Abstract
Alternative splicing of a single pre-mRNA transcript can produce protein isoforms that promote either cell growth or death. Here we show that Ro-31-8220 (Ro), an apoptotic agent that inhibits protein kinase C and activates the c-Jun N terminal kinase, decreased the proportion of the cell growth-promoting Bcl-xL splice variant. Targeted mutagenesis analyses narrowed down a critical sequence to a 16-nt G-tract element (Gt16). Transferring this element to a heterologous gene conferred Ro response on an otherwise constitutive exon. The Ro effect was reduced by okadaic acid, an inhibitor of protein phosphatases PP1 and PP2A, in a concentration-dependent manner. Search in the human genome followed by RT–PCR identified a group of genes that contain similar exonic G-tract elements and are responsive to Ro. Moreover, the Gt16 element also mediates the regulation of alternative splicing by other cell apoptosis-inducers particularly retinoic acid. Therefore, the G-tract element likely plays a role in the apoptotic agents-induced alternative splicing of a group of genes. The functions of these genes imply that this regulation will have impact on cell growth/death.
INTRODUCTION
Alternative splicing allows the generation of more than one protein isoforms from a single pre-mRNA transcript, contributing greatly to the proteomic diversity (1–4). By this way, a number of genes involved in cell growth/death generate protein isoforms that promote either cell growth or death (5,6). This regulation can be dynamically controlled by extracellular factors but rarely has a factor been coupled with a regulatory pre-mRNA element.
Alternative splicing of mammalian genes is controlled by multiple cis-acting pre-mRNA elements in either introns or exons that promote or inhibit exon inclusion (3,7). These elements control the assembly of constitutive splicing components mostly by binding trans-acting factors, including the arginine/serine-rich SR proteins (8–10), heterogeneous nuclear ribonucleoprotein particle (hnRNP) proteins and others (3,11–15). The balance between the positive and negative elements/factors controls the inclusion of exons in specific cells or during developmental stages. This balance can be changed by extracellular factors but the underlying molecular basis is less well understood than that of cell type- or developmental stage-specific splicing.
Many extracellular factors, including hormones, neurotransmitters and pharmacological agents, can regulate alternative splicing through protein kinases or phosphatases (16–18). In some cases, pre-mRNA elements and the bound splicing factors responsive to the stimuli are identified, facilitating the delineation of the involved signaling pathways. For example, in the CD44 gene, the pre-mRNA sequence for TPA (12-O-tetradecanoylphorbol 13-acetate) regulation of exon 5 is an A-rich element (19–21). This element is bound by Sam68, which is directly phosphorylated by the ERK kinase upon TPA stimulation to enhance the inclusion of exon 5 (21).
Ro-31-8220 (Ro) is a synthetic compound originally used as a PKC-inhibitor (22,23). It also activates the c-Jun N-terminal kinase (JNK) and inhibits expression of the MAP kinase phosphatase 1 MPK1 (24). It induces cell apoptosis through PKC or other pathways (25,26). The effect of Ro on alternative splicing of genes in cell growth/death has not been examined.
Bcl-x, a Bcl-2 family member critical for cell survival/apoptosis (27), generates two antagonistic isoforms Bcl-xL and Bcl-xS, through alternative inclusion of a 189-nt fragment that encodes 63 amino acids in a region highly conserved in the Bcl-2 family proteins (28,29). Bcl-xL protects cells from death (27,29–33), while as Bcl-xS inhibits Bcl-2-induced cell survival (29). The two splice variants are differentially expressed among tissues, during development and in diseases (29,34,35). The growth-promoting Bcl-xL variant is increased in tumors (35). In cultured cells, Bcl-x splicing can be regulated by ceramide, TPA or staurosporine (36–38). Due to the two variants’ antagonistic roles in cell growth/death, attempts have been made to induce cancer cell death or sensitivity to chemotherapeutic agents through modulating Bcl-x splicing (32,39,40).
Several studies have examined the molecular components controlling Bcl-x splicing. A 30-nt fragment containing G-tracts in the pre-mRNA reduces the Bcl-xL product through the hnRNP H/F proteins (41). A 361-nt fragment situated 187-nt upstream of the 5′ splice site of Bcl-xS is targeted by staurosporine to stimulate Bcl-xS production (38), likely through the PKC pathway (38); details of the regulatory element remain to be unveiled. Ceramide reduces the Bcl-xL product through two other elements (CRCE1 and CRCE2) and SAP155 (36,42,43), and protein phosphatase 1 (PP1) (36). Sam68 controls Bcl-x splicing as well but the target pre-mRNA element is unknown (44). There are also elements in the Bcl-xL downstream intron that are involved in the responses to cytokines and TPA (37). Together, these findings indicate that multiple pre-mRNA elements/factors are involved in the control of Bcl-x splicing, as for mammalian genes in general (45,46).
G-tracts are splicing regulatory pre-mRNA elements. The minimal binding and functional element is GGG and two G-tracts with a variable spacer are stronger than one copy in splicing and protein-binding assays (47–49). These elements may play a role in intron definition where the sequences they reside tend to be skipped as introns (47,50). The elements can be either splicing enhancers or silencers, by binding U1 snRNA (51), or members of the hnRNP H family, including H1(H), H2 (H′), H3 (2H9), F and GRSF-1 (41,48,49,52–55). Particularly, hnRNP H1 either enhances or represses the inclusion of target exons (41,49,55–60).
In this report, we show that Ro controls the alternative splicing of the endogenous Bcl-x gene by targeting a G-tract element through an okadaic acid-sensitive pathway. Similar G-tract element-containing exons of other genes are regulated by Ro as well. Moreover, the G-tract element appears to also mediate the control of alternative splicing by other cell apoptotic inducers particularly retinoic acid.
MATERIALS AND METHODS
Construction of splicing reporter plasmids
wDUP175 was derived from DUP175 (61,62), by replacing the 3′ splice site of the middle exon with a weaker sequence (accaccctagccatctaatcacttatacacattcattttagC, with ‘C’ as the first nt of the downstream exon). For the RARG minigene, the lengths of the upstream intron, exon and downstream intron included in the insert are 125, 326 and 294 nt, respectively. Mutations were made by polymerase chain reaction (PCR) using Pfu DNA polymerase. Minigene inserts were between the ApaI and BglII sites of DUP175, unless otherwise indicated, and confirmed by sequencing.
Cell culture, transfection and treatment
MDA-231 and BT20 cells were cultured in DMEM supplemented with 5% fetal bovine serum. HEK 293T cells were cultured in DMEM with 10% newborn calf serum. Transfections were carried out with Lipofectamine 2000 (Invitrogen) 24 h after plating according to the supplier's protocol, in 12-well plates using 0.15 μg reporter plasmid. Transfected cells were incubated with Ro for various time intervals as indicated in the text and lyzed for RNA extraction. Cells were treated in serum-free media. All phosphatase and kinase inhibitors and other chemical agents for cell treatment were purchased from Sigma–Aldrich, Oakville, Ontario, Canada, except DMSO (Fisher Sci., Ottawa, Ontario, Canada).
Semiquantitative reverse transcription (RT)-PCR
Total RNA was extracted with GenElute Mammalian Total RNA Miniprep Kit (Sigma-Aldrich, Oakville, Ontario, Canada). RT-PCR was carried out as previously described (61), except 400 ng RNA was used for 10 μl of reverse transcription reaction. PCR reaction was run at an annealing temperature of 60°C for 26 cycles. The products were resolved in 3% agarose gels containing 0.5 ug/ml of ethidium bromide in TBE buffer and documented on a UV transilluminator under a digital camera. Band intensities were quantified with the NIH Image J software 1.37v (developed at the U.S. National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/ij/). The percentages of exon-excluded or -included product in agarose gels were calculated from the actual band intensities relative to the total of spliced products (excluding the cryptically spliced ones). Agarose gel pictures in the figures are inverted digital images. The percentages of exon-excluded or -included products of the electropherograms, obtained in an automated workstation (63), were calculated from their molar numbers.
Human genome database search
Annotated human genome sequences (NCBI36) were downloaded from the ENSEMBL website (http://www.ensembl.org). A bioperl script EXON was written to extract all the exons with up to 500 nt flanking intron sequences into a whole-genome exon database. This database was searched for exons containing the G-tract element GGGGNNNNNNGGGG using another Bioperl script ExonElement to yield a database of the G-tract-containing exons in MS Excel. Exons with the same sequences were filtered out. The unique ENSEMBL gene IDs were used to obtain the HGNC symbols (whichever available) of each gene from Biomart (http://www.biomart.org/). These symbols were used to identify genes in the toxicity category with the Ingenuity Pathway Analyses, a software application that allows identification of proteins/genes clustering in the same pathway/category from a group of target genes (http://www.ingenuity.com). Exons of genes in this category were obtained by filtering the MS Excel file with the HGNC symbols/ENSEMBL gene IDs. The alternative exons were identified by aligning the exon/intron sequences using the UCSC genome database (http://www.genome.ucsc.edu/).
RESULTS
Ro decreases the proportion of the Bcl-xL product
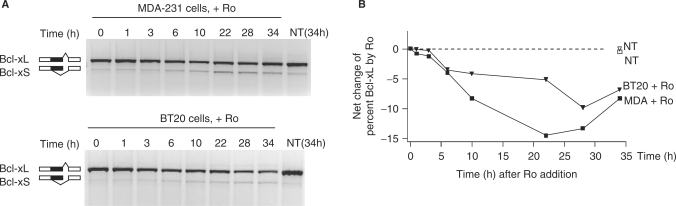
In examining the alternative splicing of the Bcl-x gene by extracellular factors, we found that Ro decreased the proportion of the Bcl-xL product in the human breast cancer cell lines MDA-231 and BT20 (Figure 1). In these cells, Bcl-xL was the predominant isoform (98%) and Bcl-xS was barely visible without treatment. Upon addition of Ro, the proportion of the Bcl-xL variant started to decrease at 6 h and was reduced about 15% at 22 h in MDA-231 cells and 10% at 28 h in BT20 cells. Similar reductions by Ro were also observed in PC12 and HEK293T cells (data not shown, and see below). Therefore, Ro decreases the proportion of the Bcl-xL product.
Figure 1.
Ro decreases the proportion of the Bcl-xL product. (A) Agarose gels of the RT-PCR products of Bcl-x from RNA samples of MDA-231 (upper) or BT20 (lower) cells incubated with Ro for different time intervals, with Bcl-x splicing patterns indicated to the left. (B) A graphed time course of the net changes of the percentages of the Bcl-xL product relative to the starting time (0 h), in MDA-231 (squares) or BT20 (triangles) cells. The dotted line marks the starting baseline level of the Bcl-xL product in the NT samples of each cell line. The final concentration of Ro was 2 µg/ml. Ro, Ro-31-8220; NT, nontreated, control for Ro, which was dissolved in DMEM.
A 16-nt G-tract element mediates Ro regulation of Bcl-x splicing
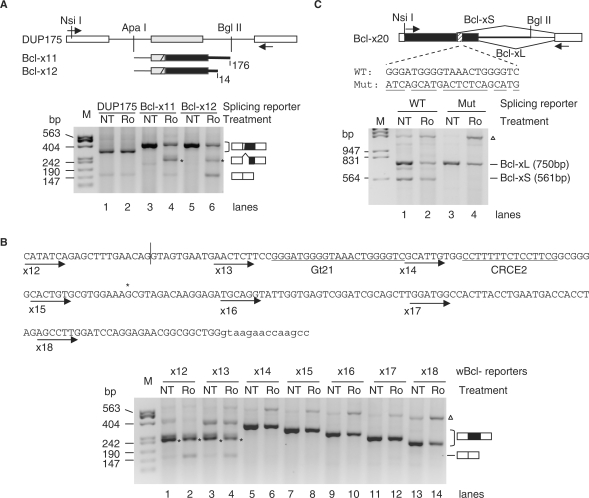
To identify the pre-mRNA sequence that can mediate the Ro effect on the reduction of the Bcl-xL product, we cloned Bcl-x fragments containing partial Bcl-x exon 2 and different lengths of the downstream intron into a splicing reporter derived from the human β-globin gene (61,62). Splicing changes of these reporters were examined by RT-PCR after transfection into HEK293T cells followed by incubation with or without Ro. The result showed that inclusion of the middle exon of the control reporter (DUP175) was not changed by Ro (Figure 2A, lanes 1 and 2). In contrast, inclusion of the Bcl-xL exon in the Bcl-x11 construct was reduced by Ro (lanes 3 and 4, 12% reduction). The Ro effect became stronger when the downstream intron was shortened in Bcl-x12 (lanes 5 and 6). Therefore, the Bcl-x12 insert is sufficient to mediate Ro-induced Bcl-xL reduction. Since the control and Bcl-x mini-genes have the same promoter, 3′ polyadenylation sites and flanking exons, the difference in their responses to Ro is likely due to their differences in the middle exon and flanking introns, suggesting that the Bcl-x inserts harbor RNA elements responsive to Ro-induced reduction of Bcl-xL.
Figure 2.
A G-tract region in Bcl-x is essential for the Ro-induced reduction of the Bcl-xL product. (A) (Upper) Diagram of the control splicing reporter DUP175 and cloned inserts of the Bcl-x minigene splicing reporters. Boxes represent exons and lines introns. Black boxes represent the Bcl-xL exon and black heavy lines Bcl-x intron. In the Bcl-x constructs, 51-nt β-globin exon sequence was fused with the exon 2 of Bcl-x containing 20-nt Bcl-xS, the 189-nt Bcl-xL exons and different lengths (nt) of the downstream introns as indicated. ApaI and BglII are the cloning sites. The arrows indicate locations of PCR primers. (Bottom) An agarosel gel of RT-PCR products from HEK293T cells transfected with the splicing reporters (as indicated above the gel) and treated with Ro or without (NT). The spliced products are diagramed to the right of the gel. Gel representative of at least three samples for each lane. (B) (Upper) Bcl-x sequence included in wBcl-x12 and other reporters. Exon sequences are in upper cases and intron in lower cases. The vertical line marks the 5′ splice site exon–intron junction for Bcl-xS. The starts of the arrow lines indicate the Bcl-x insert starting positions in each splicing reporter minigene (with reporter numbers below the arrows). In these wBcl-x reporters, the upstream 3′ splice site of the middle exons was replaced with a weaker sequence (see Materials and methods section) for a stronger Ro effect. (Bottom) Similar to the gels in (A) except with different splicing reporters as indicated above the gel. The 21-nt G-tract region (Gt21) to be examined and the CRCE2 sequence required for ceramide-regulation of Bcl-x splicing is underlined. (C) Role of the G-tract region in Ro-induced Bcl-xL reduction in its alternative 5′ splicing context. (Upper) Diagram of the Bcl-x20 splicing reporter with the Bcl-x exon 2 (black box, including the 189-nt Bcl-xL sequence) and downstream intron (514 nt, heavy line) cloned between the Nsi I and Bgl II sites of DUP175. The splicing patterns and the wild-type and mutant (mutated nts underlined) sequences of the G-tract region are as indicated. Arrows: locations of PCR primers. (Lower) An agarose gel of the RT-PCR products from cells transfected with the splicing reporters without treatment (NT) or treated with Ro (Ro) as indicated above the gel, with molecular size markers to the left and splicing products (sizes) to the right. Cells were treated with 2 µg/ml Ro or nontreated (NT) for overnight. *Product from cryptic 3′ splice site as indicated to the right of the gels or the cryptic splice position in the sequence. Gel representative of two experiments; open triangle, product resulted from pre-mRNA or plasmid; M, molecular size marker (bp).
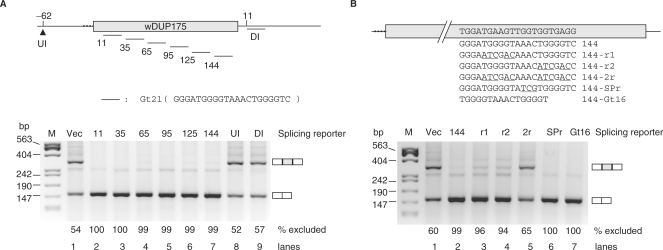
We then went on scanning this insert for RNA elements essential for the observed Ro effect. Series deletions (30-nt each) were made from the 5′ end Bcl-x sequence of a Bcl-x12 insert (wBcl-x12––x18, Figure 2B). Similar tests of these reporters in HEK293T cells showed that the Ro-induced exon exclusion disappeared completely starting from wBcl-x14 (lane 6). Replacing the 30-nt element deleted from wBcl-x13 with a β-globin sequence abolished the Ro effect as well (data not shown). These observations suggest that a sequence specifically essential for the Ro effect on splicing is within the 30-nt element, which contains the G-tracts shown to reduce the Bcl-xL product (41).
It should be noted that the wBcl-x13 also lost a substantial amount of Ro effect compared to Bcl-x12 (compare lanes 2 and 4), suggesting that the 30-nt fragment deleted from wBcl-x12 is also essential for the Ro effect. However, mutating the competing 5′ splice site in this fragment completely abolished the Ro effect (data not shown), indicating that the effect from this fragment is mainly due to the competing 5′ splice site. Therefore, instead of the competing 5′ splice site, we focused on the fragment containing the G-tracts for Ro-targeted regulatory elements.
To see whether the G-tract region is essential for the Ro effect in the context of alternative 5′ splicing of Bcl-x, we made construct Bcl-x20 and its G-tract region mutant (Figure 2C). This wild-type construct produces mainly Bcl-xL (72%), similarly as the endogenous Bcl-x gene, and responded to Ro treatment with a reduced percentage of Bcl-xL (57%) when expressed in HEK293T cells (15% reduction, lanes 1 and 2). The mutation led to almost all Bcl-xL product (lane 3), similarly to the reported effect (41). Interestingly, this mutant failed to respond to Ro (lane 4). These data suggest that the G-tract region is essential for Ro-induced reduction of the Bcl-xL product in Bcl-x's alternative 5′ splicing context.
To isolate the G-tracts’ effect on exon inclusion in a completely heterologous context, we transferred 21 nt of the G-tract fragment (Gt21) into the wDUP175 middle exon or its flanking introns (Figure 3). Replacements of exon sequences with the Gt21 at various positions lead to complete exon exclusion (Figure 3A, lanes 2–7), while Gt21 insertion or replacement mutations in introns did not substantially change the splicing pattern (lanes 8 and 9). Therefore, the Gt21 appears to be an exonic silencer in this context.
Figure 3.
The G-tract element is an exonic silencer. (A) (Upper) Diagram of Gt21 inserted into (arrowhead) or replaced (short horizontal lines) vector sequences at the indicated positions in or flanking the wDUP175 middle exon. The exonic position numbers indicate the 5′ first nt of the element, counted from the first nt of the exon. The intronic positions are relative to the last (upstream, UI) or first (downstream, DI) nt of the intron. (Bottom) An agarose gel of RT-PCR products of HEK293T cells transfected with the splicing reporters as indicated above the gel. (B) (Upper) Diagram of the splicing reporters with mutations within the Gt21 reporter 144 as in (A). Bcl-x G-tract and mutant (underlined) sequences are aligned under the replaced β-globin sequence (top, in the exon). (Bottom) Similar to that in (A) except for different reporters as indicated above the gel. Vec, wDUP175. Dotted lines refer to the weakened 3′ splice site. Gels representative of two or three samples per lane. The exon-included or -excluded products are indicated to the right of each gel. Percentages of exon-excluded products are below each lane.
To identify a shorter sequence sufficient for the silencer activity, the G tetrads in the Gt21 were mutated in the reporter 144 (Figure 3B). Mutation of each G tetrad barely relieved the repression (96 and 94% exon exclusion, lanes 3 and 4, respectively, compare with 99% for lane 2). In contrast, simultaneous mutations of both G tetrads abolished the repression, reducing the exon exclusion to a level similar to the vector's (65% exon exclusion, lane 5, compare with 60% for lane 1). Mutation in the spacer between the G tetrads did not change the repression (SPr, lane 6). Moreover, replacing the β-globin sequence with only 16 nt encompassing both G tetrads (Gt16) led to complete repression (lane 7). Therefore, the Gt16, including two G tetrads and a variable 6-nt spacer, is a minimal sequence for the silencer activity in this reporter.
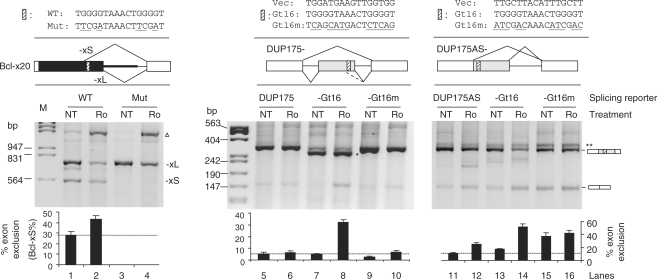
To examine the role of the G tetrads in Bcl-x response to Ro, we mutated them to TCGA in the reporter Bcl-x20 and tested the Ro effect (Figure 4, left panel). The mutations again led to almost all Bcl-xL products and abolished the Ro-induced Bcl-xL exon exclusion (compare lanes 3 and 4), as by the above mutation of the G-tract region (Figure 2C). Therefore, the G-tract nucleotides are essential for Ro-induced reduction of Bcl-xL in its alternative 5′ splicing context.
Figure 4.
The 16-nt G-tract element ‘TGGGGTAAACTGGGGT’ is essential for Bcl-xL and sufficient for a heterologous exon to be repressed by Ro. Shown are agrose gels of RT-PCR products of HEK293T cells transfected with splicing reporter minigenes with the element sequences and splicing patterns indicated above the gels. Bar graphs of the average percent (± SD, n = 3) exon exclusion for each sample are below the gels. The dotted horizontal lines mark the exon exclusion levels of the vectors. Cells were treated with 2 µg/ml Ro or nontreated (NT) for overnight. *A cryptic 5′ splice product with 38-nt deletion at the 3′ end of the middle exon (as indicated by the dotted line in the diagram above the gel); **likely heteroduplex formed between the two splice variant products; open triangle, same as in Figure 2.
To see whether the Gt16 is sufficient to confer the Ro effect on a heterologous exon that by itself does not respond to Ro, we transferred the Gt16 or its mutant to the DUP175 (Figure 4, lanes 5–10). Inclusion of the Gt16 reporter exon was reduced 27% by Ro, compared to the nontreated sample [lanes 7 and 8; the cryptic 5′ splice product has 38-nt deletion at the 3′ end of the middle exon, as described previously (64,65)]. This reduction was almost abolished by the Gt16 mutation (lanes 9 and 10), suggesting that the Ro-induced exon exclusion is indeed through the Gt16 element. Thus, the Gt16 is a Ro-responsive RNA element.
To examine the role of the Gt16 in Ro-induced alternative 5′ splice site usage as in Bcl-x, we constructed an alternative 5′ splice site reporter DUP175AS. In this reporter (Figure 4, right, lanes 11–16), the DUP175 first exon and its downstream 5′ splice site was fused with 165-nt 3′ half of the middle exon to form two identical competing 5′ splice sites. The Gt16 or its mutant sequence was transferred in between the two 5′ splice sites where they were 28 nt away from the upstream 5′ splice site. Test of these reporters showed that inclusion of the vector exon was slightly reduced by Ro (14% reduction, lanes 11 and 12). In contrast, inclusion of the Gt16 reporter exon was much more strongly reduced (34% reduction, lanes 13 and 14). Mutation of the Gt16 led to higher basal level exon exclusion (37%, lane 15) than the Gt16 reporter in nontreated cells. This higher level of the exon-excluded product was not further increased upon Ro treatment (compare lane 16 with lane 15, P = 0.31). Therefore, despite its promoting effect on the exon-excluded product, the mutant sequence does not mediate the Ro effect for further exon exclusion. The Gt16 element is thus indispensable for the strong Ro effect on the alternative 5′ splice site usage (Figure 1).
Taken together, the data from the three reporters (in Figure 4) suggest that the Gt16 element is essential and sufficient to mediate Ro-induced exclusion of both the cassette and the alternative 5′ splice exons.
The Ro-regulation of alternative splicing through the G-tract element requires phosphatases
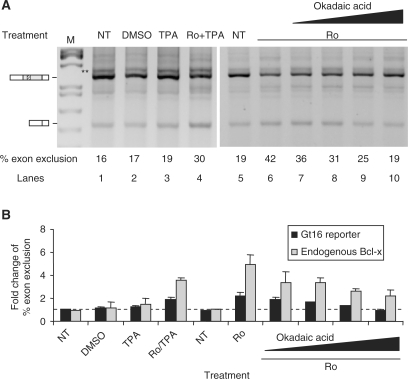
To examine the signaling pathway essential for the Ro-regulated splicing, we tested the effects of chemicals related to Ro targeted pathways (Figure 5). TPA, a PKC stimulator (66), did not change the splicing pattern of the DUP175AS-Gt16 reporter in HEK 293T cells (Figure 5A, lane 3), although it did have effect on Bcl-xL production in control PC12 cells (data not shown). Adding TPA after Ro to the HEK 293T cells slightly reduced the Ro effect (lane 4, compared to lane 6), suggesting the involvement of PKC to some extent. In contrast, okadaic acid, a phosphatase inhibitor (67), reduced the Ro-induced exon exclusion in a concentration-dependent manner (lanes 7–10), with almost complete inhibition of the Ro effect at 1000 nM. These observations suggest that okadaic acid inhibits the Ro-regulated alternative splicing through the G-tract element. Since both low (< 50 nM) and high (≥ 100 nM) concentrations of okadaic acid differentially inhibit the Ro effect, it is likely that both phosphatases 2A and PP1 are involved in the Ro regulation of splicing through the Gt16 element in HEK293T cells.
Figure 5.
Okadaic acid inhibits Ro-regulation of alternative splicing through the G-tract element. Shown are agarose gels of RT-PCR products of DUP175AS-Gt16 reporter from HEK293T cells treated with or without (NT) chemicals as indicated above the gels (A), with the average percentages of the exon-excluded products indicated under each lane. Below (B) is a bar graph of fold changes (average ± SD, n ≥ 3, relative to the nontreated NT sample for each reporter, black bars) of the percentages of exon-excluded products for each lane. Similarly obtained fold changes of the percent exon exclusion of the endogenous Bcl-x (gray bars) from the same samples are also shown beside the bars for each reporter in the graph. The dotted line marks the NT sample level. Ro: Ro-31-8220 (2 µg/ml). DMSO: vehicle for TPA. The okadaic acid concentrations, from left to right, were 20, 100, 500 and 1000 nM. TPA concentration: 120 ng/ml. **Same as in Figure 4.
Similar Ro effects were observed for the endogenous Bcl-x gene in these same samples (Figure 5B). Therefore, the phosphatases PP2A and PP1 are likely essential for the Ro-regulation of splicing of both the Gt16 reporter and the endogenous Bcl-x genes.
The G-tract element is also critical for Ro-regulation of alternative splicing of other genes
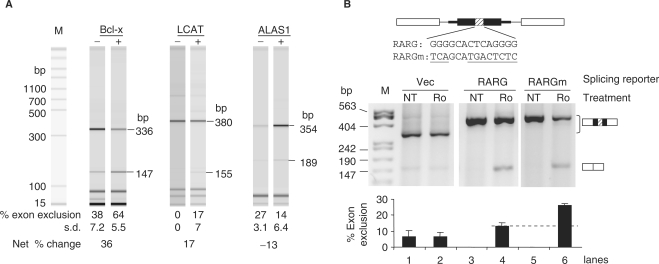
To see whether inclusion of similar G-tract-containing exons of endogenous genes can also be regulated by Ro, we first searched the human genome for exons containing the G-tract element GGGGNNNNNNGGGG, where the spacer Ns are nonessential for the silencer activity (Figure 3) and variable for both protein binding and splicing activities (47,49). This search identified 3374 exons of 2707 genes. Of these, we examined by RT-PCR 46 middle exons of genes in the Toxicity category as described by Ingenuity (http://www.ingenuity.com), with RNA samples of MDA-231 cells treated with or without Ro. We noticed that a group of genes, particularly Bcl-x, LCAT and ALAS1 (Figure 6A), responded with either decreased or increased percentages of exon-excluded product upon Ro treatment. This data suggest that a group of similar G-tract-containing exons are regulated by Ro.
Figure 6.
Ro-regulation of other G-tract-containing exons. (A) Electropherograms from an automated workstation of RT-PCR products of the G-tract-containing genes from RNA samples of MDA-231 cells treated without (–) or with (+) Ro (2 µg/ml) for 22 h. Indicated above the electropherograms are the respective gene names, below are the average percentages (± SD, n ≥ 3) of the molar amount of exon-excluded products and the net changes for each gene after Ro treatment, and to the right are the sizes (bp) of the PCR products. Molecular size (bp) markers (M) are aligned on the right side. LCAT: lecithin-cholesterol acyltransferase; ALAS1: aminolevulinate, delta-, synthase 1. (B) Agarose gels of RT-PCR assay showing the splicing changes of the reporters as indicated above the gels. RARG, retinoic acid receptor, gamma; Vec, DUP175. Below the gel is a bar graph of the average percentages (± SD, n = 3) of exon-excluded products of each lane. The dotted line marks the exon exclusion levels of the G-tract wild-type reporters induced by Ro.
To see whether the G-tracts in these exons are involved in the Ro-regulation of alternative splicing as in Bcl-xL, we then tested the Ro effect on three alternative exons in splicing reporters and found that the RARG (retinoic acid receptor, gamma) was responsive to Ro (Figure 6B), as well as a NOL1 (nucleolar protein 1) exon (data not shown). The RARG exon was completely included in cells without treatment (lane 3). Ro treatment induced exon skipping (13% exclusion, lane 4). Mutation of the G-tract element doubled the exon skipping (26% exclusion, lane 6). Therefore, the presence of the G-tract element of the RARG exon reduces the Ro-induced exon exclusion, unlike that in the Bcl-xL exon (see Discussion section), a context-dependent effect of regulatory RNA elements also seen in other systems (68,69). Nonetheless, this indicates that the G-tract elements are also involved in the Ro-regulated alternative splicing of other genes.
The G-tract element also mediates other apoptotic agents-induced alternative splicing
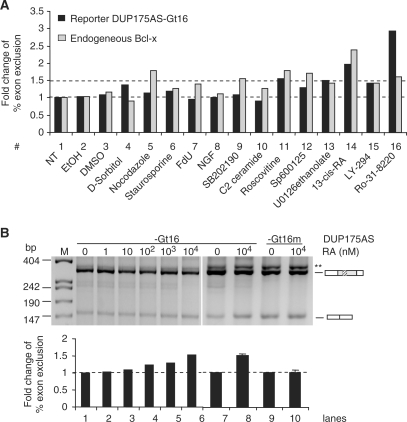
To see whether the G-tract element also mediates other stimuli to regulate splicing, we used the reporter DUP175AS-Gt16 to screen a number of chemicals related to cell cycle or cell death for splicing changes (Figure 7A). Of these chemicals, roscovitine (a cyclin-dependent kinase inhibitor), U0126 (a MEK1/2 inhibitor) and 13-cis-retinoic acid (RA) increased at least 1.5-fold of the percent exclusion of the reporter alternative exon (#11, 13 and 14), relative to the nontreated control samples. Interestingly, for the endogenous Bcl-x of the same samples, roscovitine and RA also showed similar effect (#11 and 15), as well as others including nocodazol (an inhibitor of microtubule formation), SB202190 (a p38 MAPK inhibitor) and SP600125 (a JNK inhibitor) (#5, 9 and 12). Therefore, of the multiple chemicals that modulate the Gt16 reporter and/or endogenous Bcl-x splicing, roscovitine and RA induced at least 1.5-fold changes in both the reporter and endogenous Bcl-x.
Figure 7.
The G-tract element also mediates the regulation of splicing by other apoptotic inducers. (A) A bar graph of fold changes (relative to NT sample #1) of percent exon exclusion of DUP175AS-Gt16 and endogenous Bcl-x of HEK293T cells treated with or without (NT) chemicals as indicated. The dotted lines mark the percent exon exclusion level of the NT sample (lower) or the 1.5 fold change level (upper). FdU, 5′-Fluoro-deoxyuridine; NGF, nerve growth factor; ETOH, ethanol (vehicle for #6, 9, 10 and 11); DMSO, vehicle for #5 and #12–15. The final concentrations of the chemicals are (#4–16): 400 mM, 1 µg/ml, 20 µM, 150 µM, 50 ng/ml, 10 µM, 10 µM, 20 µM, 30 µM, 10 µM, 20 µM, 50 µM and 2 µg/ml, respectively. (B) Dosage- and Gt16-dependent effect of RA on the alternative 5′ splice site usage of the DUP175AS-Gt16. Shown are agarose gels of the splicing reporters in the presence of various concentrations of RA as indicated above the gels. Below is a bar graph of the fold changes of percent exon exclusion products relative to lane 1. The dotted line marks the level of the samples without RA. Error bar: standard deviation (n = 3 and 2, for lanes 7, 8 and 9, 10, respectively). **Same as in Figure 4.
To further examine the RA effect, various concentrations of RA were applied to cells expressing the Gt16 reporter or its mutant. The result showed RA dosage-dependent increase of the percent exclusion of the alternative exons in the reporter (Figure 7B, lanes 1–10). Moreover, this regulation was not seen in the Gt16 mutant (lanes 9 and 10). Therefore, the G-tract also mediates RA regulation of splicing.
DISCUSSION
Alternative splicing of genes in cell growth/death has been examined in several cases but the control of these genes by external factors, particularly those inducing cell apoptosis, has just started to be analyzed (36–38,42,43). Our data here support a role for the G-tract element in mediating the regulation of alternative splicing of a group of genes by Ro and other apoptosis-inducers particularly RA.
G-tracts in intron definition and Ro-induced alternative splicing
In the Gt16 element, besides the two G tetrads, there are also two Ts on the two ends and six variable spacer nucleotides (Figures 3 and 4). The spacer is not essential for the silencer activity (Figure 3B, lane 6). Earlier studies with G-tracts indicate that two G triplets with 2–4 nt variable spacer nucleotides regulate alternative splicing (47). Consistently, the G nucleotide mutations (Figure 4, left panel) in the Gt16 are essential for the constitutive as well as Ro-induced repression of Bcl-xL production.
Pre-mRNA sequence regions containing G≥3 elements tend to be skipped as introns accompanying the activation of the upstream 5′ or downstream 3′ splice sites and the G-tract elements are proposed to play a role in intron definition (47,55,70). These elements are enriched close to intron ends in the genome (47,50). It is proposed that binding of these elements by hnRNP proteins may bring two splice sites in closer proximity to stimulate splicing (50). It is also enriched in pseudoexons as splicing silencers (60). Presently, the identified G-tract splicing enhancers are mostly in introns (55–57), except in one case (58), and silencers in exons or pseudo exons (59,60,70,71).
In our experiments, when the Gt16 is put in-between two identical alternative 5′ splice sites, usage of the upstream 5′ splice site was increased (Figure 4, lanes 11 and 13), consistent with similar previous observations for G-tracts and other exonic splicing silencers (47,70). When the G-tract element from Bcl-x is placed in wDUP175, the middle exon is completely excluded (Figure 3). These are consistent with the notion that G-tracts signify the sequence they reside as introns.
These experiments also support the G-tract element as a Ro-responsive RNA element in inducible alternative splicing, as shown by mutations of the G-tract element in Bcl-x and other alternative exons as well as in heterologous contexts (Figures 2, 4 and 6). Ro treatment decreases the inclusion of both cassette and alternative 5′ splice exons that contain G-tract elements. For the competing 5′ splice sites in Bcl-x and the DUP175AS-Gt16 reporter, it remains to be determined whether Ro acts by inhibiting the constitutive splicing factors for the downstream or promoting them for the upstream 5′ splice site.
The effect of the G-tract element on Ro-regulated splicing shows context-dependence. In the Bcl-x and DUP175-Gt16 and DUP175AS-Gt16 reporters, mutation of the Gt16 reduced or abolished the Ro effect (Figure 4); however, mutating the G-tract in the RARG reporter, to the same sequence as in the Bcl-x12 mutant that abolished the Ro effect (data not shown), instead enhanced the Ro effect (Figure 6B). Moreover, inclusion of the middle exon of the RARG mini-gene is repressed in HEK293T cells but its endogenous exon appears enhanced in MDA-231 cells by Ro (Figure 6, and data not shown). These different effects likely reflect the complexity of the control of alternative splicing by multiple elements/factors depending on the different sequence contexts around the regulatory element or on different splicing factors in the cell lines. Similar context-dependent effects on splicing by a regulatory element or a stimulus have also been observed in other cases of alternative splicing (14,68,69,72–75).
From other studies, it is clear that multiple pre-mRNA elements can be present in the same exon to respond to external stimuli. For example, the NMDAR1 exon 21 contains two CaRRE elements and several UAGG motifs that respond to membrane depolarization and CaMK IV (76,77). In addition to the Ro-targeted Gt16 element, there are also several other elements responsive or involved in the regulation of Bcl-x by external stimuli. The TPA- and Ro-regulated splicing is affected by elements in the Bcl-xL downstream intron (37) (Figure 2). The staurosporine-targeted element in Bcl-x lies in a 361-nt fragment 187-nt upstream of the Bcl-xS 5′ splice site. Ceramide requires another two elements CRCE1 and CRCE2 located upstream the Bcl-xS 5′ splice site and downstream the Gt16 element, respectively. The pyrimidine-rich CRCE2 element essential for the ceramide effect was left intact in the wBcl-x14 construct (Figure 2B), but this construct does not respond to Ro, suggesting that the presence of CRCE2 is not sufficient for the Ro effect. In contrast, the G-tract element itself is sufficient to do so (Figure 4), supporting a specific role for the Gt16 in responses to Ro. The relationship among these different elements in responses to the different external stimuli remains to be examined.
Factors between the apoptotic agents and the G-tract element in the inducible alternative splicing
Ro inhibits PKC and the expression of the MAPK phosphatase 1 (23,24). It also activates the JNK protein kinase (24). The PKC stimulator TPA itself did not change the splicing of the Gt16 reporter and only slightly reduced the effect of Ro on the Gt16 reporter in HEK293T cells (Figure 5, lanes 3 and 4). The Ro effect on the reporter is not sensitive to the JNK inhibitor SP600125 in HEK 293T cells (data not shown). In contrast, the Ro effect was inhibited by okadaic acid in a concentration-dependent way (Figure 5). It is thus likely that the Ro-regulation of alternative splicing through the G-tract element involves both PP2A and PP1 (67), protein phosphatases essential for splicing (78–81), and particularly PP1 is required for ceramide-induced Bcl-x splicing (36). Since the phosphatases can modulate a variety of protein kinases including PKC (82), further detailed investigation is necessary to see whether the phosphatases act with PKC or other protein kinases in the Ro-regulated splicing.
In addition to Ro, the G-tract element is also targeted by other apoptotic inducers particularly RA to control splicing (Figure 7). RA targets its nuclear steroid receptor to control gene transcription (83,84). Interestingly, co-activators of nuclear steroid receptors are known to be involved in the control of alternative splicing (85–89).
G-tract elements can directly interact with the U1 snRNA to promote the usage of the upstream 5′ splice site (51). They can also bind several members of the hnRNP H family to regulate splicing (41,48,49,52–55), particularly of the Bcl-x gene. HnRNP H family members are differentially expressed among tissues or in cancer cells (90,91), and hnRNP H1 is phosphorylated in tumor cells upon retinoic acid treatment (92). We also observed hnRNP H1/F binding to the G-tracts of the RARG exon in assays using UV crosslinking-immunoprecipitation with an anti-hnRNP H1/F antibody, and increased hnRNP H1/F phosphorylation by okadaic acid in in vivo 32P-labeling assays (data not shown), implying the involvement of hnRNP H1/F in the Ro-regulated splicing. However, since most of the hnRNP H family members are around 50 kDa, the possibility of the binding by the other members to the G-tract in UV crosslinking cannot be ruled out yet before individual members are examined and followed by loss-of-function studies.
Impact of the regulation of splicing by apoptotic agents on cell growth/death
Several apoptotic agents induce alternative splicing of genes in cell growth/death (36,38). Particularly interesting is that the Bcl-x product is switched by TPA to Bcl-xL (37), which promotes cell growth, and by Ro, ceramide and staurosporine to Bcl-xS (36,38), which promotes cell death. These changes appear consistent with the inducer effects on cell growth/death.
Of the other G-tract element-containing genes that are also responsive to Ro regulation of their exon inclusion (Figure 6), RARG is involved in the retinoic acid signaling and cell death. Taken together with the role of Bcl-x in cell growth/death, it is likely that Ro regulation of these exons will impact on a group of genes involved in cell growth/death.
The effect of the alternatively spliced variants on gene function is exemplified in the RARG gene. The G-tract-containing alternative exon of RARG encodes a segment of the NH2 terminal of the receptor. This region is phosphorylated by the JNK and CDK7 kinases in RARG or its family members (93–97), and is critical for transcriptional activities (83). Therefore, changing the inclusion level of this exon by Ro probably affects the phosphorylation and transcriptional activities of RARG.
Another potential target of G-tract-mediated regulation is deltap53, a splice variant of p53 with one exon skipped (98). Deltap53, but not p53, trans-activates the Cdk inhibitor p21 in S-phase attenuation (98). Interestingly, the alternative exon contains three G-tracts.
Taken together with the functions of the G-tract-containing exons and the apoptotic agents on cell death, it is likely that the G-tract elements play a role in controlling the generation of cell death-promoting products (e.g. Bcl-xS) through alternative splicing. More intensive analysis with whole-genome microarrays will allow us to see whether these changes are part of the program contributing to cell apoptosis induced by these agents.
ACKNOWLEDGEMENTS
We thank Robert Shiu and the CancerCare Manitoba Foundation for the Ingenuity service, and Robert Shiu and Etienne Leygue for MDA-231 and BT20 cells and helpful discussions. This study was funded by the National Cancer Institute of Canada (#016355 to J.X.). J.X. is a CIHR New Investigator and recipient of MHRC and CFI supports. Funding to pay the Open Access publication charges for this article was provided by the National Cancer Institute of Canada.
Conflict of interest statement. None declared.
REFERENCES
- 1.Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 2.Black DL. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell. 2000;103:367–370. doi: 10.1016/s0092-8674(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 3.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;27:27. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 4.Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 5.Jiang ZH, Zhang WJ, Rao Y, Wu JY. Regulation of Ich-1 pre-mRNA alternative splicing and apoptosis by mammalian splicing factors. Proc. Natl Acad. Sci. USA. 1998;95:9155–9160. doi: 10.1073/pnas.95.16.9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu JY, Tang H, Havlioglu N. Alternative pre-mRNA splicing and regulation of programmed cell death. Prog. Mol. Subcell. Biol. 2003;31:153–185. doi: 10.1007/978-3-662-09728-1_6. [DOI] [PubMed] [Google Scholar]
- 7.Roberts GC, Smith CW. Alternative splicing: combinatorial output from the genome. Curr. Opin. Chem. Biol. 2002;6:375–383. doi: 10.1016/s1367-5931(02)00320-4. [DOI] [PubMed] [Google Scholar]
- 8.Fu XD. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–580. [PMC free article] [PubMed] [Google Scholar]
- 9.Sanford JR, Longman D, Caceres JF. Multiple roles of the SR protein family in splicing regulation. Prog. Mol. Subcell. Biol. 2003;31:33–58. doi: 10.1007/978-3-662-09728-1_2. [DOI] [PubMed] [Google Scholar]
- 10.Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chabot B, LeBel C, Hutchison S, Nasim FH, Simard MJ. Heterogeneous nuclear ribonucleoprotein particle A/B proteins and the control of alternative splicing of the mammalian heterogeneous nuclear ribonucleoprotein particle A1 pre-mRNA. Prog. Mol. Subcell. Biol. 2003;31:59–88. doi: 10.1007/978-3-662-09728-1_3. [DOI] [PubMed] [Google Scholar]
- 12.Wagner EJ, Garcia-Blanco MA. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 2001;21:3281–3288. doi: 10.1128/MCB.21.10.3281-3288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ladd AN, Charlet N, Cooper TA. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol. Cell. Biol. 2001;21:1285–1296. doi: 10.1128/MCB.21.4.1285-1296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukumura K, Kato A, Jin Y, Ideue T, Hirose T, Kataoka N, Fujiwara T, Sakamoto H, Inoue K. Tissue-specific splicing regulator Fox-1 induces exon skipping by interfering E complex formation on the downstream intron of human F1gamma gene. Nucleic Acids Res. 2007;35:5303–5311. doi: 10.1093/nar/gkm569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou HL, Baraniak AP, Lou H. Role for Fox-1/Fox-2 in mediating the neuronal pathway of calcitonin/calcitonin gene-related peptide alternative RNA processing. Mol. Cell. Biol. 2007;27:830–841. doi: 10.1128/MCB.01015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stamm S. Regulation of alternative splicing by reversible protein phosphorylation. J. Biol. Chem. 2008;283:1223–1227. doi: 10.1074/jbc.R700034200. [DOI] [PubMed] [Google Scholar]
- 17.Shin C, Manley JL. Cell signalling and the control of pre-mRNA splicing. Nat. Rev. Mol. Cell. Biol. 2004;5:727–738. doi: 10.1038/nrm1467. [DOI] [PubMed] [Google Scholar]
- 18.Stamm S. Signals and their transduction pathways regulating alternative splicing: a new dimension of the human genome. Hum. Mol. Genet. 2002;11:2409–2416. doi: 10.1093/hmg/11.20.2409. [DOI] [PubMed] [Google Scholar]
- 19.Konig H, Ponta H, Herrlich P. Coupling of signal transduction to alternative pre-mRNA splicing by a composite splice regulator. EMBO J. 1998;17:2904–2913. doi: 10.1093/emboj/17.10.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matter N, Marx M, Weg-Remers S, Ponta H, Herrlich P, Konig H. Heterogeneous ribonucleoprotein A1 is part of an exon-specific splice-silencing complex controlled by oncogenic signaling pathways. J. Biol. Chem. 2000;275:35353–35360. doi: 10.1074/jbc.M004692200. [DOI] [PubMed] [Google Scholar]
- 21.Matter N, Herrlich P, Konig H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 2002;420:691–695. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- 22.Goekjian PG, Jirousek MR. Protein kinase C inhibitors as novel anticancer drugs. Expert Opin. Invest. Drugs. 2001;10:2117–2140. doi: 10.1517/13543784.10.12.2117. [DOI] [PubMed] [Google Scholar]
- 23.Dieter P, Fitzke E. RO 31-8220 and RO 31-7549 show improved selectivity for protein kinase C over staurosporine in macrophages. Biochem. Biophys. Res. Commun. 1991;181:396–401. doi: 10.1016/s0006-291x(05)81432-9. [DOI] [PubMed] [Google Scholar]
- 24.Beltman J, McCormick F, Cook SJ. The selective protein kinase C inhibitor, Ro-31-8220, inhibits mitogen-activated protein kinase phosphatase-1 (MKP-1) expression, induces c-Jun expression, and activates Jun N-terminal kinase. J. Biol. Chem. 1996;271:27018–27024. doi: 10.1074/jbc.271.43.27018. [DOI] [PubMed] [Google Scholar]
- 25.Han Z, Pantazis P, Lange TS, Wyche JH, Hendrickson EA. The staurosporine analog, Ro-31-8220, induces apoptosis independently of its ability to inhibit protein kinase C. Cell Death Differ. 2000;7:521–530. doi: 10.1038/sj.cdd.4400681. [DOI] [PubMed] [Google Scholar]
- 26.Zhu GH, Wong BC, Eggo MC, Yuen ST, Lai KC, Lam SK. Pharmacological inhibition of protein kinase C activity could induce apoptosis in gastric cancer cells by differential regulation of apoptosis-related genes. Dig. Dis. Sci. 1999;44:2020–2026. doi: 10.1023/a:1026670301787. [DOI] [PubMed] [Google Scholar]
- 27.Motoyama N, Wang F, Roth KA, Sawa H, Nakayama K, Negishi I, Senju S, Zhang Q, Fujii S, Loh DY. Massive cell death of immature hematopoietic cells and neurons in Bcl-x- deficient mice. Science. 1995;267:1506–1510. doi: 10.1126/science.7878471. [DOI] [PubMed] [Google Scholar]
- 28.Borner C. The Bcl-2 protein family: sensors and checkpoints for life-or-death decisions. Mol. Immunol. 2003;39:615–647. doi: 10.1016/s0161-5890(02)00252-3. [DOI] [PubMed] [Google Scholar]
- 29.Boise LH, Gonzalez-Garcia M, Postema CE, Ding L, Lindsten T, Turka LA, Mao X, Nunez G, Thompson CB. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 30.Gonzalez-Garcia M, Garcia I, Ding L, O'Shea S, Boise LH, Thompson CB, Nunez G. bcl-x is expressed in embryonic and postnatal neural tissues and functions to prevent neuronal cell death. Proc. Natl Acad. Sci. USA. 1995;92:4304–4308. doi: 10.1073/pnas.92.10.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reed JC. Splicing and dicing apoptosis genes. Nat. Biotechnol. 1999;17:1064–1065. doi: 10.1038/15048. [DOI] [PubMed] [Google Scholar]
- 32.Mercatante DR, Bortner CD, Cidlowski JA, Kole R. Modification of alternative splicing of Bcl-x pre-mRNA in prostate and breast cancer cells. Analysis of apoptosis and cell death. J. Biol. Chem. 2001;276:16411–16417. doi: 10.1074/jbc.M009256200. [DOI] [PubMed] [Google Scholar]
- 33.Taylor JK, Zhang QQ, Wyatt JR, Dean NM. Induction of endogenous Bcl-xS through the control of Bcl-x pre-mRNA splicing by antisense oligonucleotides. Nat. Biotechnol. 1999;17:1097–1100. doi: 10.1038/15079. [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez-Garcia M, Perez-Ballestero R, Ding L, Duan L, Boise LH, Thompson CB, Nunez G. bcl-XL is the major bcl-x mRNA form expressed during murine development and its product localizes to mitochondria. Development. 1994;120:3033–3042. doi: 10.1242/dev.120.10.3033. [DOI] [PubMed] [Google Scholar]
- 35.Xerri L, Parc P, Brousset P, Schlaifer D, Hassoun J, Reed JC, Krajewski S, Birnbaum D. Predominant expression of the long isoform of Bcl-x (Bcl-xL) in human lymphomas. Br. J. Haematol. 1996;92:900–906. doi: 10.1046/j.1365-2141.1996.423958.x. [DOI] [PubMed] [Google Scholar]
- 36.Chalfant CE, Rathman K, Pinkerman RL, Wood RE, Obeid LM, Ogretmen B, Hannun YA. De novo ceramide regulates the alternative splicing of caspase 9 and Bcl-x in A549 lung adenocarcinoma cells. Dependence on protein phosphatase-1. J. Biol. Chem. 2002;277:12587–12595. doi: 10.1074/jbc.M112010200. [DOI] [PubMed] [Google Scholar]
- 37.Li CY, Chu JY, Yu JK, Huang XQ, Liu XJ, Shi L, Che YC, Xie JY. Regulation of alternative splicing of Bcl-x by IL-6, GM-CSF and TPA. Cell Res. 2004;14:473–479. doi: 10.1038/sj.cr.7290250. [DOI] [PubMed] [Google Scholar]
- 38.Revil T, Toutant J, Shkreta L, Garneau D, Cloutier P, Chabot B. Protein kinase C-dependent control of Bcl-x alternative splicing. Mol. Cell. Biol. 2007;27:8431–8441. doi: 10.1128/MCB.00565-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villemaire J, Dion I, Elela SA, Chabot B. Reprogramming alternative pre-messenger RNA splicing through the use of protein-binding antisense oligonucleotides. J. Biol. Chem. 2003;278:50031–50039. doi: 10.1074/jbc.M308897200. [DOI] [PubMed] [Google Scholar]
- 40.Wilusz JE, Devanney SC, Caputi M. Chimeric peptide nucleic acid compounds modulate splicing of the bcl-x gene in vitro and in vivo. Nucleic Acids Res. 2005;33:6547–6554. doi: 10.1093/nar/gki960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garneau D, Revil T, Fisette JF, Chabot B. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J. Biol. Chem. 2005;280:22641–22650. doi: 10.1074/jbc.M501070200. [DOI] [PubMed] [Google Scholar]
- 42.Massiello A, Roesser JR, Chalfant CE. SAP155 Binds to ceramide-responsive RNA cis-element 1 and regulates the alternative 5′ splice site selection of Bcl-x pre-mRNA. FASEB J. 2006;20:1680–1682. doi: 10.1096/fj.05-5021fje. [DOI] [PubMed] [Google Scholar]
- 43.Massiello A, Salas A, Pinkerman RL, Roddy P, Roesser JR, Chalfant CE. Identification of two RNA cis-elements that function to regulate the 5′ splice site selection of Bcl-x pre-mRNA in response to ceramide. J. Biol. Chem. 2004;279:15799–15804. doi: 10.1074/jbc.M313950200. [DOI] [PubMed] [Google Scholar]
- 44.Paronetto MP, Achsel T, Massiello A, Chalfant CE, Sette C. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J. Cell. Biol. 2007;176:929–939. doi: 10.1083/jcb.200701005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Modafferi EF, Black DL. Combinatorial control of a neuron-specific exon. RNA. 1999;5:687–706. doi: 10.1017/s1355838299990155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 47.McCullough AJ, Berget SM. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol. Cell. Biol. 1997;17:4562–4571. doi: 10.1128/mcb.17.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dominguez C, Allain FH. NMR structure of the three quasi RNA recognition motifs (qRRMs) of human hnRNP F and interaction studies with Bcl-x G-tract RNA: a novel mode of RNA recognition. Nucleic Acids Res. 2006;34:3634–3645. doi: 10.1093/nar/gkl488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaub MC, Lopez SR, Caputi M. Members of the heterogeneous nuclear ribonucleoprotein H family activate splicing of an HIV-1 splicing substrate by promoting formation of ATP-dependent spliceosomal complexes. J. Biol. Chem. 2007;282:13617–13626. doi: 10.1074/jbc.M700774200. [DOI] [PubMed] [Google Scholar]
- 50.Martinez-Contreras R, Fisette JF, Nasim FU, Madden R, Cordeau M, Chabot B. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol. 2006;4:e21. doi: 10.1371/journal.pbio.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCullough AJ, Berget SM. An intronic splicing enhancer binds U1 snRNPs to enhance splicing and select 5′ splice sites. Mol. Cell. Biol. 2000;20:9225–9235. doi: 10.1128/mcb.20.24.9225-9235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mahe D, Mahl P, Gattoni R, Fischer N, Mattei MG, Stevenin J, Fuchs JP. Cloning of human 2H9 heterogeneous nuclear ribonucleoproteins. Relation with splicing and early heat shock-induced splicing arrest. J. Biol. Chem. 1997;272:1827–1836. doi: 10.1074/jbc.272.3.1827. [DOI] [PubMed] [Google Scholar]
- 53.Honore B, Rasmussen HH, Vorum H, Dejgaard K, Liu X, Gromov P, Madsen P, Gesser B, Tommerup N, Celis JE. Heterogeneous nuclear ribonucleoproteins H, H', and F are members of a ubiquitously expressed subfamily of related but distinct proteins encoded by genes mapping to different chromosomes. J. Biol. Chem. 1995;270:28780–28789. doi: 10.1074/jbc.270.48.28780. [DOI] [PubMed] [Google Scholar]
- 54.Caputi M, Zahler AM. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H'/F/2H9 family. J. Biol. Chem. 2001;276:43850–43859. doi: 10.1074/jbc.M102861200. [DOI] [PubMed] [Google Scholar]
- 55.Wang E, Dimova N, Cambi F. PLP/DM20 ratio is regulated by hnRNPH and F and a novel G-rich enhancer in oligodendrocytes. Nucleic Acids Res. 2007;35:4164–4178. doi: 10.1093/nar/gkm387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chou MY, Rooke N, Turck CW, Black DL. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell. Biol. 1999;19:69–77. doi: 10.1128/mcb.19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han K, Yeo G, An P, Burge CB, Grabowski PJ. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 2005;3:e158. doi: 10.1371/journal.pbio.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caputi M, Zahler AM. SR proteins and hnRNP H regulate the splicing of the HIV-1 tev-specific exon 6D. EMBO J. 2002;21:845–855. doi: 10.1093/emboj/21.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen CD, Kobayashi R, Helfman DM. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat beta-tropomyosin gene. Genes Dev. 1999;13:593–606. doi: 10.1101/gad.13.5.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sironi M, Menozzi G, Riva L, Cagliani R, Comi GP, Bresolin N, Giorda R, Pozzoli U. Silencer elements as possible inhibitors of pseudoexon splicing. Nucleic Acids Res. 2004;32:1783–1791. doi: 10.1093/nar/gkh341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie J, Black DL. A CaMK IV responsive RNA element mediates depolarization-induced alternative splicing of ion channels. Nature. 2001;410:936–939. doi: 10.1038/35073593. [DOI] [PubMed] [Google Scholar]
- 62.Dominski Z, Kole R. Selection of splice sites in pre-mRNAs with short internal exons. Mol. Cell. Biol. 1991;11:6075–6083. doi: 10.1128/mcb.11.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klinck R, Bramard A, Inkel L, Dufresne-Martin G, Gervais-Bird J, Madden R, Paquet ER, Koh C, Venables JP, Prinos P, et al. Multiple alternative splicing markers for ovarian cancer. Cancer Res. 2008;68:657–663. doi: 10.1158/0008-5472.CAN-07-2580. [DOI] [PubMed] [Google Scholar]
- 64.Treisman R, Orkin SH, Maniatis T. Specific transcription and RNA splicing defects in five cloned beta- thalassaemia genes. Nature. 1983;302:591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- 65.Krainer AR, Conway GC, Kozak D. The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell. 1990;62:35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- 66.Castagna M, Takai Y, Kaibuchi K, Sano K, Kikkawa U, Nishizuka Y. Direct activation of calcium-activated, phospholipid-dependent protein kinase by tumor-promoting phorbol esters. J. Biol. Chem. 1982;257:7847–7851. [PubMed] [Google Scholar]
- 67.Bialojan C, Takai A. Inhibitory effect of a marine-sponge toxin, okadaic acid, on protein phosphatases. Specificity and kinetics. Biochem. J. 1988;256:283–290. doi: 10.1042/bj2560283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 69.Hui J, Hung LH, Heiner M, Schreiner S, Neumuller N, Reither G, Haas SA, Bindereif A. Intronic CA-repeat and CA-rich elements: a new class of regulators of mammalian alternative splicing. EMBO J. 2005;24:1988–1998. doi: 10.1038/sj.emboj.7600677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Z, Xiao X, Van Nostrand E, Burge CB. General and specific functions of exonic splicing silencers in splicing control. Mol. Cell. 2006;23:61–70. doi: 10.1016/j.molcel.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jacquenet S, Mereau A, Bilodeau PS, Damier L, Stoltzfus CM, Branlant C. A second exon splicing silencer within human immunodeficiency virus type 1 tat exon 2 represses splicing of Tat mRNA and binds protein hnRNP H. J. Biol. Chem. 2001;276:40464–40475. doi: 10.1074/jbc.M104070200. [DOI] [PubMed] [Google Scholar]
- 72.Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol. Cell. Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jin Y, Suzuki H, Maegawa S, Endo H, Sugano S, Hashimoto K, Yasuda K, Inoue K. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goren A, Ram O, Amit M, Keren H, Lev-Maor G, Vig I, Pupko T, Ast G. Comparative analysis identifies exonic splicing regulatory sequences–The complex definition of enhancers and silencers. Mol. Cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 75.Xie JY. Control of alternative pre-mRNA splicing by Ca++ signals. Biochim. Biophys. Acta-Gene Regul. Mech. 2008 doi: 10.1016/j.bbagrm.2008.01.003. [Epub ahead of print, doi:10.1016/j.bbagrm.2008.01.003], January 17, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee JA, Xing Y, Nguyen D, Xie J, Lee CJ, Black DL. Depolarization and CaM kinase IV modulate NMDA receptor splicing through two essential RNA elements. PLoS Biol. 2007;5:e40. doi: 10.1371/journal.pbio.0050040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.An P, Grabowski PJ. Exon silencing by UAGG motifs in response to neuronal excitation. PLoS Biol. 2007;5:e36. doi: 10.1371/journal.pbio.0050036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Murray MV, Kobayashi R, Krainer AR. The type 2C Ser/Thr phosphatase PP2Cgamma is a pre-mRNA splicing factor. Genes Dev. 1999;13:87–97. doi: 10.1101/gad.13.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shi Y, Reddy B, Manley JL. PP1/PP2A phosphatases are required for the second step of Pre-mRNA splicing and target specific snRNP proteins. Mol. Cell. 2006;23:819–829. doi: 10.1016/j.molcel.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 80.Novoyatleva T, Heinrich B, Tang Y, Benderska N, Butchbach ME, Lorson CL, Lorson MA, Ben-Dov C, Fehlbaum P, Bracco L, et al. Protein phosphatase 1 binds to the RNA recognition motif of several splicing factors and regulates alternative pre-mRNA processing. Hum. Mol. Genet. 2008;17:52–70. doi: 10.1093/hmg/ddm284. [DOI] [PubMed] [Google Scholar]
- 81.Shi Y, Manley JL. A complex signaling pathway regulates SRp38 phosphorylation and pre-mRNA splicing in response to heat shock. Mol. Cell. 2007;28:79–90. doi: 10.1016/j.molcel.2007.08.028. [DOI] [PubMed] [Google Scholar]
- 82.Boudreau RT, Garduno R, Lin TJ. Protein phosphatase 2A and protein kinase Calpha are physically associated and are involved in Pseudomonas aeruginosa-induced interleukin 6 production by mast cells. J. Biol. Chem. 2002;277:5322–5329. doi: 10.1074/jbc.M108623200. [DOI] [PubMed] [Google Scholar]
- 83.Bour G, Lalevee S, Rochette-Egly C. Protein kinases and the proteasome join in the combinatorial control of transcription by nuclear retinoic acid receptors. Trends Cell. Biol. 2007;17:302–309. doi: 10.1016/j.tcb.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 84.Germain P, Chambon P, Eichele G, Evans RM, Lazar MA, Leid M, De Lera AR, Lotan R, Mangelsdorf DJ, Gronemeyer H. International Union of Pharmacology. LXIII. Retinoid X receptors. Pharmacol. Rev. 2006;58:760–772. doi: 10.1124/pr.58.4.7. [DOI] [PubMed] [Google Scholar]
- 85.Loyer P, Trembley JH, Katona R, Kidd VJ, Lahti JM. Role of CDK/cyclin complexes in transcription and RNA splicing. Cell Signal. 2005;17:1033–1051. doi: 10.1016/j.cellsig.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 86.Auboeuf D, Dowhan DH, Kang YK, Larkin K, Lee JW, Berget SM, O’Malley BW. Differential recruitment of nuclear receptor coactivators may determine alternative RNA splice site choice in target genes. Proc. Natl Acad. Sci. USA. 2004;101:2270–2274. doi: 10.1073/pnas.0308133100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Auboeuf D, Dowhan DH, Li X, Larkin K, Ko L, Berget SM, O’Malley BW. CoAA, a nuclear receptor coactivator protein at the interface of transcriptional coactivation and RNA splicing. Mol. Cell. Biol. 2004;24:442–453. doi: 10.1128/MCB.24.1.442-453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dowhan DH, Hong EP, Auboeuf D, Dennis AP, Wilson MM, Berget SM, O’Malley BW. Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERalpha and CAPERbeta. Mol. Cell. 2005;17:429–439. doi: 10.1016/j.molcel.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 89.Auboeuf D, Honig A, Berget SM, O’Malley BW. Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science. 2002;298:416–419. doi: 10.1126/science.1073734. [DOI] [PubMed] [Google Scholar]
- 90.Honore B, Vorum H, Baandrup U. hnRNPs H, H′ and F behave differently with respect to posttranslational cleavage and subcellular localization. FEBS Lett. 1999;456:274–280. doi: 10.1016/s0014-5793(99)00911-4. [DOI] [PubMed] [Google Scholar]
- 91.Honore B, Baandrup U, Vorum H. Heterogeneous nuclear ribonucleoproteins F and H/H′ show differential expression in normal and selected cancer tissues. Exp. Cell Res. 2004;294:199–209. doi: 10.1016/j.yexcr.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 92.Navakauskiene R, Treigyte G, Gineitis A, Magnusson KE. Identification of apoptotic tyrosine-phosphorylated proteins after etoposide or retinoic acid treatment. Proteomics. 2004;4:1029–1041. doi: 10.1002/pmic.200300671. [DOI] [PubMed] [Google Scholar]
- 93.Adam-Stitah S, Penna L, Chambon P, Rochette-Egly C. Hyperphosphorylation of the retinoid X receptor alpha by activated c-Jun NH2-terminal kinases. J. Biol. Chem. 1999;274:18932–18941. doi: 10.1074/jbc.274.27.18932. [DOI] [PubMed] [Google Scholar]
- 94.Rochette-Egly C, Lutz Y, Saunders M, Scheuer I, Gaub MP, Chambon P. Retinoic acid receptor gamma: specific immunodetection and phosphorylation. J. Cell. Biol. 1991;115:535–545. doi: 10.1083/jcb.115.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rochette-Egly C, Adam S, Rossignol M, Egly JM, Chambon P. Stimulation of RAR alpha activation function AF-1 through binding to the general transcription factor TFIIH and phosphorylation by CDK7. Cell. 1997;90:97–107. doi: 10.1016/s0092-8674(00)80317-7. [DOI] [PubMed] [Google Scholar]
- 96.Bour G, Gaillard E, Bruck N, Lalevee S, Plassat JL, Busso D, Samama JP, Rochette-Egly C. Cyclin H binding to the RARalpha activation function (AF)-2 domain directs phosphorylation of the AF-1 domain by cyclin-dependent kinase 7. Proc. Natl Acad. Sci. USA. 2005;102:16608–16613. doi: 10.1073/pnas.0505556102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bastien J, Adam-Stitah S, Riedl T, Egly JM, Chambon P, Rochette-Egly C. TFIIH interacts with the retinoic acid receptor gamma and phosphorylates its AF-1-activating domain through cdk7. J. Biol. Chem. 2000;275:21896–21904. doi: 10.1074/jbc.M001985200. [DOI] [PubMed] [Google Scholar]
- 98.Rohaly G, Chemnitz J, Dehde S, Nunez AM, Heukeshoven J, Deppert W, Dornreiter I. A novel human p53 isoform is an essential element of the ATR-intra-S phase checkpoint. Cell. 2005;122:21–32. doi: 10.1016/j.cell.2005.04.032. [DOI] [PubMed] [Google Scholar]