Abstract
The pancreas is derived from a pool of multipotent progenitor cells (MPCs) that co-express Pdx-1 and Ptf1a. To more precisely define how the individual and combined loss of Pdx-1 and Ptf1a affects pancreatic MPC specification and differentiation we derived and studied mice bearing a novel Ptf1aYFP allele. While the expression of Pdx-1 and Ptf1a in pancreatic MPCs coincides between E9.5–12.5 the developmental phenotypes of Pdx-1 null and Pdx-1; Ptf1a double null mice are indistinguishable, and an early pancreatic bud is formed in both cases. This finding indicates that Pdx-1 is required in the foregut endoderm prior to Ptf1a for pancreatic MPC specification. We also found that Ptf1a is neither required for specification of Ngn3-positive endocrine progenitors nor differentiation of mature β-cells. In the absence of Pdx-1 Ngn3-positive cells were not observed after E9.5. Thus, in contrast to the deletion of Ptf1a, the loss of Pdx-1 precludes the sustained Ngn3-based derivation of endocrine progenitors from pancreatic MPCs. Taken together, these studies indicate that Pdx-1 and Ptf1a have distinct but interdependent functions during pancreatic MPC specification.
Keywords: Pancreas, Islet, Development, Multipotent Progenitor Cell, Insulin, Pdx-1, Ptf1a, Ngn3, MafA, Recombinase Mediated Cassette Exchange
Introduction
The mouse pancreas originates as two evaginations of the foregut endoderm at approximately embryonic day 9 (E9) (Guz et al., 1995; Jonsson et al., 1994; Offield et al., 1996). Both the dorsal and ventral pancreatic buds contain multipotent progenitor cells (MPCs) that give rise to the endocrine, exocrine, and ductal lineages of the pancreas (Gu et al., 2002; Kawaguchi et al., 2002; Zhou et al., 2007). Recent studies suggest that a pool of pancreatic MPCs is maintained until approximately E12.5 (Zhou et al., 2007). However, the mechanisms that regulate allocation of these MPCs into transit amplifying progenitor cell subpopulations that have lineage-specific fates are not understood. In this study we explored the roles of both pancreatic and duodenal homeobox factor 1 (Pdx-1) and pancreas specific transcription factor-1a (Ptf1a/p48) in controlling acquisition of the MPC fate and how the loss of these factors affects partitioning of these cells into the pro-endocrine cell compartment as well as other non-pancreatic tissues.
Pdx-1 and Ptf1a/p48 (hereafter termed Ptf1a) have each been previously shown to play vital roles in the growth and lineage specification of pancreatic MPCs during development (Chiang and Melton, 2003; Jonsson et al., 1994; Kawaguchi et al., 2002; Krapp et al., 1998; Offield et al., 1996; Sellick et al., 2004; Stoffers et al., 1997). The overlapping expression domain of these transcription factors is thought to define the primordial pancreatic endoderm (Chiang and Melton, 2003; Kawaguchi et al., 2002). Pdx-1, which is first expressed at E8.5 in the prospective posterior foregut, demarcates both pancreatic buds by E9.5 as well as the endodermal region that produces the caudal stomach, rostral duodenum, common bile duct (CBD), gall bladder, and cystic duct (Guz et al., 1995; Jonsson et al., 1994; Offield et al., 1996). In contrast, Ptf1a, which is first expressed in the pancreas at E9.5 (Chiang and Melton, 2003; Kawaguchi et al., 2002; Krapp et al., 1998; Obata et al., 2001), is additionally expressed in the neuronal precursors of the cerebellum, spinal cord, and retina where it also performs fate determining roles (Fujitani et al., 2006b; Glasgow et al., 2005; Hoshino et al., 2005).
The developmental phenotypes of Pdx-1 and Ptf1a null mice are similar in that both fail to form acinar cells, and thus lack a normal pancreas (Jonsson et al., 1994; Krapp et al., 1998; Offield et al., 1996). However, the defects that occur in Pdx-1 null mice are more widespread and include abnormalities in the gastro-duodenal junction, submucosal Brunner’s glands, enteroendocrine cell numbers in the stomach and duodenum, and formation of peribiliary glands and mucin-producing cells in the gall bladder (Fukuda et al., 2006b; Jonsson et al., 1994; Offield et al., 1996). Moreover, Pdx-1 null animals, in contrast with Ptf1a null mice, form only a small number of short-lived insulin-producing cells as well as glucagon-producing cell clusters (Ahlgren et al., 1996; Guz et al., 1995; Offield et al., 1996). In Ptf1a mutants all islet endocrine cell types persist to late embryogenesis (Kawaguchi et al., 2002; Krapp et al., 1998).
The commitment of pancreatic MPCs to the endocrine lineage requires the expression of Neurogenin 3 (Ngn3) (Gradwohl et al., 2000; Gu et al., 2002). Ngn3-positive cells are normally detected around E9 within the pre-pancreatic endoderm and are believed to give rise to both insulin- and glucagon-expressing cells between approximately E9 and E12 (Apelqvist et al., 1999; Gradwohl et al., 2000). These early, so-called first wave, endocrine cells do not appear to populate mature islets (Herrera, 2000; Herrera et al., 1998). However, beginning around E13.5, a second wave of endocrine cell differentiation occurs during which five mono-hormone-expressing cell types are specified: β-cells (insulin), α-cells (glucagon), δ-cells (somatostatin), ε-cells (ghrelin), and PP (pancreatic polypeptide) cells (Jensen, 2004; Murtaugh and Melton, 2003; Wilson et al., 2003). The finding that the early endocrine cell lineage is still specified in both Pdx-1 and Ptf1a null mice suggests that neither factor is required for the expression of Ngn3 (Ahlgren et al., 1996; Jonsson et al., 1994; Kawaguchi et al., 2002; Krapp et al., 1998; Offield et al., 1996).
While Pdx-1 and Ptf1a clearly play vital functions in pancreatic MPCs, little is known about how each of these genes may influence each other, contribute to the maintenance of pancreatic MPCs, or are involved in fate decisions that regulate the segregation of these MPCs into specific pancreatic lineages. Given the more restricted expression of Ptf1a in the broader Pdx-1-postive foregut endoderm and its role in the specification of pancreatic MPCs, it has been hypothesized that the concurrent expression of Ptf1a and Pdx-1 elicits a commitment towards pancreatic fates instead of other foregut organs (Kawaguchi et al., 2002). Consistent with this idea, it has been reported that pan-endodermal expression of XPtf1a in Xenopus promotes ectopic pancreas formation only in sites where Pdx-1 is expressed (Afelik et al., 2006). Furthermore, in Hes1 null mice ectopic expression of Ptf1a in the CBD promotes pancreas formation (Fukuda et al., 2006a).
The findings we report here provide new insights into the interdependent roles of both Pdx-1 and Ptf1a in maintaining a pool of pancreatic MPCs. These studies were facilitated by the derivation of mice that express Yellow Fluorescent Protein (YFP) in place of Ptf1a using a strategy that involved Recombinase Mediated Cassette Exchange (RMCE) (Feng et al., 1999; Long et al., 2004). Utilizing the Ptf1aYFP allele in combination with a Pdx-1 null allele, we analyzed how the individual and combined loss of Pdx-1 and Ptf1a affects the specification and differentiation of pancreatic MPCs as well as the formation and maturation of endocrine cells.
Materials and Methods
Gene targeting and RMCE
The Ptf1aLCA targeting vector contained homology arms of 5162 (ClaI to SalI) and 2624 bp (SalI - PvuII) from a RPCI-22 BAC (clone 251-E16) (Osoegawa et al., 2000). The two homology arms flank, consecutively, a loxP site, a phosphoglycerol kinase (pgk)-driven neomycin resistance gene (neoR), a pgk-driven Herpes Simplex Virus thymidine kinase gene (tk), and a second loxP site whose orientation is inverted relative to the first site. In addition, a Diphtheria toxin A expression cassette was located outside of the short arm. The targeting vector was linearized with SacII prior to electroporation into TL-1 mouse ES cells.
The Ptf1aYFP(+HygroR) cassette exchange vector was made by inserting a 4105 bp fragment of the Ptf1a gene into a plasmid containing two inversely-oriented LoxP sites, then changing a NotI site in the 5’ UTR to an FseI site. DNA encoding Citrine (Heikal et al., 2000), an enhanced yellow fluorescent protein (YFP) variant (obtained from David W. Piston; Vanderbilt) was then inserted between the new FseI and a naturally-occurring MluI site in exon 1 of the Ptf1a gene. A pgk-driven hygromycin resistance gene (HygroR), flanked by tandem FRT sites, was inserted into a NotI site at the 3’ end of the exchange vector for positive selection during RMCE.
Gene targeting was performed using standard protocols. Homologous recombination was verified by Southern blot analysis after XmnI or EcoRI digestion. RMCE was performed as previously described (Long et al., 2004). Briefly, clone 5D12, which contains the Ptf1aLCA, was electroporated with both the Ptf1aYFP(+HygroR) exchange vector and pBS185, a Cre-expression vector. Clones surviving both hygromycin and gancyclovir were screened by PCR on both the 5’ and 3’ ends using primers: 1: 5′-CCTTCTGACTTCTCCAAGAAGGCA-3′, 2: 5′-CCTTTATGCCTGGCATTTCACTG-3′, 3: 5′-TTGACTCCCCAGGCTTGGACTGTT-3′, 4: 5′- ACCGATGGCTGTGTAGAAGTACT-3′, 5: 5′-TCTATCGCCTTCTTGACGAGTTCT-3′.
Mouse strains and husbandry
Chimeric mice were generated by microinjection of clone 5D12:1D3 ES cells into blastocysts obtained from natural matings of C57BL/6J mice then bred with other C57BL/6J mice to produce mice bearing the Ptf1aYFP(+HygroR) allele. Removal of the FRT-flanked HygroR cassette was accomplished by inbreeding with ACTB:FLPe mice (kindly provided by S.M. Dymecki, Harvard Medical School, Boston, MA). The resulting allele, Ptf1aYFP, was maintained on an outbred background. MIP:GFP transgenic mice were kindly provided by G.I. Bell (U. Chicago). Mice containing the Pdx-1LacZ ko allele have been previously described (Offield et al., 1996). All mice were maintained in a specific pathogen free state with a 12 h light-dark cycle. Experimental protocols were approved by the Vanderbilt Institutional Animal Care and Use Committee.
X-gal staining and immunodetection
For X-gal staining whole embryos or dissected gut tissues were fixed in 4% paraformaldehyde in PBS at 4° C for 30 min then washed with PBS twice followed by two washes in rinse/permeabilization buffer (2 mM MgCl2, 0.01% sodium deoxycholate, and 0.02% Nonidet P-40 in PBS). Fixed embryos or dissected gut tissues were incubated overnight at RT with 1.0 mg/mL X-gal in rinse buffer. Samples were post-fixed overnight in 4% paraformaldehyde/PBS at 4°C. Whole embryos were rinsed twice in PBS then dehydrated in methanol. A 2:1 solution of benzyl benzoate: benzyl alcohol (Sigma-Aldrich) was used to clear the dehydrated embryos. Dissected gut tissues were stored in 70% ethanol after fixation.
For routine immunodetection, embryos and dissected tissues were fixed overnight in 4% paraformaldehyde in PBS at 4°C. Tissues were transferred to 30% sucrose in PBS and equilibrated at 4°C (few hours to O/N). Embryos or tissues were embedded in O.C.T. and frozen on dry ice. 10 µm frozen sections were permeabilized with 0.2% Triton X-100 in PBS for 20 min, then washed 3 times in 0.1% Triton X-100 in PBS and 3 times in PBS alone. Sections were preincubated with 5% normal donkey serum and 1% BSA in PBS for at least 1 h at room temperature before applying primary antibody. Primary antibodies were diluted in 1% BSA in PBS as follows: rabbit anti-GFP (Molecular Probes), 1:500; chicken anti-GFP (ABCAM), 1:500; guinea pig anti-Pdx-1 (Christopher Wright), 1:1000; goat-anti Pdx-1 (Christopher Wright), 1:5000; guinea pig anti-insulin (Linco), 1:1000; rabbit anti-glucagon (Linco), 1:1000; rabbit anti-Ptf1a (Helena Edlund, Umeå U.), 1:500; guinea pig anti-Ngn3 (Maike Sander, U.C. Irvine), 1:500; rabbit anti-MafA (Bethyl Laboratories), 1:1000. The anti-Pdx-1 antibodies recognize both Pdx-1 and the LacZ-Pdx-1 fusion protein expressed from the Pdx-1LacZko allele. Tissue sections were incubated with primary antibodies overnight at 4°C then washed three times in PBS + 0.1% Triton X-100 and 3 times in PBS alone. Secondary antibodies were diluted in 1% BSA in PBS as follows: donkey anti-rabbit IgG conjugated to Cy3 (Jackson Immuno Research), 1:1000; donkey anti-rabbit IgG conjugated to Cy2 (Jackson Immuno Research), 1:500; donkey anti-guinea pig IgG conjugated to Cy3 (Jackson Immuno Research), 1:1000; donkey anti-guinea pig IgG conjugated to Cy2 (Jackson Immuno Research), 1:500; donkey anti-chicken conjugated to Cy2 (Jackson Immuno Research), 1:500; donkey anti-chicken conjugated to Cy3 (Jackson Immuno Research), 1:1000; donkey anti-goat conjugated to Alexa Fluor 647 (Invitrogen), 1:500. Sections were incubated with secondary antibodies for 1–3 hrs at room temperature and then washed three times in PBS + 0.1% Triton X-100 and 3 times in PBS alone. DRAQ5™ (Alexis) was diluted 1:1000 in PBS and used as a nuclear counterstain where noted. Cover slips were mounted using Aqua/Poly Mount (Polysciences, Inc.). Bright field and fluorescence images were acquired using either an Axioplan2 microscope (Zeiss) equipped with a QImaging RETIGA EXi camera or a Leica MZ 16 FA stereoscope with a QImaging RETIGA 4000R camera. Confocal microscopy was performed using a Zeiss LSM 510.
Statistical Analysis
The entire foregut, including the stomach, spleen, pancreas, and duodenum, at E18.5 was dissected in order to determine the percentage of MafA positive insulin-expressing cells in wild type and Ptf1a null embryos (n = 3 for each genotype). Confocal microscopy (1 µm optical slice) was performed on sections immunostained for MafA and insulin. DRAQ5™ was used to stain nuclei. The percentage of MafA positive, insulin-expressing cells was measured by counting cells (based on presence of a nucleus) in at least 20 random fields of view per embryo. Statistical significance was determined using a Student’s two tail t test.
Results
Generation of a Ptf1aLCA allele in mouse ES cells and insertion of YFP by RMCE
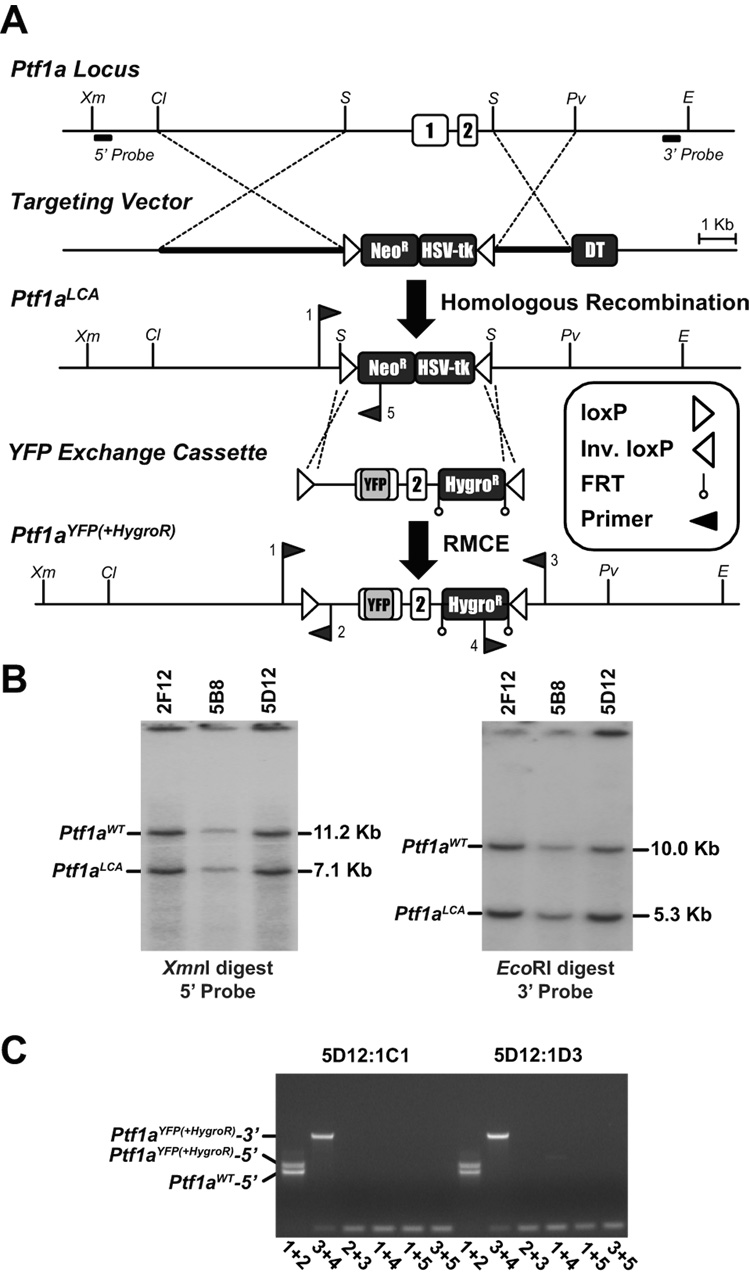
A three-step procedure was used to derive mice carrying the Ptf1aYFP allele (Fig. 1). First, gene targeting replaced a 4.1 kb region of the Ptf1a gene with two inversely-oriented loxP sites flanking both neomycin resistance (neoR) and HSV-thymidine kinase (HSV-tk) cassettes (Fig. 1A) to create a Ptf1a Loxed Cassette Acceptor (Ptf1aLCA) allele. Three of the 244 ES cell clones screened were correctly targeted as indicated by Southern blot analysis (Fig. 1B). Second, an exchange vector was designed to replace Ptf1a coding sequences with those of YFP while also replacing all but 613 bp of Ptf1a genomic DNA removed during initial gene targeting (Fig. 1A). RMCE was performed by co-electroporating the exchange vector with a Cre-expression plasmid into Ptf1aLCA ES cells. Analysis of 9 clones that survived a staggered positive-negative selection strategy (Long et al., 2004) revealed that all had undergone exchange. Clone 5D12:1D3, in which the exchange event was correctly orientated (Fig. 1C), was used to generate mice. Third, mice containing the Ptf1aYFP(+HygroR) allele were interbred with mice bearing a FLPe -expressing transgene to remove the FRT-flanked HygroR cassette.
Figure 1.
Creation of a Ptf1aLCA allele and insertion of YFP by RMCE. A) Schematic representation of the Ptf1a locus, targeting vector, Ptf1aLCA allele, Ptf1a-YFP exchange cassette, and the Ptf1aYFP(+HygroR) allele. A 4.1 kb Ptf1a fragment including both exons was replaced by two pgk-driven selection cassettes, neoR and HSV-tk, flanked by inversely orientated loxP sites during gene targeting. A pgk-driven Diphtheria toxin A cassette is located beyond the end of the short arm of DNA homology. YFP coding sequences replace 5’ UTR and Ptf1a coding sequences in exon 1. Exon 2 is retained in the exchange cassette to supply RNA splicing and poly adenylation signals with a FRT-flanked hygromycin resistance cassette located downstream of Ptf1a sequences for positive selection following RMCE. Insertion of the YFP exchange cassette into the Ptf1aLCA was accomplished by RMCE creating the Ptf1aYFP(+HygroR) allele from which mice were derived. Restriction Endonucleases: ClaI (Cl), EcoRI (E), PvuII (Pv), SalI (S), XmnI (Xm). B) Southern blot analysis of ES cell genomic DNA, using probes indicated in A, confirmed 3 ES cell clones that were correctly targeted for the Ptf1aLCA allele, 2F12, 5B8, and 5D12. Clone 5D12 was chosen for the RMCE experiment. C) PCR screening of Ptf1aYFP(+HygroR) cassette exchanged clones using primer pair combinations that detect cassettes in the forward or reverse orientation on both the 5’ and 3’ ends. Clones 5D12:1C1 and 5D12:1D3 both contain the Ptf1aYFP(+HygroR) cassette in the forward orientation. Primer locations are indicated in A and combinations are indicated in C below their respective lanes.
Ptf1aYFP expression by whole mount fluorescence microscopy
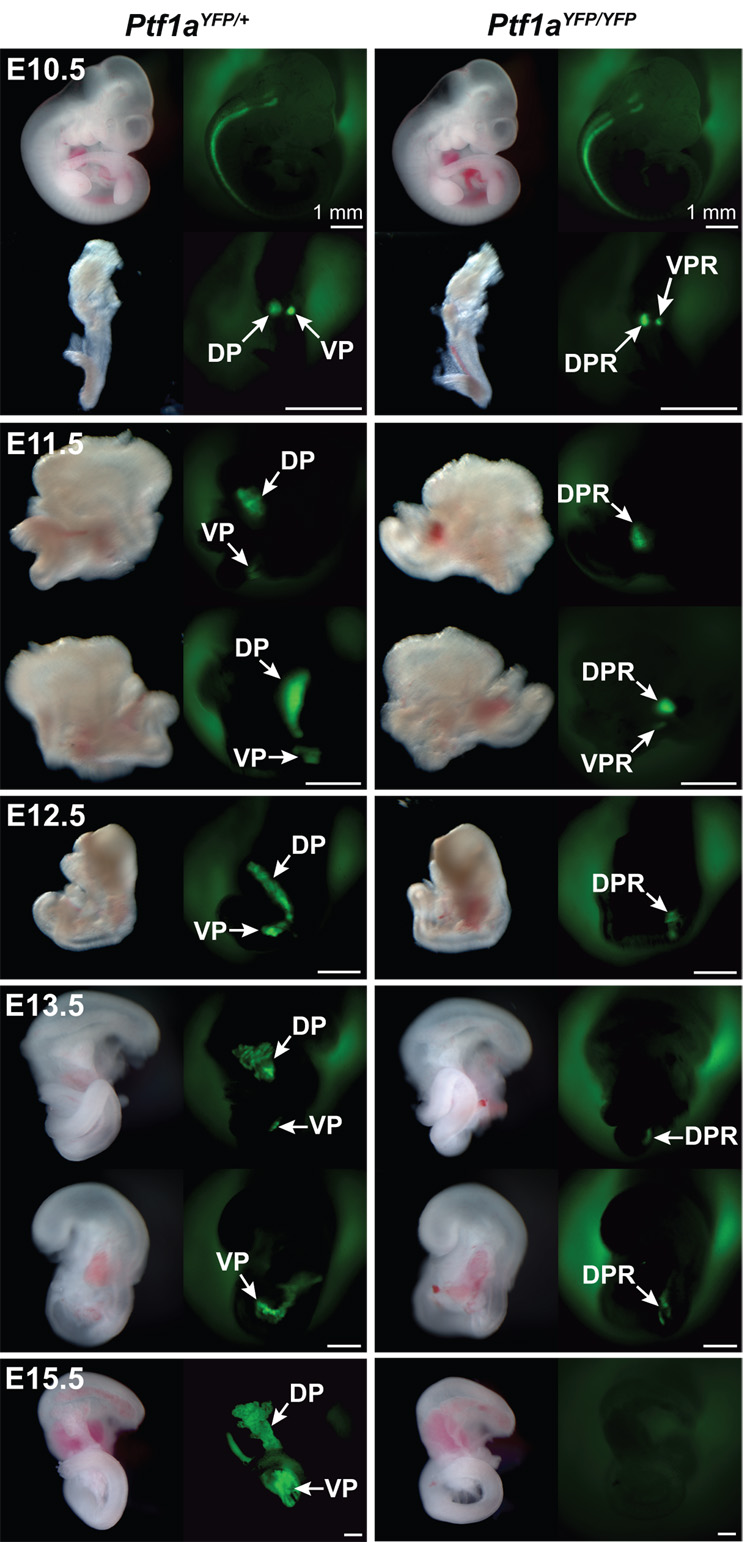
Expression of YFP from the Ptf1aYFP allele was first assessed by whole mount fluorescence microscopy. In Ptf1aYFP/+ embryos YFP fluorescence was first seen at E10.5 in the neural tube and the developing cerebellum (Fig. 2); sites of bona fide Ptf1a expression (Glasgow et al., 2005; Hoshino et al., 2005; Kawaguchi et al., 2002; Obata et al., 2001). Pancreas-specific fluorescence was observed in the dorsal and ventral pancreatic buds after dissecting the primordial gut tube away from the rest of the embryo (Fig. 2). Transcription of the Ptf1a gene is estimated to be considerably less than that of Pdx-1 at this stage of development given that fluorescence from a Pdx-1GFP allele is visible without the need to dissect out the gut tissue (Holland et al., 2006). In E11.5 embryos, Ptf1a-YFP fluorescence intensified (Fig. 2) with expression occurring throughout the pancreatic epithelium upon fusion of the dorsal and ventral pancreas at E12.5. By E13.5 pancreatic fluorescence appeared as many clusters, consistent with the expected restriction of Ptf1a expression to pro-acinar structures at this stage of development. The fluorescence intensity increased further by E15.5, corresponding with the expansion of pancreatic exocrine cells (Fig. 2). In the adult, pancreas-wide fluorescence was similar to that observed in late embryogenesis (data not shown).
Figure 2.
YFP fluorescence in Ptf1aYFP/+ and Ptf1aYFP/YFP embryos. In Ptf1aYFP/+ embryos, YFP fluorescence can be detected in all sites of bona fide Ptf1a expression by whole mount fluorescence microscopy. Uniform YFP fluorescence is seen in the neural tube as well as in the pancreatic buds at E10.5, which expand and fuse by E12.5. At E13.5, YFP fluorescence is punctate which denotes acinar restriction. At E15.5, exocrine cells have proliferated which results in increased intensity of YFP fluorescence. In Ptf1aYFP/YFP mice, dorsal and ventral Ptf1a expression is initiated at E10.5, but growth of the pancreatic buds is severely retarded by E11.5. Ventral YFP fluorescence is lost by E12.5, but can be observed in the DPR until E13.5. However, by E15.5 YFP fluorescence is extinguished in Ptf1aYFP/YFP mice. Scale bars = 50 µm unless otherwise noted. Dorsal Pancreas (DP), Ventral Pancreas (VP), Dorsal Pancreatic Remnant (DPR), Ventral Pancreatic Remnant (VPR).
Ptf1aYFP expression by immunohistochemistry
To more precisely identify cells expressing YFP from the Ptf1aYFP allele we next performed immunostaining experiments. From E9.5–11.5, Ptf1a was detected throughout the dorsal and ventral pancreatic buds, and YFP is exactly coincident with that of Ptf1a (Fig. S1). To our knowledge, this is the first report demonstrating Ptf1a protein in the pre-pancreatic endoderm at E9.5. At E12.5, when Ptf1a is expressed in MPCs residing in the distal tips of the branching epithelium at the periphery of the pancreas tissue, intense YFP immunofluorescence co-localized with Ptf1a except for some weaker, residual YFP staining towards the central part of the pancreas epithelium (Fig. S1). This is probably due to YFP having a modestly longer half-life than Ptf1a. Therefore, the perdurance of YFP transiently lineage labels cells that have recently ceased Ptf1a transcription and provides further evidence that MPCs in the distal epithelial tips give rise to the Ptf1a-negative cells found in the core, or trunk region of the pancreas tissue (Zhou et al., 2007). However, from E13.5 on, Ptf1a and YFP were entirely coincident in all acinar cells, consistent with upregulation of Ptf1a and the appearance of exocrine enzymes such as amylase at this stage of development (Chiang and Melton, 2003; Guz et al., 1995). Moreover, YFP was detected in all non-pancreatic expression domains of Ptf1a and therefore indicates Ptf1aYFP expression faithfully recapitulates that of the wild type allele (data not shown).
Ptf1a expressing progenitors are depleted by ~E13.5 in Ptf1a null embryos
Previously, lineage tracing experiments demonstrated labeling of a MPC population in the posterior foregut of Ptf1aCre/Cre mice at E10.5 (Kawaguchi et al., 2002). However, those experiments did not ascertain the duration of Ptf1a gene expression. Therefore, having shown that YFP reliably reports Ptf1a transcription, we sought to determine the spatiotemporal expression pattern of Ptf1a past E10.5 in Ptf1aYFP/YFP embryos. YFP expression was observed by whole mount fluorescent microscopy in both the dorsal and ventral pancreatic buds at E10.5 (Fig. 2). Similarly, at E11.5 YFP was seen in both the dorsal and ventral buds although expression in the ventral pancreatic remnant (VPR) disappears by E12.5 (Fig. 2). However, in contrast to the VPR, Ptf1aYFP expression in the DPR persists until E13.5, but then is no longer seen at E15.5 in the absence of Ptf1a (Fig. 2). The persistence of YFP fluorescence in the pancreatic remnants suggests that the foregut endodermal cells remain multipotent, and/or that they continue to respond to cell-autonomous and/or non-autonomous Ptf1a-inducing signals through mid-gestation. The loss of YFP fluorescence appears to be due to the dorsal Ptf1a null cells being specified to limited pancreatic fates and to the ventral Ptf1a null cells being reassigned to duodenal cell types (Kawaguchi et al., 2002; Krapp et al., 1998). This indicates that there is a limited time window, extending from ~E10.5–13.5, for the pancreatic MPCs to respond to the pro-pancreatic function of Ptf1a or to other signals that may be necessary for Ptf1aexpression.
Ventral Ptf1a-deficient cells convert to a CBD fate
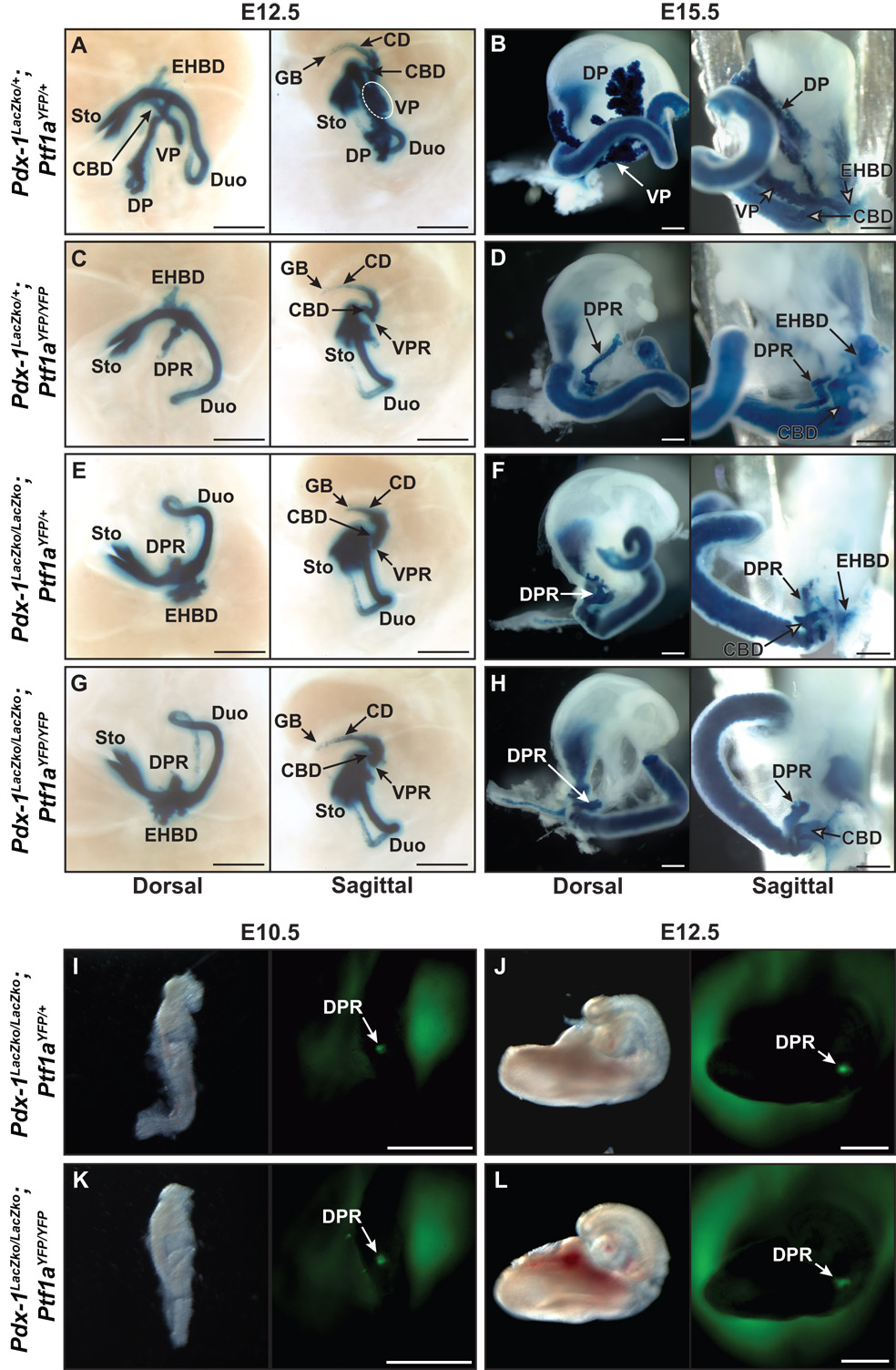
To determine how the loss of Ptf1a affects Pdx-1 xpression and pancreas morphogenesis, we next crossed mice bearing the Ptf1aYFP allele with Pdx-1LacZko/+ animals to obtain both Pdx-1LacZko/+; Ptf1aYFP/YFP and Pdx-1LacZko/+; Ptf1aYFP/+ embryos. Use of the Pdx-1LacZko allele is convenient in this setting because it allows for high resolution imaging via X-gal histochemistry of all foregut structures in wild type and mutant embryos. Double heterozygous E12.5 embryos displayed normal fusion of the dorsal and ventral pancreatic buds (Fig. 3A). The dorsal bud is normally connected to the duodenum while the ventral bud stems from the common bile duct (CBD), which itself extends from the duodenal ampulla to the liver and gall bladder. However, in Ptf1a null embryos at E12.5, fusion of the dorsal and ventral lobes did not occur and growth of the DPR was diminished with the ventral pancreatic remnant (VPR) appearing as a minute evagination of the CBD epithelium (Fig. 3C). The ventral pancreas and CBD have a common embryonic origin as the CBD progenitors in Hes1 null mutants develop into pancreatic tissue (Fukuda et al., 2006a; Sumazaki et al., 2004). Therefore we have hypothesized that in the absence of Ptf1a, ventral pancreatic MPCs contribute to CBD development. Ventral YFP fluorescence is absent in Ptf1aYFP/YFP mice at E12.5 (Fig. 2) which precludes immunofluorescence analysis of expression on sections, but at E11.5 YFP expressing cells of the VPR are detected in a Pdx-1-positive epithelial structure resembling the primordial CBD or duodenum. While our analysis does not preclude the possibility that some Ptf1a-derived cells may have already contributed to the duodenum or CBD and ceased Ptf1a transcription by E11.5, ventral Ptf1a null cells that continue to express Ptf1aYFP did not appear to coalesce into a distinct epithelial bud (Fig. 5D, third panel). This finding, when considered with the results of previous studies, suggests that ventral pancreatic MPCs are converted to duodenal and/or CBD progenitor cells in the absence of Ptf1a (Kawaguchi et al., 2002). In contrast, Ptf1a null dorsal pancreatic MPCs organize into a distinct epithelial bud (Fig. 5D).
Figure 3.
Pancreas morphogenesis in Pdx-1 null, Ptf1a null, and Pdx-1, Ptf1a double null embryos. A–H) Histological analysis of β-galactosidase expression from the Pdx-1LacZko allele in Pdx-1 null (Pdx-1LacZko/LacZko; Ptf1aYFP/+), Ptf1a null (Pdx-1LacZko/+; Ptf1aYFP/YFP), and Pdx-1, Ptf1a double null (Pdx-1LacZko/LacZko; Ptf1aYFP/YFP) embryos at E12.5 and E15.5. A) In Pdx-1LacZko/+; Ptf1aYFP/+ E12.5 embryos, the dorsal pancreas is associated with the duodenum, the ventral pancreas is evaginating from the CBD epithelium and the two lobes have fused near the duodenum at their proximal ends. B) At E15.5 in Ptf1aYFP/+; Pdx-1LacZko/+ embryos, normal epithelial branching and lobulation indicates acinar development. X-gal staining also denotes Pdx-1 expression in the CBD, extrahepatic biliary ducts, cystic duct, and gall bladder. C) In Ptf1a null embryos at E12.5, the epithelium of the DPR is growth deficient and does not branch while the VPR is barely discernable. D) In Ptf1a null embryos at E15.5, LacZ expression is observed in the DPR which is minimally branched and extends from the duodenum towards the spleen. The absence of a VPR indicates assimilation of ventral pancreatic MPCs into the CBD and/or duodenum epithelium. E) Pdx-1 null embryos at E12.5 have a considerably smaller DPR than in Ptf1a null embryos at the same stage while they display a similar structure resembling a VPR contiguous with the CBD. F) By E15.5 the DPR of Pdx-1LacZko/LacZko; Ptf1aYFP/+ embryos undergoes limited branching and X-gal staining is more intense in the gall bladder and cystic duct, due to homozygosity of the Pdx-1LacZko alleles, although no gross abnormalities are observed. As in Ptf1a null mice, there appears to be no VPR associated with the CBD in Pdx-1 null mice at E15.5 which suggests ventral pancreatic cells have contributed to the epithelium of the CBD and/or duodenum. G, H) The morphological phenotype of Pdx-1, Ptf1a double null embryos is identical to Pdx-1 null embryos. I–L) Whole mount fluorescence microscopy of YFP (Ptf1aYFP) in Pdx-1 null and Pdx-1, Ptf1a double null embryos at E10.5 and E12.5. I, J) In Pdx-1 null embryos, YFP can be observed in the DPR from E10.5–12.5. YFP is not detectable at E13.5 which indicates a loss of dorsal pancreatic progenitor cells and that Ptf1a expression may be dependent on Pdx-1 after E12.5. K–L) YFP fluorescence in double null embryos indicates that neither Pdx-1 nor Ptf1a is necessary for dorsal expression of Ptf1a or epithelial budding. Scale bars = 50 µm. Duodenum (Duo), Stomach (Sto), Extrahepatic Biliary Ducts (EHBD), Common Bile Duct (CBD), Cystic Duct (CD), Gall Bladder (GB), Dorsal Pancreatic Remnant (DPR), Ventral Pancreatic Remnant (VPR).
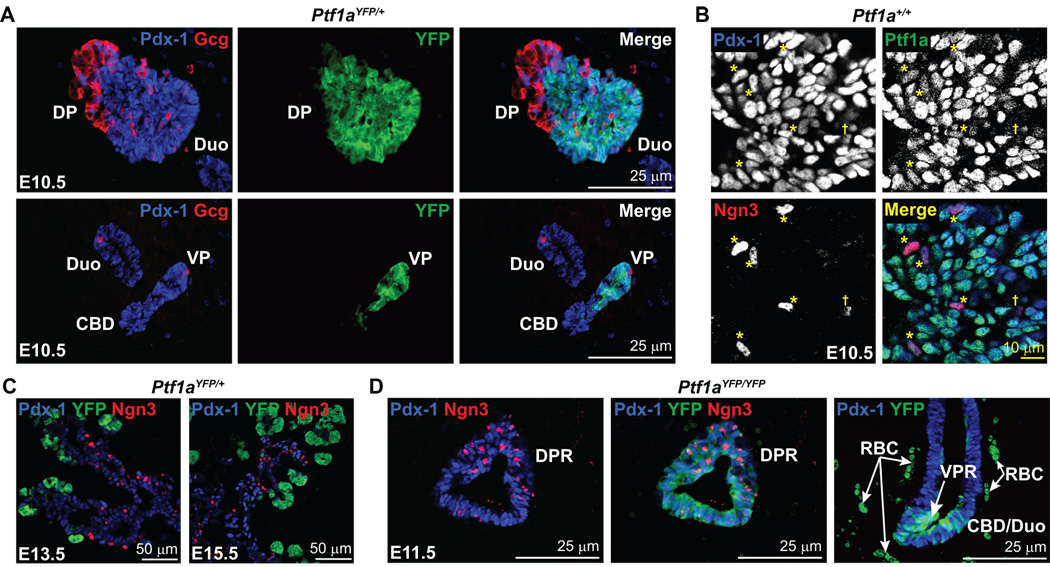
Figure 5.
Developmental compartmentalization of Pdx-1, Ptf1a, and Ngn3. A) Immunofluorescent analysis of Pdx-1 and Ptf1aYFP expression at E10.5 indicates their coincidence throughout the dorsal and ventral pancreatic epithelium with the exception of the first wave endocrine cells. The duodenum and CBD are only Pdx-1-positive and do not express Ptf1a. B) Confocal immunofluorescence analysis for Pdx-1, Ptf1a, and Ngn3 in pancreatic progenitor cells of the dorsal pancreatic bud at E10.5. Ptf1a, Pdx-1, and Ngn3 coincide in scattered cells throughout the pancreatic epithelium at E10.5 (*) although Ptf1a staining in these cells is relatively weak compared to those that do not express Ngn3. Some Ngn3-positive cells are Ptf1a-negative (†), but all Ptf1a-positive and Ngn3-positive cells express Pdx-1. C) Compartmentalization of Ptf1aYFP and Ngn3 expression. By E13.5 and thereafter, Ngn3-positive cells reside in the Pdx-1-positive ductal cords of the pancreas, but do not coincide with YFP which is restricted to pro-acinar structures. Thus, early Ngn3-positive cells express Ptf1a while subsequent generations do not. D) Ptf1a is not necessary for Ngn3 expression. Ngn3-positive cells are found throughout the DPR of Ptf1aYFP/YFP embryos at E11.5 along with Pdx-1 and YFP. Also, YFP-positive cells are observed in a columnar epithelium near the site where the ventral pancreas arises which illustrates a lack of coalescence of Ptf1a-expressing cells into a distinct epithelial bud. This suggests that these cells are already specified to a duodenal and/or CBD fate. Dorsal Pancreas (DP), Ventral Pancreas (VP), Dorsal Pancreatic Remnant (DPR), Ventral Pancreatic Remnant (VPR), Common bile duct (CBD), Duodenum (Duo), Red blood cell (RBC).
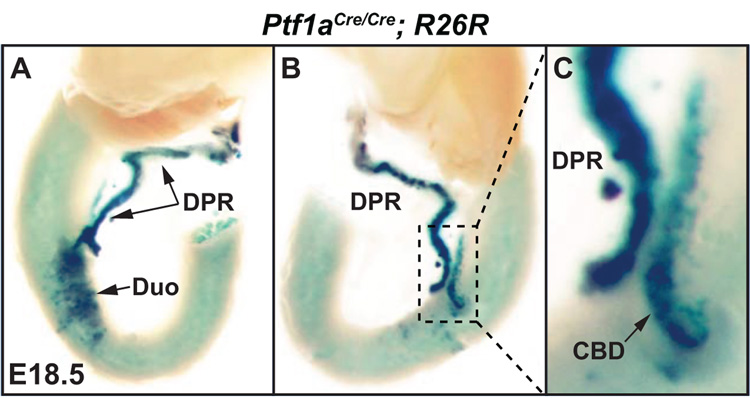
In E15.5 embryos that are null for Ptf1a (Pdx-1LacZko/+; Ptf1aYFP/YFP) the DPR was seen to have undergone some outgrowth from the duodenum towards the spleen, although it was noticeably smaller and less branched when compared to double heterozygous controls (compare Figs. 3B and 3D). Furthermore, a VPR could not be discerned. Thus, we hypothesized that these cells had become integrated into the CBD epithelium and/or duodenum (Fig. 3D). For this reason we further examined gut tissue from day E18.5 embryos that were previously obtained from a Cre-based lineage tracing experiment (Kawaguchi et al., 2002). As was previously shown, Ptf1a-deficient MPCs contribute to the development of the DPR and duodenum (Fig. 4A). In addition, cells forming the CBD were also seen that expressed LacZ thereby indicating that Ptf1a-deficient ventral MPCs do indeed become CBD progenitor cells (Figs. 4B,C).
Figure 4.
Lineage tracing in Ptf1aCre/Cre; R26R embryos at E18.5. A) Dorsal view of the dissected gut from a Ptf1a null embryo at E18.5. X-gal histochemistry was used to detect cells in which the R26R allele had been recombined by the expression of Cre from the Ptf1a locus (Kawaguchi et al., 2002). This analysis indicates as previously reported that Ptf1a-expressing cells contribute to the epithelium of the DPR as well as the duodenum in the absence of a functional Ptf1a allele. B, C) Sagittal views of the dissected gut shown in A. Ptf1a-lineage labeled cells are found in the DPR, duodenum and the CBD. This indicates that the CBD lineage is indeed available to ventral Ptf1a null pancreatic MPCs. Duodenum (Duo), Dorsal Pancreatic Remnant (DPR), Common Bile Duct (CBD).
Dorsal expression of Ptf1a is independent of Pdx-1
We next examined the interdependency of Pdx-1 and Ptf1a in embryos lacking Pdx-1 but retaining expression of Ptf1a (Pdx-1LacZko/LacZko; Ptf1aYFP/+). Similar to prior results from Ptf1a lineage tracing experiments in mice homozygous for a hypomorphic Pdx-1 allele (Fujitani et al., 2006a), at E10.5 YFP was observed only in the DPR in Pdx-1 null mice (Fig. 3I). This finding indicates that mechanisms that establish dorsal Ptf1a expression are independent of Pdx-1. Moroever, by immunostaining we observed Ptf1a in the foregut endoderm of Pdx-1 null embryos as early as E9.5 (Fig. S4). Unlike Ptf1aYFP/YFP mice in which YFP fluorescence is observed until E13.5, expression of the Ptf1aYFP allele is apparent until only E12.5 in Pdx-1 null embryos (Fig. 3J and data not shown). This result implies that maintenance of Ptf1a expression requires Pdx-1 or that fate allocation of Pdx-1-deficient MPCs occurs earlier than in absence of Ptf1a because they can no longer respond to signals that induce Ptf1a transcription, which are present until at least E13.5.
As previously demonstrated in Pdx-1 null mice, outgrowth of the DPR is markedly diminished (Offield et al., 1996). Similar to Ptf1a null embryos at E12.5, we observed that discernment of a VPR from the adjacent CBD epithelium is questionable in Pdx-1 null embryos (Fig. 3E). The lack of ventral YFP fluorescence indicates , as previously reported (Fujitani et al., 2006a), not just the absence of ventral pancreatic cells at mid-gestational stages but also supports the idea that Pdx-1 is required for the production and/or survival of ventral Ptf1a-expressing pancreatic MPCs (Fig. 3I,J). Expression of Ptf1a is not reactivated at later embryonic stages because YFP is not detected in mice lacking Pdx-1 at E13.5 or 15.5 (data not shown), and the DPR that is derived from Ptf1a-expressing cells undergoes only limited expansion and branching during this time (Fujitani et al., 2006a; Kawaguchi et al., 2002). However, as in the Ptf1a null mice at E15.5, a VPR is not evident in the Pdx-1 null animals (Fig. 3F) suggesting that Pdx-1, like Ptf1a, is required for maintenance of the ventral pancreatic fate. Thus, in the absence of Pdx-1, ventral foregut epithelial cells appear to default to either CBD or duodenal fates although lineage tracing in Pdx-1−/−; Ptf1aCre/+ embryos would be required to unequivocally establish the lineages that Pdx-1 null pancreatic MPCs adopt.
Pdx-1 is more fundamental than Ptf1a to pancreas specification
To determine the effect of a combined inactivation of Pdx-1 and Ptf1a on pancreas development we also generated Pdx-1LacZko/LacZko; Ptf1aYFP/YFP embryos. The phenotype of these embryos was indistinguishable from those lacking only Pdx-1. YFP was detected at E10.5 only dorsally, an important result which indicates that dorsal pancreatic bud formation is independent of Pdx-1 and Ptf1a (Fig. 3K). Also, the developmental potency of Pdx-1; Ptf1a double null progenitor cells is similarly limited as in Pdx-1 null mice since the ability to respond to Ptf1a inducing signals is present until ~E12.5 (Fig. 3L and data not shown). Analysis of Pdx-1 driven β-galactosidase expression between E12.5–15.5 showed that tissue morphogenesis in the double null was identical to that of Pdx-1 null mutants (compare Figs. 3G,H with 3E,F). By E15.5 there was no VPR, and a variably branched, cyst-like structure that was indistinguishable from that observed in Pdx-1 null mice was observed (Fig. 3H). Therefore, the limited expansion and/or branching of the DPR in Pdx-1 null embryos is not caused by the residual presence of Ptf1a.
Pancreatic MPCs are defined by the co-expression of Pdx-1 and Ptf1a between E9.5 and E12.5
Although Pdx-1 and Ptf1a are expressed in pancreatic progenitors, the spatial and temporal coincidence of their corresponding proteins has not been examined previously (Gu et al., 2002; Kawaguchi et al., 2002). As expected from previous Ptf1a lineage tracing studies (Kawaguchi et al., 2002), immunodetection of Pdx-1 and Ptf1a-driven YFP in E10.5 Ptf1aYFP/+ embryos identified their coincidence only in the pancreatic epithelium, and not in other regions of Pdx-1 expression such as the CBD or duodenum (Fig. 5A). Since YFP perdurance in Ptf1a-negative cells could be misconstrued as indicating Ptf1a expression [due to the slightly longer half-life of YFP than Ptf1a, as shown in Fig. S1] we examined Ptf1a protein expression directly using an antibody that produces no signal on Ptf1a−/− mutant tissue (Fig. S2A,B). Ptf1a was found to coincide with Pdx-1 in the vast majority of pancreatic epithelial cells from E9.5 to 10.5 (Fig. 5B, S2C, S3A–D). Furthermore, at E11.5 Pdx-1 and Ptf1a still coincide in almost all pancreatic epithelial cells (data not shown). To our knowledge this is the first report of the concurrent expression of both Pdx-1 and Ptf1a at the protein level in the pre-pancreatic endoderm at E9.5 as well as their persistence in pancreatic MPCs past E10.5. By E12.5, Ptf1a has begun to compartmentalize to the distal tips of the pancreatic epithelium (Fig. S1, S2A), a MPC pool that is also Pdx-1-positive (Fig. S2A) (Zhou et al., 2007). Thus, a hallmark of pancreatic MPCs appears to be the coincidence of Pdx-1 and Ptf1a protein from E9.5 to around E12.5. This finding is consistent with a recent report that suggests this stage of embryonic development is a crucial period during which a fixed number of pancreatic progenitors is specified since eliminating cells from this progenitor pool reduces the final size of the organ (Stanger et al., 2007). However, by about E13 Ptf1a-expressing cells appear to become unipotent and, thereafter, only give rise to acinar cells (Zhou et al., 2007).
Pdx-1, but not Ptf1a, is required to sustain pancreatic Ngn3 expression past E9.5
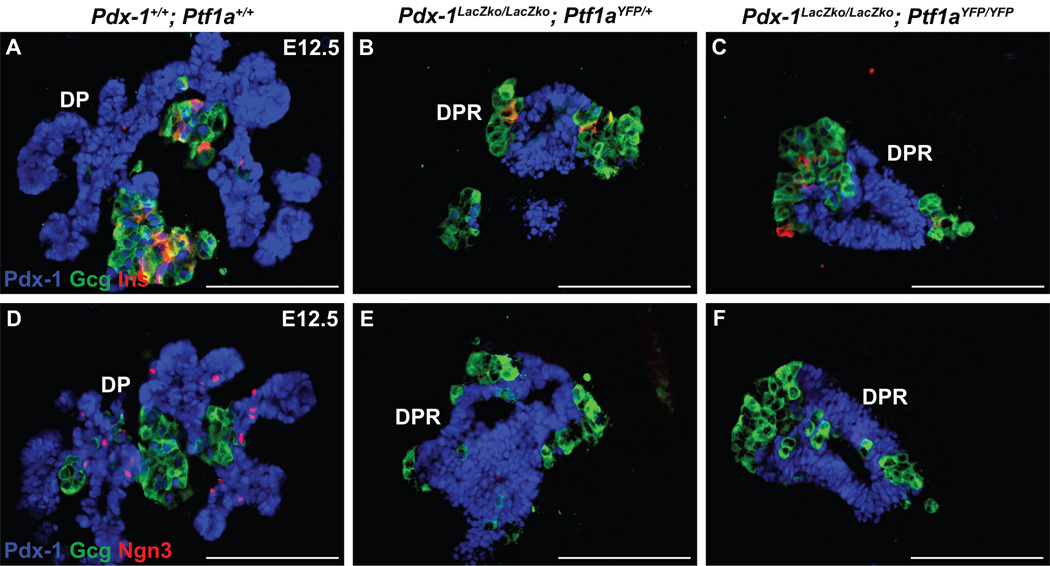
Previous single-cell PCR analysis of early pancreas bud cells at E10.5 suggests that Ngn3 is expressed in a subset of cells that also express Pdx-1 and Ptf1a (Chiang and Melton, 2003). For this reason, we sought to determine how the individual or combined loss of Pdx-1 and Ptf1a affects Ngn3 expression and endocrine cell specification. By immunostaining, Ngn3-positive cells were detected in the pre-pancreatic endoderm at E9.5, although they only appeared to express Pdx-1 and not Ptf1a (Fig. S3A–D). However, cells positive for Pdx-1, Ptf1a, and Ngn3 protein were observed to be dispersed throughout the pancreatic epithelium of E10.5 wild type embryos (Fig. 5B, S2C). The Ptf1a signal, while present above background, was weaker in Ngn3-positive cells and some Ngn3-postive cells did not stain for Ptf1a. Thus, as cells commit to Ngn3 expression, there appears to be an associated down-regulation of Ptf1a. At later embryonic stages (E13.5 and E15.5), Ptf1a no longer coincides with Ngn3 and instead is restricted to acinar cells (Fig. 5C).
As would be predicted by the presence of endocrine cells in Ptf1a null mice, Ngn3 is present in Ptf1aYFP/YFP embryos at E11.5 (Fig. 5D). In contrast, although insulin- and glucagon-expressing cells were present in Pdx-1 null (Pdx-1LacZko/LacZko;Ptf1aYFP/+) and Pdx-1; Ptf1a double null (Pdx-1LacZko/LacZko; Ptf1aYFP/YFP) embryos obtained between E10.5–12.5 (Fig. 6B,C and Fig. S3I,J), detectable levels of Ngn3 protein were not observed anywhere within the pancreatic epithelium (Fig. 6E,F and S3I,J). However, Ngn3-positive cells were present in the pre-pancreatic endoderm at E9.5 in the absence of either Pdx-1 or both Pdx-1 and Ptf1a (Fig. S3F,G). These results indicate that both first-wave endocrine cells and their progenitors can be specified independently of both Pdx-1 and Ptf1a. In addition, given that Ngn3-positive endocrine progenitors were not observed at E10.5 in Pdx-1 null embryos, these findings suggest that the Ngn3-positive cells present at E9.5 are rapidly depleted as they and/or their progeny give rise to the first wave endocrine cells.
Figure 6.
Early endocrine cells develop independently of Pdx-1 and Ptf1a. Immunofluorescence analysis for insulin, glucagon and Pdx-1. A) First wave insulin-and glucagon-expressing cells are associated with the pancreatic epithelium of wild type mice at E12.5. These endocrine cells are also present in the DPR of Pdx-1 null (B), and Pdx-1; Ptf1a double null (C) mice which indicates that neither transcription factor is necessary for their formation. D) Ngn3 is observed in the dorsal pancreas of wild type embryos at E12.5 but not in Pdx-1 null (E) or Pdx-1; Ptf1a double null (F) embryos, which suggests that Pdx-1 is required to sustain the specification of pancreatic Ngn3–expressing endocrine progenitors from an MPC pool. Scale bars = 25 µm.
Ptf1a is not required for β-cell maturation
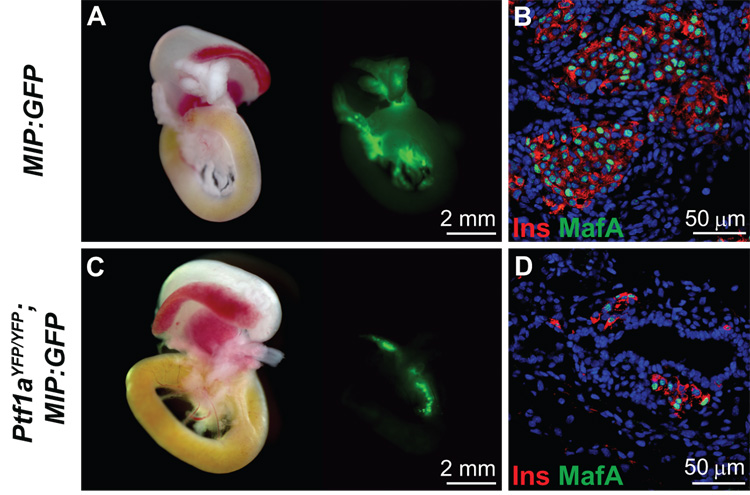
Finally, we sought to determine whether Ptf1a is necessary for β-cell maturation by examining expression of both insulin and MafA in Ptf1a null mice. MafA was examined since it is currently considered to represent a marker of the terminal stages of normal beta cell differentiation (Matsuoka et al., 2004). In contrast to previous reports (Kawaguchi et al., 2002; Krapp et al., 1998), splenic endocrine cells were not observed in Ptf1a null mice by immunofluorescence. Thus, to identify more readily the presence of insulin-producing cells we intercrossed mice bearing the Ptf1aYFP allele with MIP:GFP transgenic animals (Hara et al., 2003). At E18.5, Ptf1aYFP/YFP; MIP:GFP embryos did not exhibit any YFP fluorescence while GFP-positive cells were sparsely distributed in the DPR but not in the spleen (Fig. 7C). β-cell mass appeared to be greatly reduced in Ptf1aYFP/YFP; MIP:GFP embryos compared to MIP:GFP mice (Fig. 7A and 7C). In Ptf1a null mice at E18.5, 83.0 ± 2.0% of insulin producing cells are MafA-positive and appear as single cells and in small clusters associated with the cystic, ductal epithelium of the DPR (Fig. 7D). MafA/insulin co-positive cells had more intense Pdx-1 staining than the MafA-negative insulin-expressing cells (data not shown). In wild type mice, insulin-producing cells were clustered in larger islet-like structures and 91.0 ± 2.0% were positive for MafA (Fig. 7B). While the reduced proportion of MafA-expressing insulin-positive cells in Ptf1a null tissue was statistically significant (P<0.01), the majority of the insulin-producing cells were MafA-positive thereby suggesting that Ptf1a function is not required for β-cell maturation.
Figure 7.
MafA is expressed in insulin-producing cells of Ptf1a null embryos. A) Whole mount fluorescence microscopy of gut sections from E18.5 MIP:GFP embryos indicates β-cell specific GFP fluorescence. B) Confocal immunofluorescence analysis of insulin and MafA in wild type embryos at E18.5. DRAQ5™ was used as a nuclear counterstain. 91.0 ± 2.0% of the insulin-producing cells express MafA in wild type mice indicating that they are mature β-cells. C) Whole mount fluorescence of the MIP:GFP transgene in Ptf1aYFP/YFP embryos at E18.5 allows direct observation of insulin-producing cells since YFP is not expressed past ~E13.5 in Ptf1a null mice. In contrast to previous reports, we do not observe pancreatic endocrine cells in the spleen. Rather they are localized to the DPR. D) Confocal immunofluorescence analysis of insulin and MafA in Ptf1aYFP/YFP embryos at E18.5. DRAQ5™ was used as a nuclear counterstain. 83.0 ± 2.0% of the insulin-producing cells in Ptf1a null embryos express MafA which indicates that the majority also possess properties of mature β-cells. Therefore Ptf1a does not directly contribute to endocrine cell maturation. The ~8.5% decrease of MafA expression in insulin-producing cells is statistically significant (P<0.01), however the apparent reduction in β-cell mass is the more dominant phenotype (compare A and C).
Discussion
Derivation of a Ptf1aLCA allele and insertion of YFP by RMCE
Mice carrying a Ptf1aYFP allele were generated by a combined gene targeting and RMCE strategy in order to facilitate detection and analysis of cells expressing Ptf1a. The Ptf1aYFP allele closely mimicked expression of the endogeneous allele except for a small increase in YFP perdurance compared to Ptf1a in pancreatic tissue. However, the extended detectability of YFP may, in fact, be a useful trait in certain circumstances as it is able to identify cells that have recently terminated Ptf1a transcription. In addition, ES cells carrying the Ptf1aLCA allele will have continuing utility since they enable other mutations and reporters to be rapidly inserted into the Ptf1a gene locus by RMCE.
Pdx-1-positive foregut endodermal cells respond to Ptf1a inducing signals from E10.5 to E13.5
Previous studies using Flk1 null embryos have implicated endothelial cells in the activation of Ptf1a gene transcription in the dorsal, but not the ventral pancreatic bud. This is consistent with the observation that normal aortic endothelium, when co-cultured with Flk1 null dorsal foregut endoderm, restores Ptf1a expression (Yoshitomi and Zaret, 2004). Although the factor(s) that induce Ptf1a expression remain unidentified, our studies of Ptf1aYFP/YFP embryos allow us to propose that the signaling mechanisms that initiate expression of Ptf1a remain active in the dorsal and ventral pre-pancreatic endodermal regions until ~E13.5 and ~E11.5, respectively. The loss of Ptf1aYFP expression after this time is consistent with the pancreatic MPC population having either undergone a change in fate to become some other tissue (e.g., duodenum, CBD) or having entered into a subsequent phase of pancreatic lineage diversification and differentiation. In other words, we propose that posterior foregut endoderm cells that express Pdx-1 have a limited window of competence in which to respond to pro-pancreatic ,signals, and that this interval is shorter in ventral than dorsal foregut endoderm. We consider it likely that the DPR in Ptf1a null embryos is derived from pancreatic MPCs that escape signaling mechanisms which drive alternate fate specifications. Indeed, given that Stanger et al. have shown that depletion of pancreatic MPCs reduces final organ size (Stanger et al., 2007), we suggest that the reduced numbers of pancreatic MPCs contributing to the DPR in Ptf1a null embryos may be due, at least in part, to cell fate conversions, thereby accounting for its abrogated expansion. However, further studies will be necessary to determine whether decreased proliferation and/or increased apoptosis of pancreatic MPCs may also contribute to the diminished size of the DPR in Ptf1a null embryos.
Ptf1a and Pdx-1 are independently required for specification of the ventral pancreas
In E15.5 Ptf1a null embryos there is no VPR visible whereas a DPR is clearly evident. Since, normally, the ventral pancreas buds off from and remains associated with the CBD we hypothesized that the absence of Ptf1a leads to adoption of CBD fates by ventral cells that should have become pancreas. This led us to further examine the results of a previous lineage tracing experiment (Kawaguchi et al., 2002) and to find that ventral pancreas MPCs that lack Ptf1a do indeed contribute to the development of the CBD, as well as the duodenum. This conclusion is consistent with recent studies establishing a common origin for cells of the ventral pancreas and CBD (Fukuda et al., 2006a; Sumazaki et al., 2004).
In Pdx-1 null embryos at E15.5 the DPR is much smaller (Offield et al., 1996) than in Ptf1a null mice at the same age. Additionally, these embryos lack a VPR. As is the case for the Ptf1a null animals it seems that ventral “prospective pancreatic” endodermal cells appear to integrate into the CBD epithelium and become committed to this alternate developmental program at a very early stage. In fact, the specification, production, and/or survival of ventral MPCs appears to be altogether deficient in Pdx-1 null embryos since we never observed ventral Ptf1aYFP expression beyond E10.5.
Neither Pdx-1 nor Ptf1a is required to initiate a dorsal pancreatic program
Recently, Ptf1a lineage tracing in Pdx-1 hypomorphic embryos was used to show that Ptf1a transcription requires higher Pdx-1 expression levels in ventral foregut endoderm compared to dorsal foregut endoderm (Fujitani et al., 2006a). However, here we report for the first time that dorsal Ptf1a expression is independent of Pdx-1 since Ptf1aYFP is regionally activated in the absence of Pdx-1 between E10.5–12.5. Moreover, in Pdx-1−/−; Ptf1aYFP/+ embryos, YFP fluorescence is lost after E12.5, a full day earlier than in Ptf1a null embryos. Because the signals inducing Ptf1aYFP expression are present until at least E13.5 (as described above) pancreatic MPCs may undergo lineage diversification more rapidly in the absence of Pdx-1 than of Ptf1a. Alternatively, the loss of Ptf1aYFP expression could also reflect either the decreased proliferation and/or increased apoptosis of Pdx-1 null pancreatic MPCs. In any case, the earlier depletion of pancreatic MPCs in Pdx-1 null embryos seems likely to account for the smaller size of the DPR compared to Ptf1a null mice.
The analysis of Pdx-1; Ptf1a double homozygous null embryos showed that Ptf1a is initially expressed in the dorsal, but not ventral, pancreatic remnant, which undergoes abrogated expansion and branching in a phenotype identical to that of Pdx-1 null embryos. Therefore, the limited expansion of the Pdx-1 null DPR does not involve Ptf1a, and strongly supports the idea that Pdx-1 is necessary to render the posterior foregut endoderm competent to respond to Ptf1a. Furthermore, the highly localized expression of Ptf1a in double null mutant embryos indicates that there must be an underlying parallel molecular genetic program leading to the specification and outgrowth of the dorsal pancreatic bud.
The co-expression of Pdx-1 and Ptf1a identify pancreatic MPCs
Indirect lineage-labeling methods have shown that both Ptf1a and Pdx-1 are expressed in pancreatic MPC-type cells (Gu et al., 2002; Kawaguchi et al., 2002). Here, we show that both factors are co-expressed in most, if not all, pancreatic MPCs between E9.5–12.5. Given that Ptf1a can induce ectopic pancreas formation from the CBD epithelium in Hes1 null mice (Fukuda et al., 2006a) our observations further support the notion that Pdx-1/Ptf1a co-production is obligatory for specification of pancreatic MPCs. Sox9, which has also recently been shown to play an important role in pancreatic MPCs as evidenced by pancreatic hypoplasia in the absence of this transcription factor, may complement the function of both Pdx-1 and Ptf1a by also promoting the maintenance of multipotency, survival and/or proliferation of pancreatic MPCs (Seymour et al., 2007).
As pancreatic development proceeds, Ptf1a-expression becomes compartmentalized to the periphery of the pancreas epithelium between E11.5–12.5. The recent work of Zhou et al., which complements our findings, also concluded that cells in the distal tips of the pancreas epithelium that express Pdx-1, Ptf1a, cMyc and Cpa1 are tripotent pancreatic MPCs with the ability to differentiate into endocrine, acinar and duct cells (Zhou et al., 2007). At approximately E13, Ptf1a-positive cells appear to switch to a pro-acinar transit amplifying pool (Zhou et al., 2007) and, shortly thereafter, acinar markers begin to be expressed (Chiang and Melton, 2003; Guz et al., 1995). Our findings are also consistent with those of Stanger et al. (Stanger et al., 2007) and Masui et al. (Masui et al., 2007). Indeed, the latter study has indicated that a change in the subunit composition of the trimeric Ptf1 complex occurs at this time.
Although the early pancreatic MPC pool appears homogeneous with respect to the coincidence of both Pdx-1 and Ptf1a, these cells are likely to be heterogeneous with respect to the expression of other genes. At least six types of progenitor cells have previously been identified in the E10.5 pancreas epithelium based on the expression combinations of Pdx-1, Nkx2.2, Nkx6.1, Ptf1a, Ngn3, Pax4, Pax6, Isl1, NeuroD, Gcg, Ins, Sst, and PP as determined by single-cell RT-PCR analysis (Chiang and Melton, 2003). Indeed, one class of cells that was identified express Ptf1a, Pdx-1, and Ngn3. Our results confirm the coincidence of Pdx-1, Ptf1a, and Ngn3 in a subset of pancreatic epithelial cells from E10.5–11.5. However, the precise identification and functional characterization of all pancreatic MPC subtypes will be necessary for understanding pancreas development and how lineage specifications occur in a spatiotemporally regulated manner.
Endocrine cell differentiation and the role of Ngn3
While the ontogeny, fate, and function of endocrine cells formed before E13.5 remains unclear, Ngn3 is required for their formation (Gradwohl et al., 2000). In this study we found pancreatic insulin- and glucagon-producing cells in Pdx-1 null and Pdx-1; Ptf1a double null embryos from E10.5–12.5, which do not express detectable levels of Ngn3. However, Ngn3-positive cells were found in the pre-pancreatic endoderm at E9.5 in the absence of Pdx-1. This suggests that Pdx-1 is required for the sustained derivation of endocrine progenitor cells from a pancreatic MPC pool past E9.5. Given this, it is important to understand how Pdx-1 mediates the specification of pancreatic endocrine progenitors in vivo to increase the efficiency of deriving Ngn3-expressing cells from human ES cells in vitro.
In contrast to Pdx-1 deficiency, Ptf1a null pancreatic tissue contained close to wild-type numbers of readily detectable Ngn3-producing cells at E11.5, a finding that agrees with the study of Flk-1−/− embryos where dorsal pancreatic Ngn3 transcription was activated in the absence of Ptf1a induction (Yoshitomi and Zaret, 2004). Although one previous study of Ptf1a null mice reported the identification of endocrine cells in close proximity to the pancreatic region until ~E17.5, at which time they were inferred to migrate into the spleen (Krapp et al., 1998), we were unable to identify such splenic endocrine cells, even in Ptf1aYFP/YFP embryos that were analyzed just prior to parturition. Instead, by using a MIP:GFP transgene to facilitate the identification of pancreatic β-cells, we found these cells scattered along the DPR. While Kawaguchi et al. also reported finding splenic endocrine cells in Ptf1a null embryos (Kawaguchi et al., 2002), review of the primary data indicates that the former conclusion can no longer be supported. The faint immunostaining observed previously by Kawaguchi et al. appears to have been non-specific since, in the current study, GFP-positive cells were not observed in the spleens of Ptf1aYFP/YFP; MIP:GFP embryos at E18.5.
The insulin-producing cells found in the Ptf1a null mice were found to express MafA, a marker of late-stage β-cell maturation (Matsuoka et al., 2004). While the physiological function of these β-cells could not be tested directly because Ptf1a null animals die perinatally, this result supports the previously proposed hypothesis that the normal pancreas has both Ptf1a-dependent and Ptf1a-independent pancreatic endocrine lineages, wherein only 95% of adult duct and insulin cell types, and 75% of glucagon cells, were labeled in Ptf1a lineage-tracing studies (Kawaguchi et al., 2002). The expression of MafA in the insulin-producing cells of Ptf1a null mice further supports the notion for there being more than one natural pathway for generating mature β-cells in vivo. A corollary of these findings is that these different developmental programs might have specific properties that make them more or less amenable to large-scale recapitulation in vitro. It therefore remains possible that, although Ptf1a is not absolutely required for the generation of mature β-cells in vivo, the induction of a Ptf1a-expressing MPC state may be necessary for the efficient derivation of β-cells.
Finally, our results may have direct bearing on the derivation of insulin-expressing cells from human ES cells (D'Amour et al., 2006). If pancreatic MPCs have a very restricted proliferative capacity, as several studies now suggest, greater knowledge of the factors that determine both the specification and maintenance of this important pool of cells may in time enable the development of novel strategies for either regenerating specific pancreatic lineages, such as β-cells, in vivo, or generating such cells ex vivo through the directed differentiation of human ES cells, or other such pluripotent cells.
Supplementary Material
Acknowledgments
This work was supported by NIH grants DK42502 and DK72473. We thank Susan Hipkens for help performing the RMCE experiment and Jill Lindner for assistance with mouse husbandry. We also thank the staff of the Vanderbilt Transgenic/ES Cell Shared Resource for performing the gene targeting and blastocyst microinjections and the Vanderbilt Cell Imaging Shared Resource for help acquiring the confocal images. We also thank Ray MacDonald for reading the manuscript and for providing helpful comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afelik S, et al. Combined ectopic expression of Pdx1 and Ptf1a/p48 results in the stable conversion of posterior endoderm into endocrine and exocrine pancreatic tissue. Genes Dev. 2006;20:1441–1446. doi: 10.1101/gad.378706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlgren U, et al. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in IPF1/PDX1-deficient mice. Development. 1996;122:1409–1416. doi: 10.1242/dev.122.5.1409. [DOI] [PubMed] [Google Scholar]
- Apelqvist A, et al. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- Chiang MK, Melton DA. Single-cell transcript analysis of pancreas development. Dev Cell. 2003;4:383–393. doi: 10.1016/s1534-5807(03)00035-2. [DOI] [PubMed] [Google Scholar]
- D'Amour KA, et al. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- Feng YQ, et al. Site-specific chromosomal integration in mammalian cells: highly efficient CRE recombinase-mediated cassette exchange. J Mol Biol. 1999;292:779–785. doi: 10.1006/jmbi.1999.3113. [DOI] [PubMed] [Google Scholar]
- Fujitani Y, et al. Targeted deletion of a cis-regulatory region reveals differential gene dosage requirements for Pdx1 in foregut organ differentiation and pancreas formation. Genes Dev. 2006a;20:253–266. doi: 10.1101/gad.1360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitani Y, et al. Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development. 2006b;133:4439–4450. doi: 10.1242/dev.02598. [DOI] [PubMed] [Google Scholar]
- Fukuda A, et al. Ectopic pancreas formation in Hes1 -knockout mice reveals plasticity of endodermal progenitors of the gut, bile duct, and pancreas. J Clin Invest. 2006a;116:1484–1493. doi: 10.1172/JCI27704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda A, et al. Loss of the major duodenal papilla results in brown pigment biliary stone formation in pdx1 null mice. Gastroenterology. 2006b;130:855–867. doi: 10.1053/j.gastro.2005.11.031. [DOI] [PubMed] [Google Scholar]
- Glasgow SM, et al. Ptf1a determines GABAergic over glutamatergic neuronal cell fate in the spinal cord dorsal horn. Development. 2005;132:5461–5469. doi: 10.1242/dev.02167. [DOI] [PubMed] [Google Scholar]
- Gradwohl G, et al. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu G, et al. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- Guz Y, et al. Expression of murine STF-1, a putative insulin gene transcription factor, in beta cells of pancreas, duodenal epithelium and pancreatic exocrine and endocrine progenitors during ontogeny. Development. 1995;121:11–18. doi: 10.1242/dev.121.1.11. [DOI] [PubMed] [Google Scholar]
- Hara M, et al. Transgenic mice with green fluorescent protein-labeled pancreatic beta -cells. Am J Physiol Endocrinol Metab. 2003;284:E177–E183. doi: 10.1152/ajpendo.00321.2002. [DOI] [PubMed] [Google Scholar]
- Heikal AA, et al. Molecular spectroscopy and dynamics of intrinsically fluorescent proteins: coral red (dsRed) and yellow (Citrine) Proc Natl Acad Sci U S A. 2000;97:11996–12001. doi: 10.1073/pnas.97.22.11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera PL. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development. 2000;127:2317–2322. doi: 10.1242/dev.127.11.2317. [DOI] [PubMed] [Google Scholar]
- Herrera PL, et al. Two transgenic approaches to define the cell lineages in endocrine pancreas development. Mol Cell Endocrinol. 1998;140:45–50. doi: 10.1016/s0303-7207(98)00028-8. [DOI] [PubMed] [Google Scholar]
- Holland AM, et al. A mouse carrying the green fluorescent protein gene targeted to the Pdx1 locus facilitates the study of pancreas development and function. Genesis. 2006;44:304–307. doi: 10.1002/dvg.20214. [DOI] [PubMed] [Google Scholar]
- Hoshino M, et al. Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron. 2005;47:201–213. doi: 10.1016/j.neuron.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Jensen J. Gene regulatory factors in pancreatic development. Dev Dyn. 2004;229:176–200. doi: 10.1002/dvdy.10460. [DOI] [PubMed] [Google Scholar]
- Jonsson J, et al. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, et al. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- Krapp A, et al. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 1998;12:3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q, et al. Efficient DNA cassette exchange in mouse embryonic stem cells by staggered positive-negative selection. Genesis. 2004;39:256–262. doi: 10.1002/gene.20053. [DOI] [PubMed] [Google Scholar]
- Masui T, et al. Early pancreatic development requires the vertebrate Suppressor of Hairless (RBPJ) in the PTF1 bHLH complex. Genes Dev. 2007 doi: 10.1101/gad.1575207. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka TA, et al. The MafA transcription factor appears to be responsible for tissue-specific expression of insulin. Proc Natl Acad Sci U S A. 2004;101:2930–2933. doi: 10.1073/pnas.0306233101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh LC, Melton DA. Genes, signals, and lineages in pancreas development. Annu Rev Cell Dev Biol. 2003;19:71–89. doi: 10.1146/annurev.cellbio.19.111301.144752. [DOI] [PubMed] [Google Scholar]
- Obata J, et al. p48 subunit of mouse PTF1 binds to RBP-Jkappa/CBF-1, the intracellular mediator of Notch signalling, and is expressed in the neural tube of early stage embryos. Genes Cells. 2001;6:345–360. doi: 10.1046/j.1365-2443.2001.00422.x. [DOI] [PubMed] [Google Scholar]
- Offield MF, et al. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Osoegawa K, et al. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 2000;10:116–128. [PMC free article] [PubMed] [Google Scholar]
- Sellick GS, et al. Mutations in PTF1A cause pancreatic and cerebellar agenesis. Nat Genet. 2004;36:1301–1305. doi: 10.1038/ng1475. [DOI] [PubMed] [Google Scholar]
- Seymour PA, et al. SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc Natl Acad Sci U S A. 2007;104:1865–1870. doi: 10.1073/pnas.0609217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanger BZ, et al. Organ size is limited by the number of embryonic progenitor cells in the pancreas but not the liver. Nature. 2007 doi: 10.1038/nature05537. [DOI] [PubMed] [Google Scholar]
- Stoffers DA, et al. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat Genet. 1997;15:106–110. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- Sumazaki R, et al. Conversion of biliary system to pancreatic tissue in Hes1-deficient mice. Nat Genet. 2004;36:83–87. doi: 10.1038/ng1273. [DOI] [PubMed] [Google Scholar]
- Wilson ME, et al. Gene expression cascades in pancreatic development. Mech Dev. 2003;120:65–80. doi: 10.1016/s0925-4773(02)00333-7. [DOI] [PubMed] [Google Scholar]
- Yoshitomi H, Zaret KS. Endothelial cell interactions initiate dorsal pancreas development by selectively inducing the transcription factor Ptf1a. Development. 2004;131:807–817. doi: 10.1242/dev.00960. [DOI] [PubMed] [Google Scholar]
- Zhou Q, et al. A multipotent progenitor domain guides pancreatic organogenesis. Dev Cell. 2007;13:103–114. doi: 10.1016/j.devcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.