This is consistent with the in-line mapping observation, in which the linker region undergoes a significant conformational change in the presence of activated HutP (Figures 2b and 4a). It was not possible to determine which 21-mer is U1–U21 or U35–U55, because of the symmetry problem mentioned above. To clarify the directionality of the 55-mer RNA bound to HutP, we tentatively assigned the fragment on the right side in Figure 7c as U1–U21 (site I), and thus U35–U55 (site II) is on the left side. The full-length of the RNA in the crystals were also confirmed by dissolving the quaternary complex (HutP-l-his–Mg2+-55-mer RNA) crystals in water, after washing them a few times in reservoir solution, and then isolated and analyzed the RNA, using reverse transcriptase. These analyses indicated that the RNA present in the crystals was, indeed, the 55-mer RNA (Supplementary Figure 6).
Figure 7.
The overall structure of the HutP quaternary complex (HutP-l-his–Mg2+-55-mer RNA). (a) Ribbon model (HutP, green; RNA, orange) viewed along the 3-fold axis. The bound l-histidine and the Mg2+ ions are represented by blue stick and magenta cpk (Corey–Pauling–Kolturn) models, respectively. (b) The 2Fo–Fc electron density map contoured at the 1.0 sigma level, showing the continuous electron density for the entire 21-mer RNA. The RNA is shown as a stick model. The region beyond 21 nt is enlarged and shown in a stereo view (dotted box). The electron densities of this region contain those of the symmetry-related region of U35P. For details please see the text. (c) Transparent representation of the molecular surface of the hexameric HutP and the bound 55-mer RNA. The U1 and U35 residues are colored blue. The bound RNA is shown as a stick model, and the disordered linker region of the RNA is shown as blue dashed lines.
RNA–HutP interactions
The structure revealed that the U1–U21 and U35–U55 fragments are bound to HutP in a manner such that the bases interact with the surface of HutP, while the phosphates point outward, thus providing structural evidence for the sequence-dependent nature of the recognition. The observed specific interactions of the A and G nucleotides of the UAG sequence are exactly the same as those of the previous complex (15). Similar to the HutP, the TRAP is also known to bind specifically to the multiple XAG residues located within the 203-nt untranslated trp operon leader transcript (26), however the overall conformations of the bound RNAs are very different from each other (Supplementary Figure 7). The interactions of the U7 nucleotide (also U14 and U21) are also the same. The bases of U1, U2, U6, U8, U9, U13, U15, U16 and U20 are exposed to the solvent. Although the bases of U3, U10 and U17 have no direct hydrogen bonds to HutP, the O2 atoms of these bases closely contact the N7 atoms of the adjacent bases A4, A11 and A18, respectively (Figure 7b). Thus, the bases of the spacer U nucleotides weakly contact HutP, suggesting that these interactions may be non-specific, consistent with the finding that the spacer nucleotides are sequence independent. Another interesting point is the backbone conformation presented by the torsional angles (α, β, γ, δ, ε and ζ), and (χ) of the bond between the ribose ring and the base (Supplementary Table 2). The (α,γ) pairs of U2, U7–U9, and U14–16 show the extended conformation of the spacer nucleotides between the UAG sequences. The unusual values of ε in U7 (23.0°) and U14 (56.5°) are coupled with the unusual values of ζ in U7 (74.1°) and U14 (71.8°), respectively, forming a further extended structure with U8 and U15 at the 3′ sides, respectively. The pseudotorsional angles are also informative to extract the backbone chain conformation of the RNA (27), where η and θ are defined by C4′i-1-Pi-C4′-Pi+1 and Pi-C4′-Pi+1-C4′+1, respectively (Supplementary Table 2). The θ values of U2, U9 and U16 are in an unusual gauche conformation. This conformation permits a τ angle favorable for an in-line orientation (Supplementary Figure 2), and leads to efficient cleavage (Figure 2b). The (η, θ) pairs of those bases are (241.1, 53.8), (207.1, 75.7) and (211.2, 54.2), which are in the range of the ‘stack’ switching region (27). Thus, the nucleotides U2, U9 and U16 play the role of the ‘apex’ to form a triangular shape, and function to facilitate the bending of the RNA for recognition of the next XAG-binding site on the surface of the HutP hexamer. Nevertheless, the present crystal structure of the quaternary complex of HutP revealed, consistent with the in-line mapping and gel-shift analyses, that HutP indeed binds to two sites within the 55-model RNA, using its dual-RNA-binding surfaces. Thus, the binding manner of the 55-model RNA to HutP is expected to reflect the actual binding of the native hut terminator on the activated HutP. The present complex structure revealed that the XAG motifs in site I bind in a triangular fashion on one surface of HutP, and the following linker nucleotides carry the remaining XAG motifs in site II to the backside of HutP, so as to bind in the same fashion as in site I (Figure 7c). As a result, the hut terminator appears to wrap the dual RNA-binding surfaces of the HutP hexamer in a 5′ to 3′ direction.
DISCUSSION
We revealed in the present study that HutP recognizes two XAG-rich sites (I and II) within the hut terminator region. Occupying these sites by the HutP flanking to the G-C-rich stem region of the terminator, the formation of the terminator RNA can be prevented, possibly upon HutP binding to the 5′ site (site I) would allow binding to the 3′ site (site II). These studies also suggest that the 5′ XAG-binding region is in a single-stranded region, and that the 3′ XAG region is in a paired hairpin. Thus, as stated, HutP undoubtedly binds to the 5′ XAG region as soon as it is free from the polymerase, and when the 3′ XAG region is initially synthesized, it too can bind to this segment as soon as it escapes from the transcribing polymerase. Alternatively, HutP may bind to the RNA while polymerase is paused, after synthesizing the hairpin. Each surface of hexameric HutP recognizes three XAG motifs within the RNA, and the bound RNA is arranged in a novel triangular fashion (Figure 7b). For efficient recognition of the XAG motifs by HutP, the hut terminator RNA possesses a 12–17-nt linker. The HutP residues specifically recognize A and G bases, in agreement with our previous findings (15). Additionally, we have shown which XAG motifs of sites I and II are important, and the conformational changes that occur in the terminator region upon HutP binding, using in-line mapping assays. These studies suggested that substitutions in the first three or second three XAG motifs markedly affect the binding to HutP. Since the model RNA does not assume any specific secondary structure, it is possible that the substitutions in these model RNAs, for generating mutant 1 and mutant 2, did not reflect secondary structural variations, but rather the loss of important functional groups in this region. Substitutions of either the first three XAG motifs in the site I region or the last three XAG motifs in site II seemed to abolish the overall contacts necessary for wrapping the RNA, including the conformational change in the linker region. We also observed that the spacer region in the hut terminator RNA is more accessible for metal ion-mediated cleavage upon binding to hexameric activated HutP (Figure 4a), suggesting that activated HutP is able to destabilize the secondary structure of the terminator region. On the other hand, unlike the results with the model 55-mer RNA, substitutions in the first XAG motif alone in the native hut terminator abolished the binding to the activated HutP. This analysis confirms that the first XAG motif is important for the recognition of the 5′ end of the terminator RNA. In addition, it is noteworthy to mention that in the native RNA, the first two XAG motifs appear to be readily accessible, as they are located upstream from the stable stem. If these motifs are not accessible, then the activated HutP may not be able to contact the other XAG-rich regions in the hut terminator. These studies also imply that the anti-termination process initiates on the hut terminator RNA from the 5′ to 3′ direction. To support this notion, our in vitro and in vivo analyses revealed that the first XAG residues (site I) in the hut terminator RNA provide an important contact for HutP to initiate the anti-termination process (Figures 5b and 6b). A similar observation was also reported in another anti-termination protein, TRAP, in which the activated-protein binding proceeds in a 5′ to 3′ direction (28). This directionality is probably set, as a default, in both terminator RNAs to provide accessible XAG sites at the 5′end of the RNA that allow the protein to roll onto the terminator. Interestingly, both TRAP and HutP regulate transcription in a similar way by conserving recognition of repeating RNA triplets, even though the three-dimensional structures are very different.
In short, based on the above findings and our previous results, we propose that HutP becomes activated initially by the l-his and Mg2+ ions, recognizes the first two XAG motifs near the 5′ end of the terminator RNA, and then rolls over the terminator toward the third XAG motif. Once HutP recognizes the third XAG motif, the adjacent G-C-rich base-pair stem will start to melt, because of steric hindrance, thus forcing the RNA to undergo a conformational change. Consequently, the second XAG-rich region is exposed and approaches to bind onto the other surface of HutP, resulting in the disruption of the stable stem region (Figure 8). Taken together, the present studies, for the first time, revealed how the anti-termination protein, HutP, destabilizes the hut terminator RNA structure by accessing two XAG-rich sites. Interestingly, these XAG residues strategically flank the highly stable stem region (G-C-rich region) of the terminator, so that HutP can use these two sites effectively to destabilize the terminator RNA, in order to allow the polymerase access for transcription.
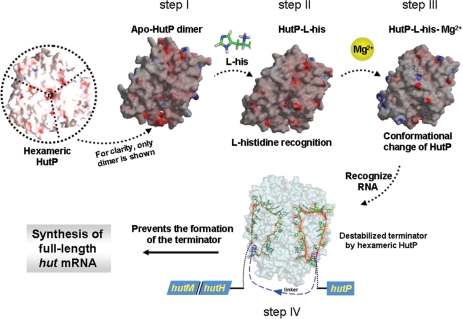
Figure 8.
Proposed snap-shots for the HutP-mediated antitermination process of transcription, to regulate the hut operon in Bacillus species. HutP is shown as a hexamer viewed down to noncrystallographic 3-fold axis. In step I to step III, only one dimer is shown for clarity during activation process (a view down close to crystallographic 2-fold axis). Step I, existence of hydrophobic pocket; step II, selection of l-histidine and discrimination of other amino acids; step III, overall conformational changes in the presence of Mg2+ before binding to RNA. Step IV, HutP binds to two XAG-rich regions within the hut terminator RNA and prevents the formation of anti-terminator RNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by funds from the National Institute of Industrial Science and Technology (AIST) to P.K.R.K. S.C.B.G was partly supported by the Japan Society for the Promotion of Science (JSPS). T.K. thanks Prof. S. Yokoyama for his encouragement and support. Funding to pay the Open Access publication charges for this article was provided by Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Babitzke P, Yanofsky C. Reconstitution of Bacillus subtilis trp attenuation in vitro with TRAP, the trp RNA-binding attenuation protein. Proc. Natl Acad. Sci. USA. 1993;90:133–137. doi: 10.1073/pnas.90.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Houman F, Diaz-Torres MR, Wright A. Transcriptional antitermination in the bgl operon of E. coli is modulated by a specific RNA binding protein. Cell. 1990;62:1153–1163. doi: 10.1016/0092-8674(90)90392-r. [DOI] [PubMed] [Google Scholar]
- 3.Aymerich S, Steinmetz M. Specificity determinants and structural features in the RNA target of the bacterial antiterminator proteins of the BglG/SacY family. Proc. Natl Acad. Sci. USA. 1992;8:10410–10414. doi: 10.1073/pnas.89.21.10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnaud MD, Debarbouille M, Rapoport G, Saier M.H., Jr., Reizer J. In vitro reconstitution of transcriptional attenuation by the SacT and SacY proteins of Bacillus subtilis. J. Biol. Chem. 1996;271:18966–18972. doi: 10.1074/jbc.271.31.18966. [DOI] [PubMed] [Google Scholar]
- 5.Lu Y, Turner RJ, Switzer RL. Function of RNA secondary structures in transcriptional attenuation of the Bacillus subtilis pyr operon. Proc. Natl Acad. Sci. USA. 1996;93:14462–14467. doi: 10.1073/pnas.93.25.14462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alpert CA, Siebers U. The lac operon of Lactobacillus casei contains lacT, a gene coding for a protein of the Bg1G family of transcriptional antiterminators. J. Bacteriol. 1997;179:1555–1562. doi: 10.1128/jb.179.5.1555-1562.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glatz E, Nilsson RP, Rutberg L, Rutberg B. A dual role for the Bacillus subtilis glpD leader and the GlpP protein in the regulated expression of glpD: antitermination and control of mRNA stability. Mol. Microbiol. 1996;19:319–328. doi: 10.1046/j.1365-2958.1996.376903.x. [DOI] [PubMed] [Google Scholar]
- 8.Oda M, Katagai T, Tomura D, Shoun H, Hoshino T, Furukawa T. Analysis of the transcriptional activity of the hut promoter in Bacillus subtilis and identification of a cis-acting regulatory region associated with catabolite repression downstream from the site of transcription. Mol. Microbiol. 1992;6:2573–2582. doi: 10.1111/j.1365-2958.1992.tb01434.x. [DOI] [PubMed] [Google Scholar]
- 9.Wray LV, Jr., Fisher SH. Analysis of Bacillus subtilis hut operon expression indicates that histidine-dependent induction is mediated primarily by transcriptional antitermination and that amino acid repression is mediated by two mechanisms:regulation of transcription initiation and inhibition of histidine transport. J. Bacteriol. 1994;176:5466–5473. doi: 10.1128/jb.176.17.5466-5473.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oda M, Kobayashi N, Ito A, Kurusu Y, Taira K. cis-Acting regulatory sequences for antitermination in the transcript of the Bacillus subtilis hut operon and histidine-dependent binding of HutP to the transcript containing the regulatory sequences. Mol. Microbiol. 2000;35:1244–1254. doi: 10.1046/j.1365-2958.2000.01795.x. [DOI] [PubMed] [Google Scholar]
- 11.Kumarevel TS, Mizuno H, Kumar P.KR. Allosteric activation of HutP protein, that regulates transcription of hut operon in Bacillus subtilis, mediated by various analogs of histidine. Nucleic Acids Res. Suppl. 2003;3:199–200. doi: 10.1093/nass/3.1.199. [DOI] [PubMed] [Google Scholar]
- 12.Kumarevel TS, Fujimoto Z, Karthe P, Oda M, Mizuno H, Kumar P.KR. Crystal structure of activated HutP: an RNA binding protein that regulates Transcription of the hut operon in Bacillus subtilis. Structure. 2004;12:1269–1280. doi: 10.1016/j.str.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 13.Kumarevel TS, Gopinath S.CB, Nishikawa S, Mizuno H, Kumar P.KR. Identification of important chemical groups of the hut mRNA for HutP interactions that regulate the hut operon in Bacillus subtilis. Nucleic Acids Res. 2004;32:3904–3912. doi: 10.1093/nar/gkh725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumarevel TS, Mizuno H, Kumar P.KR. Characterization of the metal ion binding site in the anti-terminator protein, HutP, of Bacillus subtilis. Nucleic Acids Res. 2005;33:5494–5502. doi: 10.1093/nar/gki868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumarevel TS, Mizuno H, Kumar P.KR. Structural basis of HutP-mediated anti-termination and roles of the Mg2+ ion and L-histidine ligand. Nature. 2005;434:183–191. doi: 10.1038/nature03355. [DOI] [PubMed] [Google Scholar]
- 16.Krol A, Carbon P. A guide for probing native small nuclear RNA and ribonucleoprotein structures. Methods Enzymol. 1989;180:212–227. doi: 10.1016/0076-6879(89)80103-x. [DOI] [PubMed] [Google Scholar]
- 17.Gopinath SCB, Matsugami A, Katahira M, Kumar PKR. Human vault-associated non-coding RNAs bind to mitoxantrone, a chemotherapeutic compound. Nucleic Acids Res. 2005;33:4874–4881. doi: 10.1093/nar/gki809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson GA, Bott KF. Nutritional factors influencing the development of competence in the Bacillus subtilis transformation system. J. Bacteriol. 1968;95:1439–1449. doi: 10.1128/jb.95.4.1439-1449.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chasin L, Magasanik B. Induction and repression of the Histidine-degrading enzymes of Bacillus subtilis. J. Biol. Chem. 1968;243:5165–5178. [PubMed] [Google Scholar]
- 20.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography and NMR system (CNS): a new software system for macromolecular structure determination. Acta Cryst. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 21.Oldfield TJ. A number of real-space torsional angle refinement techniques for proteins, nucleic acids, ligands and solvent. Acta Cryst. D. 2001;57:82–94. doi: 10.1107/s0907444900014098. [DOI] [PubMed] [Google Scholar]
- 22.DeLano WL. San Carlos, CA, USA: DeLano Scientific; 2002. [(18April2008,datelastaccessed)]. The PyMOL Molecular Graphics System. http://www.pymol.org. [Google Scholar]
- 23.Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Ribo- switches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 25.Eda S, Hoshino T, Oda M. A novel mutation of the Bacillus subtilis hut operon that relieves both catabolite repression and amino acid repression. Appl. Microbiol. Biotechnol. 1999;51:85–90. doi: 10.1007/s002530051367. [DOI] [PubMed] [Google Scholar]
- 26.Antson AA, Dodson EJ, Dodson G, Greaves RB, Chen X.-P, Gollnick P. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature. 1999;401:235–242. doi: 10.1038/45730. [DOI] [PubMed] [Google Scholar]
- 27.Duarte CM, Pyle AM. RNA structure comparison, motif search and discovery using a reduced representation of RNA conformational space. J. Mol. Biol. 1998;284:1465–1478. doi: 10.1093/nar/gkg682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbolina MV, Li X, Gollnick P. Bacillus subtilis TRAP binds to its target by a 5′ to 3′ directional mechanism. J. Mol. Biol. 2005;345:667–679. doi: 10.1016/j.jmb.2004.10.071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.