Abstract
Spemann’s organizer develops in response to dorsal determinants that act via maternal components of the wnt pathway. The function of siamois, a wnt-inducible homeobox gene, in Spemann’s organizer development was examined by fusion of defined transcriptional regulatory domains to the siamois homeodomain. Similar to native siamois, a VP16 activator fusion induced axis formation, indicating that siamois functions as a transcriptional activator in axis induction. Fusion of the engrailed repressor generated a dominant inhibitor that blocked axis induction by Xwnt8, β-catenin, and siamois, and repressed wnt activation of the goosecoid promoter. Dorsal injection of the engrailed-siamois fusion resulted in complete inhibition of dorsal development and organizer gene expression, an effect rescued by siamois, but not by Xwnt8 or β-catenin. Thus, as a zygotic mediator of maternal dorsal signals, siamois function is required for development of Spemann’s organizer.
In Xenopus, maternal factors establish dorsoventral pattern in the cleavage embryo, resulting in formation of Spemann’s organizer at the gastrula stage (reviewed in ref. 1). Maternal dorsal determinants, localized to the vegetal pole at fertilization, are displaced by cortical rotation to the future dorsal domain of the cleavage embryo (2–6), a region defined functionally as the Nieuwkoop center. An early response to these determinants is the nuclear accumulation, in dorsal blastomeres, of β-catenin, a component of the wnt pathway required for dorsal development (7–10). These observations suggest that stimulation of a maternal wnt pathway upstream of, or at, β-catenin results in dorsal development. While the identified components of the wnt pathway are maternally expressed (11), transcriptional targets that respond to maternal signals and are zygotic effectors of dorsal development are not yet defined. A strong candidate for a zygotic effector of maternal dorsal signals is the wnt-inducible factor siamois (12).
The homeobox gene siamois was isolated in a functional screen for factors with axis-inducing activity (12). Siamois is expressed in dorsal blastomeres at the mid-blastula transition, and ventral injection of siamois mRNA results in complete axial duplications. In contrast to other zygotic axis-inducers, siamois is induced by components of the wnt signaling pathway (Xwnt8, frizzled, dishevelled, dominant-negative GSK3β, APC, and β-catenin), but not by other factors regulating axial or mesodermal development (13–17). The induction of siamois by wnt signaling is unaffected by cycloheximide (unpublished data), consistent with a direct transcriptional activation, possibly mediated by a nuclear complex of β-catenin and LEF-1/XTcf-3 (18–20). In animal explants, siamois activates expression of organizer-specific genes, in the absence of mesodermal gene expression or differentiation (13, 16). The results suggest a role for siamois in organizer formation, whereas other zygotic factors such as noggin, chordin, and goosecoid are likely to play a role in organizer function.
In this paper, I examine the transcriptional activity of siamois resulting in axis induction and, using a dominant inhibitory form of siamois, determine the requirement for siamois activity in endogenous axis formation and in the response to described axis inducers. The results indicate that siamois functions as a transcriptional activator to induce organizer gene expression and axis formation, and that inhibition of endogenous siamois fully blocks formation of Spemann’s organizer, axial development, and the inducing activity of the wnt signaling pathway. Furthermore, siamois activates a wnt-responsive element of the goosecoid promoter, suggesting that siamois may coordinately regulate organizer gene expression. Therefore, as a zygotic mediator of maternal wnt signaling, siamois is essential for formation of Spemann’s organizer and subsequent axial development.
MATERIALS AND METHODS
Embryos and Microinjection.
Embryos were collected, fertilized, injected, and cultured as previously described (21), and embryonic stage was determined according to Nieuwkoop and Faber (22). Dorsal and ventral blastomeres were identified by pigmentation differences (23). Capped, in vitro transcribed RNA was synthesized by using a Megascript kit (Ambion, Austin, TX) programmed with linearized DNA template, and 5–10 nl of RNA solution was injected.
Construction of Siamois Fusions and Mutagenesis.
Siamois fusions were constructed from individual domains produced by PCR amplification for the siamois homeodomain and VP16 activator or restriction fragment isolation for the engrailed repressor (Fig. 1A). Ligation products were subcloned into pCS2+ (24). Sequencing and in vitro translation were used to verify the constructs. For engrailed-siamois, the engrailed initiator methionine is used, and for VP16-siamois, PCR amplification resulted in incorporation of methionine at the N terminus of the VP16 activator. In addition, the siamois coding region was amplified by PCR and subcloned into pCS2+. Mutagenesis of siamois and engrailed-siamois was performed with the QuikChange site-directed mutagenesis kit (Stratagene), using oligonucleotides complementary to bases 572–598 of siamois with a single mismatch at position 584 (C → A for the lysine mutant and C → G for the glutamate mutant). Sequencing and in vitro translation were used to verify the mutants.
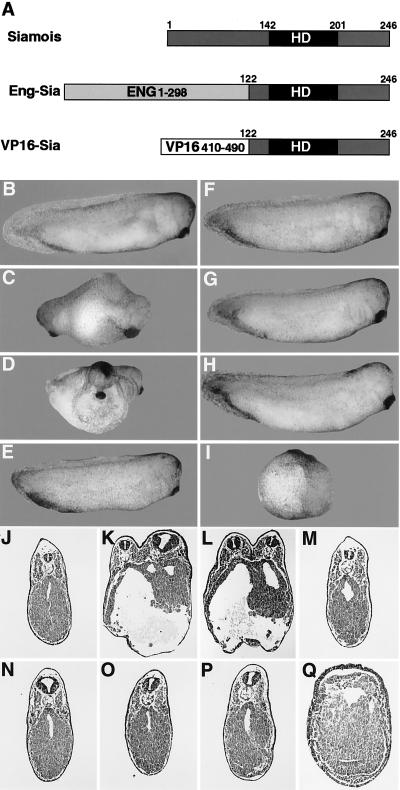
Figure 1.
Axis formation is affected by siamois fusions. (A) Schematic of the siamois fusion constructs. A C-terminal region of siamois (residues 122–246), containing the homeodomain (HD), was fused to the engrailed repressor (residues 1–298) (Eng-Sia) or the VP16 activator (residues 410–490) (VP16-Sia). At the four-cell stage, one ventral (B–E, J–M) or two dorsal (F–I, N–Q) blastomeres were injected with 30 pg of β-galactosidase (B, F, J, N), siamois (C, G, K, O), VP16-Sia (D, H, L, P), or Eng-Sia (E, I, M, Q) mRNA. (J–Q) Transverse sections. See Table 1 for quantitation.
Histology, in Situ Hybridization, and Reverse Transcription (RT)–PCR Assay.
For histology, samples were fixed in MEMFA (25) and processed for Paraplast sectioning and hematoxylin/eosin staining. For in situ hybridization, gastrula stage embryos were fixed in MEMFA and probed with an antisense, digoxigenin-labeled RNA as previously described (25). For the RT-PCR assay, RNA extraction, cDNA synthesis, and gel electrophoresis were as previously described (26). PCR conditions and primers for EF1α, brachyury, Xwnt8, noggin, chordin, and cerberus were previously described (26–28). Primers for Xnr3 were U-ATCCAACTAACTACATCG and D-TAGTGGGACAAGAAGTGC (28 cycles; U and D indicating upstream and downstream) and for siamois were U-AACTTTCTCCAGAACC and D-GTCAGTGTGGTGATTC (30 cycles). The siamois primers detect endogenous siamois but not injected RNAs.
Luciferase Assays.
Injected embryos were harvested at the gastrula stage and assayed with the Luciferase Assay System (Promega). Duplicate samples of 10 embryos were lysed in 100 μl of reporter lysis solution and 20 μl of cleared lysate was combined with 100 μl of luciferase assay substrate. Light output was measured for 15 sec after a 3-sec delay by using a Turner Designs (Sunnyvale, CA) luminometer.
RESULTS
Siamois is composed of a C-terminal paired-type homeodomain and N-terminal sequences unrelated to previously described transcriptional regulatory domains, so it is unclear whether siamois functions as a transcriptional activator or repressor. To define the transcriptional activity of siamois that results in axis induction, well characterized regulatory domains, the herpes simplex virus VP16 activator (29, 30) or the Drosophila engrailed repressor (31–33) was fused to the siamois homeodomain, and the axis-inducing activity of the fusion proteins was determined (Fig. 1A). At the four-cell stage, a single ventral blastomere was injected with mRNA encoding siamois, the VP16-siamois fusion (VP16-Sia), or the engrailed-siamois fusion (Eng-Sia), and axial development was assessed at the tailbud stage (Fig. 1 B–E). Ventral injection of VP16-Sia induced complete axial duplication at a frequency similar to siamois, and Eng-Sia did not induce axis formation (Fig. 1 J–M and Table 1). This result indicates that siamois functions as a transcriptional activator in inducing axial development.
Table 1.
Effects of siamois fusions on axial development
| mRNA injected | Axis induction
|
Axis inhibition
|
||
|---|---|---|---|---|
| n/N | % | n/N | % | |
| β-Galactosidase | 0/63 | 0 | 0/35 | 0 |
| Siamois | 52/71 | 72 | 0/41 | 0 |
| VP16-Sia | 49/62 | 79 | 0/40 | 0 |
| Eng-Sia | 0/46 | 0 | 35/38 | 92 |
| Sia-HD | 0/22 | 0 | 0/25 | 0 |
| SiaΔHD | 0/26 | 0 | 0/20 | 0 |
| VP16 | 0/21 | 0 | 0/20 | 0 |
| Eng | 0/31 | 0 | 0/49 | 0 |
At the four-cell stage, a single ventral blastomere (axis induction columns) or both dorsal blastomeres (axis inhibition columns) were injected with 30 pg of mRNA encoding siamois, VP16-Sia, or Eng-Sia or 100 pg of mRNA encoding β-galactosidase, the siamois homeodomain (Sia-HD), siamois with a deletion of the homeodomain (SiaΔHD), the VP16 activator domain (VP16), or the engrailed repressor domain (Eng). Embryos were scored for axis formation at the tailbud stage. n, Injected embryos with secondary axis formation (axis induction) or reduction of axis formation resulting in dorsoanterior index (DAI) ≤ 2 (axis inhibition); N, total number of injected embryos; %, percent affected embryos. The data presented are representative of more than three experiments.
The demonstration that siamois functions as a transcriptional activator suggested the possibility that Eng-Sia may antagonize endogenous siamois by targeting an active repressor to siamois-binding sites. To assess the potential dominant inhibitory activity of Eng-Sia, both dorsal blastomeres of four-cell stage embryos were injected with siamois, VP16-Sia, or Eng-Sia (Fig. 1 F–I). Dorsal injection of Eng-Sia resulted in a severe or complete loss of axial development. Greater than 90% of injected embryos displayed a reduction or loss of anterior and axial structures, resulting in the absence of somitic muscle, notochord, and neural tube. Eng-Sia-injected embryos do initiate and complete gastrulation, similar to ventralization observed in response to UV irradiation, suggesting that axis inhibition is not due to nonspecific gastrulation defects. In contrast, dorsal injection of siamois or VP16-Sia had no effect on axis formation (Fig. 1 N–Q and Table 1). Complete inhibition of axial development was obtained with doses of Eng-Sia similar to those required for axial duplication by siamois or VP16-Sia. The reciprocal response to VP16-Sia and Eng-Sia suggests that the activator function of siamois is required for axial development and that Eng-Sia inhibits axis formation by repressing transcriptional targets of endogenous siamois. These results were obtained by mRNA injection, providing a maternal-type expression, not the zygotic expression characteristic of endogenous siamois. Zygotic expression of siamois, VP16-Sia, and Eng-Sia, by injection of expression plasmids, resulted in effects on axis formation identical to those observed with mRNA injection (data not shown). In control experiments, embryos were injected with the individual protein domains used to construct the fusions, including the VP16 activator, the engrailed repressor, and the siamois homeodomain, as well as siamois lacking the homeodomain. In no case did these individual domains alter axial development (Table 1).
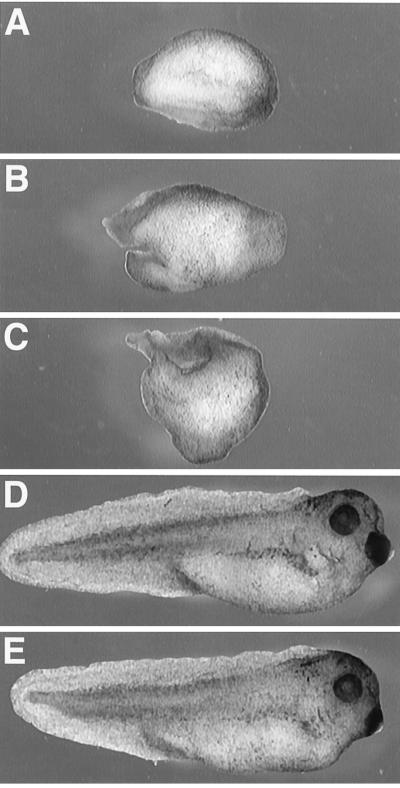
The inhibitory action of Eng-Sia is predicted to result from a specific block of endogenous siamois function. However, it is possible that the siamois homeodomain, as part of an overexpressed fusion protein, may bind targets not normally regulated by siamois and inhibit additional transcriptional regulators, particularly other homeodomain proteins. To assess the specificity of axis inhibition by Eng-Sia, the ability of Xwnt8 (34, 35), β-catenin (36), and siamois (12) to rescue axis formation in Eng-Sia-injected embryos was examined. At the four-cell stage, both dorsal blastomeres were coinjected with Eng-Sia and β-galactosidase, Xwnt8, β-catenin, or siamois (Fig. 2). No rescue of axial development was observed with coinjection of Eng-Sia and Xwnt8 or β-catenin, consistent with a role for siamois downstream of the wnt pathway. Failure to rescue axis formation was observed at doses of Xwnt8 and β-catenin that induced complete axial duplication in ventral injection of control embryos (data not shown). In contrast, a 3-fold excess of siamois resulted in complete rescue of axial development, indicating that the effects of Eng-Sia are due to a block of endogenous siamois (Table 2).
Figure 2.
Rescue of axial defects resulting from Eng-Sia injection. At the four-cell stage, both dorsal blastomeres were injected with 30 pg of Eng-Sia (A–D) in combination with 1 ng of β-galactosidase (A), 5 pg of Xwnt8 (B), 1 ng of β-catenin (C), or 100 pg of siamois (D) mRNA. (E) As a control, both dorsal blastomeres were injected with 1 ng of β-galactosidase mRNA. See Table 2 for quantitation. In a parallel experiment, ventral injection with Xwnt8, β-catenin, or siamois mRNA induced axis formation (data not shown).
Table 2.
Rescue of axis inhibition resulting from Eng-Sia injection
| mRNA injected | Axis inhibition
|
|
|---|---|---|
| n/N | % | |
| β-gal | 1/55 | 2 |
| Eng-Sia + β-gal | 69/76 | 91 |
| Eng-Sia + Xwnt8 | 22/26 | 85 |
| Eng-Sia + β-catenin | 28/29 | 97 |
| Eng-Sia + siamois | 5/56 | 9 |
At the four-cell stage, both dorsal blastomeres were injected with 30 pg of Eng-Sia mRNA or 1 ng of β-galactosidase (β-gal), 5 pg of Xwnt8, 1 ng of β-catenin, or 100 pg of siamois mRNA. Embryos were scored for axis formation at the tailbud stage. n, Injected embryos with reduction of axis formation resulting in DAI ≤ 2; N, total number of injected embryos; %, percent affected embryos. In a parallel experiment, ventral injection of Xwnt8, β-catenin, and siamois induced secondary axis formation (data not shown). The data presented are representative of more than three experiments.
Eng-Sia is likely to function by targeting the repressor domain to specific sequences bound by the siamois homeodomain and, therefore, altering the DNA-binding properties of Eng-Sia should lessen or abolish activity. The requirement for specific DNA-binding by siamois and Eng-Sia was examined by generating mutations in residue 50 (amino acid 9 of helix 3) of the homeodomain, a key amino acid in determining DNA-binding specificity (refs. 37 and 38, reviewed in ref. 39). The glutamine at position 50 of the siamois homeodomain was mutated to lysine, found in the bicoid homeodomain and predicted to alter DNA-binding specificity, or glutamate, predicted to diminish DNA binding. The mutations resulted in a severe reduction (lysine) or loss (glutamate) of axis induction by siamois and axis inhibition by Eng-Sia (Table 3). Therefore, both the dorsalizing activity of siamois and the ventralizing activity of Eng-Sia are dependent on appropriate sequence-specific binding. In support of the requirement for siamois DNA-binding specificity, constructs containing the goosecoid homeodomain fused to the VP16 activator (VP16-Gsc) or the engrailed repressor (Eng-Gsc) did not induce or inhibit axis formation, respectively (unpublished data).
Table 3.
Effect of homeodomain mutations on the activity of siamois and Eng-Sia
| mRNA injected | Axis induction
|
Axis inhibition
|
||
|---|---|---|---|---|
| n/N | % | n/N | % | |
| None | 0/37 | 0 | 0/34 | 0 |
| Siamois | 33/40 | 83 | 0/33 | 0 |
| SiamoisQ191K | 5/36 | 14 | 0/40 | 0 |
| SiamoisQ191E | 1/43 | 2 | 0/21 | 0 |
| Eng-Sia | 0/26 | 0 | 19/24 | 79 |
| Eng-SiaQ191K | 0/29 | 0 | 2/28 | 7 |
| Eng-SiaQ191E | 0/22 | 0 | 1/30 | 3 |
Site-specific mutations were generated in siamois and Eng-Sia that converted a glutamine at position 50 of the homeodomain (Q191) into a lysine (Q191K) or glutamate (Q191E). At the four-cell stage, a single ventral blastomere (axis induction columns) or both dorsal blastomeres (axis inhibition columns) were injected with 30 pg of siamois or Eng-Sia mRNA or 100 pg of the siamois or Eng-Sia mutant mRNAs. Embryos were scored for axis formation at the tailbud stage. n, Injected embryos with secondary axis formation (axis induction) or reduction of axis formation resulting in DAI ≤ 2 (axis inhibition); N, total number of injected embryos; %, percent affected embryos. The data presented are representative of three experiments.
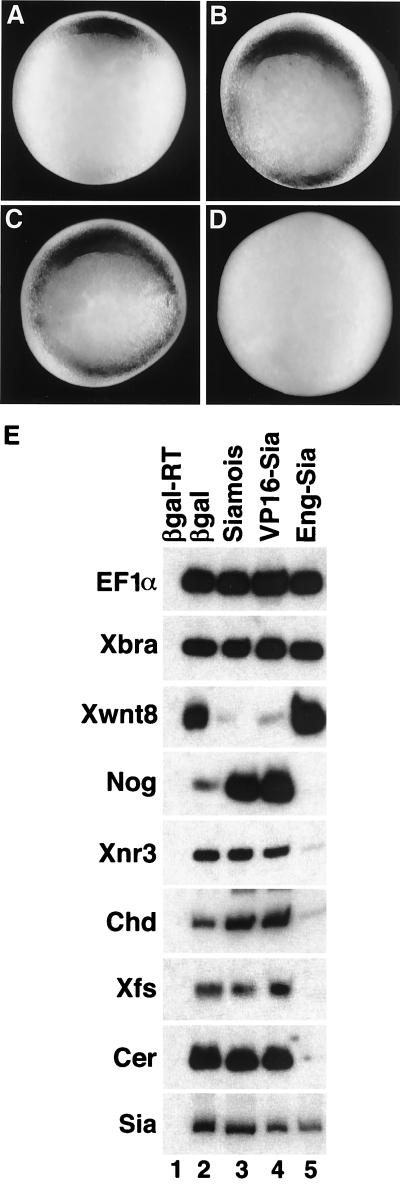
Dorsal injection of Eng-Sia resulted in ventralized embryos with morphological features indistinguishable from UV-irradiated embryos (DAI = 0), suggesting a complete loss of Spemann’s organizer. To assess organizer formation, both blastomeres of two-cell stage embryos were injected with β-galactosidase, siamois, VP16-Sia, or Eng-Sia mRNA, and organizer-specific genes were examined in gastrulae (Fig. 3). In situ hybridization analysis for goosecoid mRNA (40) indicated that siamois and VP16-Sia induced goosecoid expression throughout the marginal zone, consistent with the dorsalizing activity of these mRNAs. In contrast, Eng-Sia fully inhibited goosecoid expression, in agreement with an absence of organizer (Fig. 3 A–D). Additional markers were examined by reverse transcription–PCR (Fig. 3E). The organizer markers noggin (41), Xnr3 (42), chordin (43), follistatin (27), and cerberus (28) were expressed at elevated or normal levels in response to siamois and VP16-Sia, and levels were greatly reduced or undetectable in response to Eng-Sia. In contrast, the ventrolateral marker Xwnt8 (44) was repressed by siamois and VP16-Sia and was elevated in response to Eng-Sia. Expression of panmesodermal brachyury (45), as well as endogenous siamois, was unaffected by siamois, VP16-Sia, or Eng-Sia. The results indicate that siamois and VP16-Sia induce an expansion of organizer, resulting in dorsalization, whereas Eng-Sia blocked organizer formation without interfering with general mesoderm induction. Consistent with gastrula marker expression, at the tailbud stage siamois and VP16-Sia blocked expression of a blood marker, α-globin (ventral fate), whereas Eng-Sia blocked muscle actin and panneural neural cell adhesion molecule (NCAM) (dorsal fates), without inhibiting α-globin (data not shown).
Figure 3.
Organizer formation is affected by siamois fusions. At the two-cell stage, both blastomeres were injected with 30 pg of β-galactosidase, siamois, VP16-Sia, or Eng-Sia mRNA, and embryos were harvested at stage 10.25 (early gastrula). (A–D) Vegetal view of injected embryos analyzed by in situ hybridization for goosecoid expression. (A) β-Galactosidase had no effect on the organizer-specific expression of goosecoid (n = 24). Siamois (B) and VP16-Sia (C) resulted in expansion of goosecoid expression in 79% (n = 24) and 88% (n = 24) of embryos, respectively. (D) Eng-Sia resulted in a reduction or loss of goosecoid expression in 83% (n = 24) of embryos. (E) Embryos were harvested at the gastrula stage and processed for reverse transcription–PCR analysis of the organizer genes noggin (Nog), Xnr3, chordin (Chd), follistatin (Xfs), and cerberus (Cer), the ventrolateral gene Xwnt8, the panmesodermal gene brachyury (Xbra), endogenous siamois (Sia), and the ubiquitous EF1α. Injection of siamois (lane 3) or VP16-Sia (lane 4) inhibited Xwnt8 expression and enhanced expression of noggin and chordin, without affecting Xnr3, follistatin, cerberus, siamois, or brachyury expression. Eng-Sia (lane 5) inhibited expression of noggin, Xnr3, chordin, follistatin, and cerberus, enhanced Xwnt8 expression, and had no effect on siamois or brachyury. EF1α is a control for RNA recovery and loading. β-Galactosidase-injected embryos (lane 2) served as a positive control, while an identical reaction without reverse transcriptase controlled for PCR contamination (lane 1).
The inducing activity of siamois, the induction of siamois by the wnt pathway, and the inability of Xwnt8 and β-catenin to rescue axis inhibition by Eng-Sia suggest that siamois acts as a transcriptional mediator of wnt signaling. To test this idea, the ability of Eng-Sia to inhibit axis induction by the “upstream” factors, Xwnt8 and β-catenin, and the “downstream” factors, noggin and chordin, as well as siamois itself, was examined. At the four-cell stage, a single ventral blastomere was injected with Xwnt8, β-catenin, siamois, noggin, or chordin mRNA in combination with β-galactosidase or Eng-Sia mRNA. While axial duplication in response to noggin and chordin was unaffected by Eng-Sia, axis induction by Xwnt8, β-catenin, and siamois (at a dose equal to Eng-Sia) was fully blocked (Fig. 4 and Table 4). The results indicate that siamois mediates transcriptional responses to the wnt pathway that are required for axis induction. In support of this conclusion, Eng-Sia does not block nuclear accumulation of β-catenin in dorsal blastomeres or in ventral blastomeres injected with Xwnt8. In addition, siamois does not stimulate nuclear accumulation of β-catenin (data not shown). Furthermore, axis induction by noggin and chordin in the presence of Eng-Sia suggests that these factors act downstream of siamois, consistent with the ability of siamois to induce organizer-specific genes, including noggin and chordin.
Figure 4.
Eng-Sia inhibits axis induction by the wnt pathway. At the four-cell stage, one ventral blastomere was injected with 30 pg of β-galactosidase (A–F) or Eng-Sia (G–L) in combination with 5 pg of Xwnt8 (B, H), 1 ng of β-catenin (C, I), 30 pg of siamois (D, J), 200 pg of noggin (E, K), or 200 pg of chordin (F, L) mRNA. Axis induction was observed in response to Xwnt8, β-catenin, siamois, noggin, and chordin. Eng-Sia blocked axis induction by Xwnt8, β-catenin, and siamois but not by noggin and chordin. Neither β-galactosidase nor Eng-Sia induced axis formation. See Table 4 for quantitation. In a parallel experiment, dorsal injection of Eng-Sia inhibited axis formation (data not shown).
Table 4.
Inhibition of axis-inducing factors by Eng-Sia
| mRNA injected | Axis induction
|
|||
|---|---|---|---|---|
| +β-gal
|
+Eng-Sia
|
|||
| n/N | % | n/N | % | |
| β-gal | 0/29 | 0 | 0/28 | 0 |
| Xwnt8 | 18/20 | 90 | 1/19 | 5 |
| β-Catenin | 15/18 | 83 | 0/20 | 0 |
| Siamois | 16/19 | 89 | 0/21 | 0 |
| Noggin | 16/22 | 73 | 13/18 | 72 |
| Chordin | 19/21 | 90 | 15/20 | 75 |
At the four-cell stage, a single ventral blastomere was injected with 1 ng of β-galactosidase (β-gal), 5 pg of Xwnt8, 1 ng of β-catenin, 30 pg of siamois, 200 pg of noggin, or 200 pg of chordin mRNA in combination with 30 pg of β-gal or Eng-Sia mRNA. Embryos were scored for axis formation at the tailbud stage. n, Injected embryos with secondary axis formation; N, total number of injected embryos; %, percent affected embryos. The data presented are representative of more than three experiments.
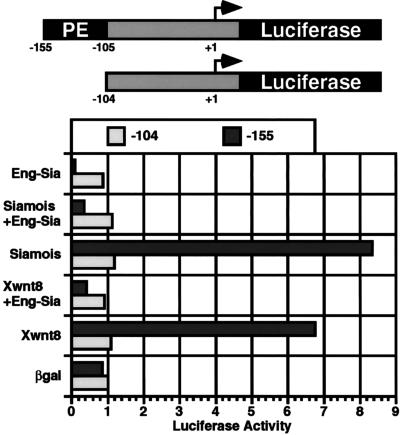
The potential role for siamois as a transcriptional mediator of wnt signaling was directly tested by using a defined 50-bp wnt-responsive goosecoid promoter element (−155 to −105). A luciferase reporter containing 155 bp of goosecoid promoter sequence responded strongly to Xwnt8, whereas a reporter containing 104 bp of promoter sequence was unresponsive (46). Induction of the reporter constructs (−155 or −104) by Xwnt8, siamois, and Eng-Sia, as well as mixtures of the mRNAs, was tested by injecting a single ventral blastomere at the four-cell stage and assaying luciferase activity at the gastrula stage (Fig. 5). The −155 reporter was induced 8- to 10-fold by siamois and 6- to 8-fold by Xwnt8. In contrast, Eng-Sia repressed the −155 reporter, resulting in a 6- to 7-fold decrease in basal activity. Coinjection of Eng-Sia with Xwnt8 or siamois repressed induction of the −155 reporter, resulting in activity below basal levels. The −104 reporter, lacking the wnt-response element, was unresponsive to Xwnt8, siamois, or Eng-Sia. Activation of the wnt-response element by siamois and the ability of Eng-Sia to block activation by Xwnt8 suggest that siamois directly mediates transcriptional responses to wnt signaling. This is confirmed by preliminary studies showing direct binding of siamois to the wnt-response element (unpublished data). Furthermore, the results support the conclusion that siamois functions as a transcriptional activator and that Eng-Sia can repress transcriptional targets of siamois.
Figure 5.
Siamois activates the goosecoid promoter via a wnt-responsive regulatory element. A 50-bp wnt-responsive proximal element (PE) is located between bases −155 and −105 of the goosecoid promoter (46). Luciferase reporter constructs containing 155 bp of promoter sequence (including PE) or the 104-bp minimal promoter were tested for responsiveness to Xwnt8, siamois, Eng-Sia, or a mixture of mRNAs. At the four-cell stage, one ventral blastomere was injected with 200 pg of the −155 or −104 reporter plasmid in combination with 50 pg of β-galactosidase, Xwnt8, siamois, or Eng-Sia mRNA, or mixtures of Eng-Sia and Xwnt8 or siamois mRNAs. At the gastrula stage, extract was prepared and luciferase activity was measured. Basal activity of uninjected embryos was subtracted for all values, averages were determined for duplicate samples, and values were normalized to the activity of the −104 reporter coinjected with β-galactosidase. The data presented are representative of three experiments.
DISCUSSION
The results demonstrate that siamois is required for development of Spemann’s organizer and subsequent axis formation. The ability of siamois to rescue axis inhibition by Eng-Sia indicates a specific block of endogenous siamois. However, other transcriptional activators with similar DNA-binding specificity may also be inhibited by Eng-Sia. In addition, the cooperative binding of paired-type homeodomain proteins suggests that Eng-Sia may indirectly influence factors that interact with siamois at target promoters (ref. 47, reviewed in ref. 39). This latter possibility is supported by the in vitro interaction of siamois with Mix.1, a paired-type homeodomain protein implicated in ventral development (48). At the late blastula stage, expression of siamois and Mix.1 overlaps and interactions may regulate dorsoventral pattern, a process potentially influenced by Eng-Sia.
Siamois is induced by all components of the wnt pathway, including β-catenin, and this induction occurs in the presence of cycloheximide, indicating that preexisting maternal components directly activate siamois transcription. This suggests that siamois may mediate transcriptional responses to wnt signaling, and two observations support this proposal. First, the effects of Eng-Sia injection and antisense ablation of β-catenin on axial development are indistinguishable (7). Second, axis inhibition by Eng-Sia is not rescued by Xwnt8 or β-catenin, consistent with a dependence of wnt dorsalizing activity on siamois function. The ability of β-catenin to enter the nucleus as a complex with LEF-1 (18–20), a maternal transcription factor, points to the potential role of a β-catenin–LEF-1 complex in directly activating siamois at the mid-blastula transition. In agreement with this idea, siamois transcripts are present in dorsal cells containing nuclear β-catenin, and siamois can rescue axis formation after antisense depletion of β-catenin (49). Alternatively, undescribed maternal components of the wnt pathway, acting either downstream of, or in a complex with, β-catenin, may regulate siamois transcription. It should be noted that dorsal cells containing nuclear β-catenin are present in a broad domain along the animal–vegetal axis, and only a subset of these cells express siamois, suggesting that additional signals may play a role in regulating siamois expression.
In Xenopus, dorsal determinants are displaced from the vegetal pole to future dorsal regions by cortical rotation, establishing dorsoventral polarity that results in formation of Spemann’s organizer (refs. 2–5, reviewed in ref. 6). Although the identity of the dorsal determinants is undefined, their position corresponds to the site of nuclear β-catenin and siamois transcription. In UV-irradiated embryos, dorsal determinants remain at the vegetal pole, resulting in vegetal cells containing nuclear β-catenin and siamois transcripts (14). This suggests that in mediating the transcriptional response to wnt signaling, siamois is regulating zygotic events that have their origin in maternal dorsal determinants. However, vegetal expression of siamois is not sufficient for axis formation, suggesting that vegetal cells are not competent to respond to siamois, or that signals not present in vegetal cells act in conjunction with siamois to regulate organizer formation.
The described expression pattern of siamois suggests an indirect regulation of organizer formation. In the late blastula, siamois is expressed in dorsal cells positioned vegetally to brachyury-expressing marginal cells, suggesting that siamois-expressing cells induce organizer in a distinct group of marginal cells (12). However, siamois binds and activates the goosecoid promoter, demonstrating that at least one organizer gene may be directly regulated by siamois. Consistent with a direct mechanism, goosecoid expression overlaps extensively with siamois (unpublished), a result that contrasts with the nonoverlapping brachyury expression. Whether additional organizer genes are directly regulated by siamois remains to be determined.
The requirement for siamois function in the development of Spemann’s organizer in Xenopus suggests that siamois homologs may play a similar role in the development of other vertebrate organizers. Support for this proposal awaits the isolation and analysis of siamois homologs from fish, mouse, and chick. Furthermore, the interacting components of the wnt pathway constitute a conserved signaling system that functions in diverse biological processes in both vertebrates and invertebrates (reviewed in ref. 11). A conserved role for siamois, or siamois-like factors in other wnt signaling events, such as neural patterning or limb development, is an intriguing possibility to pursue.
Acknowledgments
I am grateful to D. Goldhamer, P. Labosky, and M. Mullins for critical reading of the manuscript; P. Lemaire and J. Gurdon for siamois; J. Jaynes for Drosophila engrailed; K. Cho for the goosecoid reporters; T. Kadesch for herpes simplex virus VP16; and G. Moorer and B. Munson for excellent technical assistance. This work was supported in part by grants from the National Institutes of Health (HD 35159) and the Pew Scholars Program.
ABBREVIATIONS
- VP16-Sia
VP16-siamois fusion
- Eng-Sia
engrailed-siamois fusion
- DAI
dorsoanterior index
Note
After submission of this study a related paper was published (50) that supports the conclusions presented here. It has recently been reported that a complex of β-catenin and LEF-1/Xtcf-3 can directly activate siamois transcription (51).
References
- 1.Kessler D S, Melton D A. Science. 1994;266:596–604. doi: 10.1126/science.7939714. [DOI] [PubMed] [Google Scholar]
- 2.Fujisue M, Kobayakawa Y, Yamana K. Development. 1993;118:163–170. doi: 10.1242/dev.118.1.163. [DOI] [PubMed] [Google Scholar]
- 3.Holowacz T, Elinson R P. Development. 1993;119:277–285. doi: 10.1242/dev.119.1.277. [DOI] [PubMed] [Google Scholar]
- 4.Sakai M. Development. 1996;122:2207–2214. doi: 10.1242/dev.122.7.2207. [DOI] [PubMed] [Google Scholar]
- 5.Kikkawa M, Takano K, Shinagawa A. Development. 1996;122:3687–3696. doi: 10.1242/dev.122.12.3687. [DOI] [PubMed] [Google Scholar]
- 6.Elinson E P, Kao K R. Dev Growth Differ. 1989;31:423–430. doi: 10.1111/j.1440-169X.1989.00423.x. [DOI] [PubMed] [Google Scholar]
- 7.Heasman J, Crawford A, Goldstone K, Garner-Hamrick P, Gumbiner B, McCrea P, Kintner C, Noro C Y, Wylie C. Cell. 1994;79:791–803. doi: 10.1016/0092-8674(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 8.Schneider S, Steinbeisser H, Warga R M, Hausen P. Mech Dev. 1996;57:191–198. doi: 10.1016/0925-4773(96)00546-1. [DOI] [PubMed] [Google Scholar]
- 9.Larabell C A, Torres M, Rowning B A, Yost C, Miller J R, Wu M, Kimelman D, Moon R T. J Cell Biol. 1997;136:1123–1136. doi: 10.1083/jcb.136.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller J R, Moon R T. Genes Dev. 1996;10:2527–2539. doi: 10.1101/gad.10.20.2527. [DOI] [PubMed] [Google Scholar]
- 11.Moon R T, Brown J D, Torres M. Trends Genet. 1997;13:157–162. doi: 10.1016/s0168-9525(97)01093-7. [DOI] [PubMed] [Google Scholar]
- 12.Lemaire P, Garrett N, Gurdon J B. Cell. 1995;81:85–94. doi: 10.1016/0092-8674(95)90373-9. [DOI] [PubMed] [Google Scholar]
- 13.Carnac G, Kodjabachian L, Gurdon J B, Lemaire P. Development. 1996;122:3055–3065. doi: 10.1242/dev.122.10.3055. [DOI] [PubMed] [Google Scholar]
- 14.Brannon M, Kimelman D. Dev Biol. 1996;180:344–347. doi: 10.1006/dbio.1996.0306. [DOI] [PubMed] [Google Scholar]
- 15.Yang-Snyder J, Miller J R, Brown J D, Lai C J, Moon R T. Curr Biol. 1996;6:1302–1306. doi: 10.1016/s0960-9822(02)70716-1. [DOI] [PubMed] [Google Scholar]
- 16.Fagotto F, Guger K, Gumbiner B M. Development. 1997;124:453–460. doi: 10.1242/dev.124.2.453. [DOI] [PubMed] [Google Scholar]
- 17.Vleminckx K, Wong E, Guger K, Rubinfeld B, Polakis P, Gumbiner B M. J Cell Biol. 1997;136:411–420. doi: 10.1083/jcb.136.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Behrens J, von Kries J P, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Nature (London) 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 19.Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann B G, Kemler R. Mech Dev. 1996;59:3–10. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- 20.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 21.Thomsen G H, Melton D A. Cell. 1993;74:433–441. doi: 10.1016/0092-8674(93)80045-g. [DOI] [PubMed] [Google Scholar]
- 22.Nieuwkoop P D, Faber J. Normal Table of Xenopus laevis (Daudin) Amsterdam: North Holland; 1967. [Google Scholar]
- 23.Klein S L. Dev Biol. 1987;120:299–304. doi: 10.1016/0012-1606(87)90127-8. [DOI] [PubMed] [Google Scholar]
- 24.Rupp R A, Snider L, Weintraub H. Genes Dev. 1994;8:1311–1323. doi: 10.1101/gad.8.11.1311. [DOI] [PubMed] [Google Scholar]
- 25.Hemmati-Brivanlou A, Frank D, Bolce M E, Brown R D, Sive H L, Harland R M. Development. 1990;110:325–330. doi: 10.1242/dev.110.2.325. [DOI] [PubMed] [Google Scholar]
- 26.Wilson P A, Melton D A. Curr Biol. 1994;4:676–686. doi: 10.1016/s0960-9822(00)00152-4. [DOI] [PubMed] [Google Scholar]
- 27.Hemmati-Brivanlou A, Kelly O G, Melton D A. Cell. 1994;77:283–295. doi: 10.1016/0092-8674(94)90320-4. [DOI] [PubMed] [Google Scholar]
- 28.Bouwmeester T, Kim S-H, Sasai Y, Lu B, De Robertis E M. Nature (London) 1996;382:595–601. doi: 10.1038/382595a0. [DOI] [PubMed] [Google Scholar]
- 29.Sadowski I, Ma J, Triezenberg S, Ptashne M. Nature (London) 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 30.Triezenberg S J, Kingsbury R C, McKnight S L. Genes Dev. 1988;2:718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- 31.Jaynes J B, O’Farrell P H. EMBO J. 1991;10:1427–1433. doi: 10.1002/j.1460-2075.1991.tb07663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han K, Manley J L. EMBO J. 1993;12:2723–2733. doi: 10.1002/j.1460-2075.1993.tb05934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Badiani P, Corbella P, Kioussis D, Marvel J, Weston K. Genes Dev. 1994;8:770–782. doi: 10.1101/gad.8.7.770. [DOI] [PubMed] [Google Scholar]
- 34.Sokol S, Christian J L, Moon R T, Melton D A. Cell. 1991;67:741–752. doi: 10.1016/0092-8674(91)90069-b. [DOI] [PubMed] [Google Scholar]
- 35.Smith W C, Harland R M. Cell. 1991;67:753–765. doi: 10.1016/0092-8674(91)90070-f. [DOI] [PubMed] [Google Scholar]
- 36.Funayama N, Fagotto F, McCrea P, Gumbiner B M. J Cell Biol. 1995;128:959–968. doi: 10.1083/jcb.128.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanes S D, Brent R. Cell. 1989;57:1275–1283. doi: 10.1016/0092-8674(89)90063-9. [DOI] [PubMed] [Google Scholar]
- 38.Treisman J, Gonczy P, Vashishtha M, Harris E, Desplan C. Cell. 1989;59:553–562. doi: 10.1016/0092-8674(89)90038-x. [DOI] [PubMed] [Google Scholar]
- 39.Mann R S. BioEssays. 1995;17:855–863. doi: 10.1002/bies.950171007. [DOI] [PubMed] [Google Scholar]
- 40.Cho K W Y, Morita E A, Wright C V E, De Robertis E M. Cell. 1991;65:55–64. doi: 10.1016/0092-8674(91)90407-p. [DOI] [PubMed] [Google Scholar]
- 41.Smith W C, Knecht A K, Wu M, Harland R M. Nature (London) 1993;361:547–549. doi: 10.1038/361547a0. [DOI] [PubMed] [Google Scholar]
- 42.Smith W C, McKendry R, Ribisi S, Jr, Harland R M. Cell. 1995;82:37–46. doi: 10.1016/0092-8674(95)90050-0. [DOI] [PubMed] [Google Scholar]
- 43.Sasai Y, Lu B, Steinbeisser H, Geissert D, Gont L K, De Robertis E M. Cell. 1994;79:779–790. doi: 10.1016/0092-8674(94)90068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Christian J L, Gavin B J, McMahon A P, Moon R T. Dev Biol. 1991;143:230–234. doi: 10.1016/0012-1606(91)90073-c. [DOI] [PubMed] [Google Scholar]
- 45.Smith J C, Price B M J, Green J B A, Weigel D, Herrmann B G. Cell. 1991;67:79–87. doi: 10.1016/0092-8674(91)90573-h. [DOI] [PubMed] [Google Scholar]
- 46.Watabe T, Kim S, Candia A, Rothbacher U, Hashimoto C, Inoue K, Cho K W. Genes Dev. 1995;9:3038–3050. doi: 10.1101/gad.9.24.3038. [DOI] [PubMed] [Google Scholar]
- 47.Wilson D, Sheng G, Lecuit T, Dostatni N, Desplan C. Genes Dev. 1993;7:2120–2134. doi: 10.1101/gad.7.11.2120. [DOI] [PubMed] [Google Scholar]
- 48.Mead P E, Brivanlou I H, Kelley C M, Zon L I. Nature (London) 1996;382:357–360. doi: 10.1038/382357a0. [DOI] [PubMed] [Google Scholar]
- 49.Wylie C, Kofron M, Payne C, Anderson R, Hosobuchi M, Joseph E, Heasman J. Development. 1996;122:2987–2996. doi: 10.1242/dev.122.10.2987. [DOI] [PubMed] [Google Scholar]
- 50.Fan M J, Sokol S Y. Development. 1997;124:2581–2589. doi: 10.1242/dev.124.13.2581. [DOI] [PubMed] [Google Scholar]
- 51.Brannon M, Gomperts M, Sumoy L, Moon R T, Kimelman D. Genes Dev. 1997;11:2359–2370. doi: 10.1101/gad.11.18.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]