Abstract
Plasmodium falciparum, the agent of malignant malaria, is one of mankind’s most severe scourges. Efforts to develop preventive vaccines or remedial drugs are handicapped by the parasite’s rapid evolution of drug resistance and protective antigens. We examine 25 DNA sequences of the gene coding for the highly polymorphic antigenic circumsporozoite protein. We observe total absence of silent nucleotide variation in the two nonrepeated regions of the gene. We propose that this absence reflects a recent origin (within several thousand years) of the world populations of P. falciparum from a single individual; the amino acid polymorphisms observed in these nonrepeat regions would result from strong natural selection. Analysis of these polymorphisms indicates that: (i) the incidence of recombination events does not increase with nucleotide distance; (ii) the strength of linkage disequilibrium between nucleotides is also independent of distance; and (iii) haplotypes in the two nonrepeat regions are correlated with one another, but not with the central repeat region they span. We propose two hypotheses: (i) variation in the highly polymorphic central repeat region arises by mitotic intragenic recombination, and (ii) the population structure of P. falciparum is clonal—a state of affairs that persists in spite of the necessary stage of physiological sexuality that the parasite must sustain in the mosquito vector to complete its life cycle.
Keywords: malaria, circumsporozoite protein, genetic polymorphism, mitotic recombination, clonality
There are 300–500 million clinical cases of malaria per year, more than 1 million children die in Sub-Saharan Africa, and more than 2 billion people are at risk throughout the world (1). Plasmodium falciparum is the agent of malignant malaria, the most fatal version of the disease. Malaria has been an elusive target for medical intervention. Epidemiological control efforts first were directed against the Anopheles mosquito vectors, which soon evolved resistance to massively applied insecticides. Current medicine seeks development of protective vaccines or remedial drugs directly against the parasite. These exertions are handicapped, however, by the parasite’s rapid evolution of multidrug resistance and multiple protective antigens. Underlying this evolution is a wealth of genetic variation that seemingly recombines rapidly to generate ever newly protected phenotypes. The human active form of the parasite is haploid, but diploidy occurs in the mosquito vector, where fertilization takes place, and newly haploid organisms (sporozoites) are formed that are transmitted from the mosquito’s salivary glands to human blood vessels.
Protective immunity against P. falciparum was demonstrated in the 1970s by immunization of human patients with irradiated sporozoites (2). Parasite genes that code for antigenic determinants subsequently have been isolated and characterized. One of these genes, coding for the circumsporozoite protein, has been extensively investigated and chosen as the target for vaccine development (e.g., ref. 3). The success of efforts for developing an effective malaria vaccine is contingent on determining the extent of diversity of the gene of the circumsporozoite protein (Csp), but also on identifying the mechanisms by which this variation is generated and persists in populations of P. falciparum.
Numerous studies indicate that Csp and other antigenic genes are polymorphic and that their multiple allelic forms differ in their ability to abrogate recognition by the host’s immune response (4–6). These data have been interpreted as instantiation of widespread polymorphism throughout the genome. Yet, we have investigated allelic variation in a diverse set of nine gene loci and found a complete absence of silent site polymorphism (unpublished results), which most likely is because of a recent derivation (within a few thousand years) of all extant P. falciparum populations from a single propagule. It seems, therefore, paradoxical that Csp and other P. falciparum genes would be so highly polymorphic, because these genes must have shared the recent allelic homogenization caused by the population bottleneck. A hypothesis that would reconcile the recent origin of the widely dispersed populations of P. falciparum with the rich polymorphism of the antigenic genes is that P. falciparum has specific mechanisms for rapidly generating antigenic variability. We assess this hypothesis by investigating the DNA sequence polymorphisms of known allelic variants of the P. falciparum Csp. We seek to ascertain not only the pattern and process by which variation arises in this important antigen, but also their bearing on the population structure of P. falciparum.
MATERIALS AND METHODS
DNA Sequences.
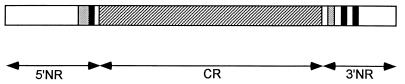
The Csp of P. falciparum consists of an amino terminus coding region [5′nonrepeat region (NR)], a central region of tandem repeats (CR), and a carboxyl-terminus coding region (3′NR) (Fig. 1). The CR consists of numerous repeats coding for 4-aa-long motifs, which in our sample of P. falciparum are of two kinds: NANP (Asn, Ala, Asn, and Pro), repeated 36–49 times per gene, and NVDP (Asn, Val, Asp, and Pro), repeated 2–4 times per gene. In the chimpanzee parasite, P. reichenowi, these two repeats occur 26 and five times, respectively, and a third motif, NVNP (Asn, Val, Asn, and Pro) is repeated four times. The CR is clearly delineated by invariant 12-nt-long sequences at the 5′ (AATCCTGATCCA) and 3′ (AATAAAAACAAT) boundaries.
Figure 1.
Structure of the P. falciparum Csp. 5′NR and 3′NR brace the CR (hatched) made up of a variable number of tandem repeats encoding 4-aa-long motifs. The light gray boxes represent B-cell epitopes; the dark boxes represent T-cell epitopes. The gene length ranges 1,194–1,413 nucleotides because of two 5′ NR indels and a variable number of CRs.
Table 1 lists the 25 complete coding sequences of the P. falciparum Csp (obtained from GenBank) and the strains’ geographic origins, representative of the major malarial regions. The GenBank accession numbers hereafter will be used to refer to the sequences. The P. reichenowi sequence is used for outgroup comparisons.
Table 1.
P. falciparum strains, geographic origin, accession numbers, and source references
| Strain | Origin | Accession no. | Reference |
|---|---|---|---|
| WELLCOME | West Africa | M15505 | (34) |
| T9/94 | Thailand | M83173 | (35) |
| 806 | Thailand | M83149 | (35) |
| 807 | Thailand | M83150 | (35) |
| 827 | Thailand | M83156 | (35) |
| 834a | Thailand | M83158 | (35) |
| 835b | Thailand | M83161 | (35) |
| 836 | Thailand | M83163 | (35) |
| 837 | Thailand | M83164 | (35) |
| 838 | Thailand | M83165 | (35) |
| 841 | Thailand | M83166 | (35) |
| 842 | Thailand | M83167 | (35) |
| 843 | Thailand | M83168 | (35) |
| 844 | Thailand | M83169 | (35) |
| 946 | Thailand | M83170 | (35) |
| K1 | Thailand | M83174 | (35) |
| T4 | Thailand | M19752 | (36) |
| MAD20 | Papua New Guinea | M83172 | (35) |
| IMTM22 | Brazil | K02194 | (35) |
| T9-101 | Thailand | M57499 | (37) |
| Sal-1 | Santa Lucia | U20969 | ∗ |
| CVD1 | Netherlands | M83886 | (38) |
| NF54 | Netherlands | M22982 | (39) |
| 3D7 | Netherlands | X15363 | (40) |
| T9-98 | Thailand | M57498 | (37) |
| P. reichenowi | M60972 | (41) |
, S. H. Qari and A. A. Lal, personal communication.
Alignment and Phylogenetic Analysis.
We align each of the three gene regions separately by means of a progressive multiple-sequence alignment algorithm, by using the clustalw computer program (7), with corrections made by eye. For the phylogenetic analysis of the CR we use the branch-and-bound search method with maximum parsimony optimization of paup (8). The sequences are aligned by creating gaps as needed so that identical amino acid motifs align with one another; the corresponding nucleotides are the characters.
Recombination Tests.
We estimate Rm, the minimum number of recombination events, within and between the 5′NR and 3′NR by means of the four-gamete test of ref. 9, by using the dnasp2 program (10). Rm tends to underestimate the number of recombination events that have occurred during the history of the sequences represented in the sample.
We test for linkage disequilibrium between the 5′NR and 3′NR by using the D statistic of ref. 11. Singleton polymorphic sites (those in which only a single sequence contains the variant nucleotide) are not included in the analysis. The intervening CR is variable in length owing to variation in the number of repeats; therefore, we specify nucleotide distances between the 5′NR and 3′NR according to M15505. The statistical significance of D is determined with Fisher’s exact test. Linkage tests are performed with the dnasp2.51 program (10).
RESULTS
The amino acid polymorphisms present in the NRs are shown in Table 2. No silent substitutions are in these regions, and all amino acid replacements occur in the segments identified as B-cell or T-cell epitopes (Fig. 1), which are involved in the parasite’s evasion of the human host’s immune system (12, 13). By comparison with the P. reichenowi Csp, we determine that there are two indels: a 30-bp insertion (present in M15505 and M83173) and a 57-bp deletion (absence in M83886, M22982, X15363, and M57498) directly adjacent to each other, and proximal to the 5′NR putative B-cell epitope located at amino acid sites 118–132 (14). Two singleton polymorphisms occur within the deletion (at amino acid sites 96 and 97). Two other polymorphisms occur within the 5′NR B-cell epitope, at sites 114 (a singleton) and 127 (variant only in Santa Lucia, one Thailand, and the three Netherlands sequences). In the 3′NR there is a B-cell epitope (amino acid sites 334–348) and two T-cell epitopes (sites 363–373 and 398–407). Several amino acid polymorphisms occur in these epitopes (Table 2).
Table 2.
Amino acid polymorphisms of the 5′NR and 3′NR in the Csp of P. falciparum
| Strain | Codon position
|
||
|---|---|---|---|
| 5′NR
|
[repeat] | 3′NR
|
|
| 1111111111 | 33333333334444 | ||
| 47888888888899999999990000000012 | 34466677790000 | ||
| 59012345678901234567890123456747 | 47834701380357 | ||
| M15505 | TDNDNGNNNNGNNNNGDNGREGKDEDKRDGKG | DNAEQKQNLDPQDE | |
| M83173 | I............................... | .............. | |
| M83149 | I----------..................... | .........N.E.. | |
| M83150 | I----------..................... | ...TE....GSE.. | |
| M83156 | I----------..................... | ..D....Y...... | |
| M83158 | I----------..................... | .........N.E.. | |
| M83161 | I----------..................... | ...KET..IG.EE. | |
| M83163 | I----------..................... | ...TE....GSE.. | |
| M83164 | I----------..................... | ...TE....GSE.. | |
| M83165 | I----------..................... | .........N.E.. | |
| M83166 | I----------..................... | .........N.E.. | |
| M83167 | I----------..................... | ..D....Y...... | |
| M83168 | I----------..................... | .........N.E.. | |
| M83169 | I----------..................... | .........N.E.. | |
| M83170 | I----------..................... | .........N.E.. | |
| M83174 | I----------..................... | ..D....Y...... | |
| M19752 | .----------.......N...........T. | .........N.E.. | |
| M83172 | I----------..................... | .............. | |
| K02194 | .----------..................... | ......K.IN.E.. | |
| M57499 | .----------..................... | .........N.E.. | |
| U20969 | .----------........G...........A | N........GSE.. | |
| M83886 | .-----------------------------.A | .S.KEN...N.E.A | |
| M22982 | .-----------------------------.A | .S.KEN...N.E.A | |
| X15363 | .-----------------------------.A | .S.KEN...N.E.A | |
| M57498 | .-----------------------------.A | ......K..N.E.A | |
Dots (.) indicate identity with M15505, dashes (-) represent gaps.
We tested first for recombination events within and between the 5′NR and 3′NR. The four-gamete test (9) manifests a minimum of four recombinant events among the 25-gene sequences of P. falciparum (see Table 2). One recombination event is within the 5′NR (somewhere between amino acid site 45 and the 30-bp insertion—the lack of precision is because of the identity of all sequences between sites 46 and 78), two others are within the 3′NR (between sites 363 and 364 and somewhere between 373 and 398); the fourth recombination event (somewhere between amino acid sites 127 and 363) spans the CR.
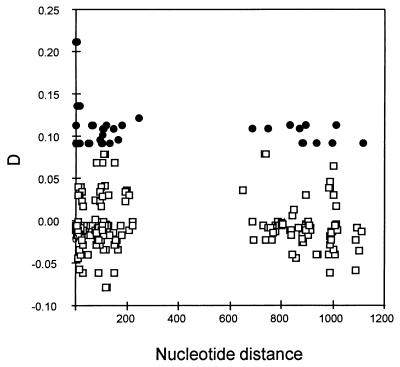
Next, we tested for linkage disequilibrium within and between the 5′NR and 3′NR. We made 253 pairwise comparisons involving 23 polymorphic sites (we treat the two indels as individual polymorphisms), five in the 5′NR and 18 in the 3′NR. Forty-two of the comparisons reveal significant correlation between sites. The number of pairwise comparisons within the 5′NR or within the 3′NR amount to 163, of which 29 (18.5%) yield significant disequilibrium values (P < 0.05); the distance between sites in these comparisons is ≤ 247 nucleotides. The remaining 90 comparisons are between the 5′NR and 3′NR, 13 (13.8%) of which are significantly correlated (P < 0.05); the distance between sites in these comparisons is ≥ 649 nucleotides. Fig. 2 plots the value of D (which measures linkage disequilibrium, ref. 11) against nucleotide distance. The magnitude of D is independent of nucleotide distance—the linkage is as strong between the 5′NR and 3′NR as it is between proximal sites within the same region.
Figure 2.
Linkage disequilibrium (D) as a function of nucleotide distance. Comparisons are made within and between 5′NR and 3′NR sites. Significant D values (P < 0.05) are represented by •; nonsignificant values are represented by □.
The immunodominant CR is recognized as the most polymorphic domain of the Csp. This region typically has not been included in analyses of nucleotide diversity because of difficulties in obtaining an appropriate alignment. Only two amino acid motifs occur among the 25 Csp sequences of P. falciparum: NANP (1 in Table 3) is present 1,032 times (41.28 ± 3.1 per sequence) and NVDP (2 in Table 3) is present 88 times (3.5 ± 0.6 per sequence). At the nucleotide level there are 10 variants of the NANP motif (plus one more in P. reichenowi) and four variants of NVDP, which occur with vastly different frequencies (Table 4). We refer to these variant nucleotide sequences as the repeat allotypes (RATs).
Table 3.
Composition of the CR of the Csp and its association with the NRs
| Sequence | Repeat motifs | Type
|
Number of repeats
|
|||
|---|---|---|---|---|---|---|
| 5′NR | 3′NR | 1 | 2 | 3 | ||
| M15505 | 1212111111111111111211111111111111111111111111 | III | III | 43 | 3 | 0 |
| M83173 | 1212111111111111111211111111111111111111111111 | IV | III | 43 | 3 | 0 |
| M83149 | 12121211111111111111111111111111111111111111 | I | I | 41 | 3 | 0 |
| M83150 | 12121111111111111112111111111111111111111111111 | I | IV | 44 | 3 | 0 |
| M83156 | 121211111111111111111111111111111111111111111111111 | I | VIII | 49 | 2 | 0 |
| M83158 | 1212121211111111111111111111111111111111111111 | I | I | 42 | 4 | 0 |
| M83161 | 1212121111111111111111111211111111111111111 | I | VII | 39 | 4 | 0 |
| M83163 | 12121111111111111112111111111111111111111111111 | I | IV | 43 | 3 | 0 |
| M83164 | 12121111111111111112111111111111111111111111111 | I | IV | 46 | 3 | 0 |
| M83165 | 1212121111111111111111111111111111111111111111 | I | I | 43 | 3 | 0 |
| M83166 | 1212121211111111111111111111111111111111111111 | I | I | 42 | 4 | 0 |
| M83167 | 12121211111111111111111111111111111111111111111 | I | VIII | 46 | 3 | 0 |
| M83168 | 1212121211111111111111111111111111111111111111 | I | I | 42 | 4 | 0 |
| M83169 | 12121211111111111111111111111111111111111111 | I | I | 41 | 3 | 0 |
| M83170 | 1212121211111111111111111111111111111111111111 | I | I | 42 | 4 | 0 |
| M83174 | 1212121111111111211111111111111111111111111 | I | VIII | 39 | 4 | 0 |
| M19752 | 12121211111111111111111111111111111111111111 | V | I | 41 | 3 | 0 |
| M83172 | 121212111111111111111121111111111111111111 | I | III | 38 | 4 | 0 |
| K02194 | 12121211111111111111121111111111111111111 | VI | VI | 37 | 4 | 0 |
| M57499 | 12121212111111111111111111111111111111111111 | VI | I | 40 | 4 | 0 |
| U20969 | 1212121111111111111112111111111111111111 | VII | V | 36 | 4 | 0 |
| M83886 | 121212111111111111111112111111111111111111 | II | II | 38 | 4 | 0 |
| M22982 | 12121211111111111111111112111111111111111111 | II | II | 40 | 4 | 0 |
| X15363 | 12121211111111111111111112111111111111111111 | II | II | 40 | 4 | 0 |
| M57498 | 12121211111111111111121111111111111111111 | II | IX | 37 | 4 | 0 |
| P. reichenowi | 12121212131213131311111111111111111 | — | — | 26 | 5 | 4 |
The repeat motifs NANP, NVDP, and NVNP are represented by 1, 2, and 3, respectively. The Roman numbers refer to different NR amino acid sequences (see Table 2).
Table 4.
Amino acid and nucleotide sequences of the RATs
| RAT | Motif
|
% | Number | |
|---|---|---|---|---|
| Amino acid* | Nucleotide | |||
| A | NANP | aat gca aac cca | 55.1 | 566 |
| B | NANP | ... ... ..t ..t | 16.1 | 165 |
| C | NANP | ... ... ..t ... | 7.6 | 78 |
| D | NANP | ... ..c ..t ..a | 6.2 | 64 |
| E | NANP | ... ... ... ..c | 6.2 | 64 |
| F | NANP | ..c ... ... ..c | 5.1 | 52 |
| G | NANP | ... ..c ... ... | 3.1 | 32 |
| H | NANP | ... ... ... ..t | 0.3 | 3 |
| I | NANP | ..c ... ... ... | 0.2 | 2 |
| J | NANP | ... ..c ... ..c | 0.1 | 1 |
| Z† | NANP | ... ... ..t ..c | — | — |
| M | NVDP | ... .t. g.t ... | 52.3 | 46 |
| N | NVDP | ... .t. g.t ..c | 31.8 | 28 |
| O | NVDP | ..c .t. g.t ..t | 14.8 | 13 |
| P | NVDP | ... .t. g.t ..t | 1.1 | 1 |
| X† | NVNP | ... .t. ..t ..c | — | — |
% refers to the RAT incidence within its amino acid motif.
The composition of the amino acid motifs is NANP: Asn, Ala, Asn, Pro; NVDP: Asn, Val, Asp, Pro; NVNP: Asn, Val, Asn, Pro.
RATs found exclusively in P. reichenowi.
The NVDP motif is always preceded and followed by NANP. It seems as if the CR is made of two repeating units, the doublet 1–2 and the singlet 1. (In P. reichenowi the repeating units appear to be 1–2, 1–3, and 1.) Changes in the number of repeats within the CR can be accounted for by a process of duplication and deletion of these repeating units. The RAT composition of the P. falciparum sequences is displayed in Fig. 3, where identical RATs have been aligned. There is considerable conservation of organization along the sequences, with variations in length readily accountable as random duplications (e.g., seven extra A’s in M83156; the set BDCAF repeated in eight sequences), or deletions (e.g., the doublet BE or the singlet E in M15505, M83173, M83150, M83161, M83163, M83164, and M83174).
Figure 3.
Alignment of the RATs of the CR of the Csp. Letter codes for RATs are given in Table 4. The amino acid motif NVDP is shaded; NANP is unshaded. Discordant sites are shown in open boxes.
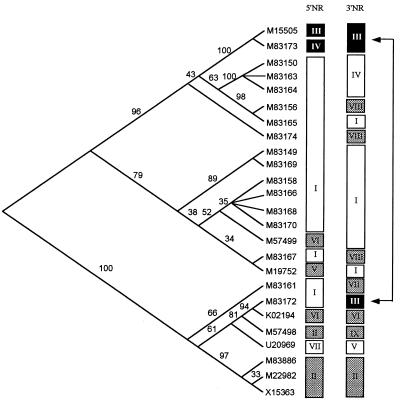
Fig. 4 shows a cladogram of the 25 RAT sequences, showing their association with the seven distinct haplotypes of the 5′NR and nine haplotypes of the 3′NR.
Figure 4.
Maximum parsimony tree based on the nucleotide sequence of the CR, by using the alignment in Fig. 3. Displayed is the association of the CR with the 5′ and 3′NR types (see Table 3); shadings are for clarity. Bootstrap values are given for each branch. Arrows indicate a possible recombinant event.
DISCUSSION
Polymorphisms in the Csp of P. falciparum are not randomly distributed along the gene sequence, but rather they are restricted to the B- and T-cell epitopes, used by the parasite in evading the immune defense of the human host. All segregating nucleotide sites in the NRs of the gene are found in nonsynonymous sites; silent polymorphisms are totally absent. The absence of amino acid variation in the nonantigenic regions of the protein and of silent polymorphism has been attributed to strong selective constraints on the circumsporozoite protein (15). Our survey of all complete coding sequences available in GenBank has not manifested any segregating silent sites in the 5′NR or 3′NR. The only third codon position sites that are polymorphic occur in codons 45, 367, and 405 (Table 2), but they are associated with, or result in amino acid replacements, namely, ACT (Thr)→ATC (Ile), AAG (Lys)→AAC (Asn), and GAT (Asp)→GAA (Glu), respectively.
The polymorphisms in the 5′NR and 3′NR most likely arise by a process of nucleotide point mutation followed by selection. In a retrospective study of Csp alleles originating from 50-year-old natural infections of P. falciparum, Qari et al. (12) conclude that these point mutations have multiple independent origins. This conclusion is supported by the observation that particular mutations do not follow the descent relationships of the strains that possess them, i.e., they are of independent origin (“homoplasic,” ref. 16). Furthermore, the amount of allelic variation in these NRs that is found in an endemic village is typically equal to the observed global antigenic diversity (12), which suggests, moreover, that these alleles are maintained by positive selection (see ref. 17).
It has been estimated that the allelic variants of the Csp NRs of P. falciparum are about 2 million years old, and thus may have coevolved with the human host’s immune factors that target their particular epitopes (18). The time estimate is based on a nonsynonymous substitution rate (3.0 × 10−9 per site per year) derived from comparisons between P. falciparum and rodent Plasmodium species, assuming a clock-like behavior for circumsporozoite protein. However, strong selection is known to act on the circumsporozoite protein (3, 4), which is most effective in large populations—millions of humans are infected by P. falciparum, and one single patient may harbor 1010 parasites (19). The evolution of nonsynonymous substitutions therefore may be quite inconsistent with a molecular clock. Furthermore, the epitope targets of immune selection may vary along the gene sequence from one to another Plasmodium species. Thus, the B- or T-cell epitope polymorphisms of the P. vivax gene that are likely to be maintained by balancing selection have homologous regions that in P. falciparum are monomorphic and seem unrelated to any antigenic determinant (20).
We propose, instead, the hypothesis that antigenic variation in the P. falciparum Csp is of recent evolutionary origin and has arisen in response to strong selection acting on large parasite populations. Silent nucleotide polymorphisms are more likely to escape selection and evolve as a molecular clock than nonsynonymous polymorphisms. We have found a total absence of silent nucleotide variation in nine P. falciparum gene loci and have calculated that all extant populations of P. falciparum derive from a single propagule that lived less than 70,000 years ago (unpublished results), even though P. falciparum diverged several million years ago from its closest known relative, P. reichenowi (21). Selective constraints directed against silent variation cannot account for its absence in P. falciparum, because silent sites are extensively variable among Plasmodium species. The absence of silent variation in the 5′NR and 3′NR of Csp also supports the recent origin of P. falciparum world populations. Therefore, the nonsilent polymorphisms of the 5′NR and 3′NR are likely to have arisen recently, within the last several thousand years, probably in response to strong selection imposed by the host’s immune response. The observation that linkage disequilibrium between polymorphic sites is not smaller between than within the 5′NR and 3′NR suggests that the entire region is tightly linked and would have a similar history; i.e., the variation in the CR has a recent origin as well.
If the world populations of P. falciparum share recent ancestry, we need to account for the occurrence of silent and nonsilent polymorphisms in the CR. There are a number of silent substitutions among the 10 distinct nucleotide sequences (RATs) that encode the NANP amino acid motif of P. falciparum; others exist among the four RATs that encode the NVDP motif (Table 4). The substantial variability that occurs within the CR is largely accounted for by the variation in the number of repeats and their sequence (Table 3). The alignment shown in Fig. 3 indicates that the pattern of CR variation can be readily generated by shuffling and insertion/deletion through intragenic recombination between RATs. The persistence of only two amino acid motifs and their distinctive CR arrangements suggests that their integrity is maintained by selective pressure.
The RAT alignments displayed in Fig. 3 make possible to see how silent polymorphism would be present in the CR, vis-à-vis its absence in the 5′NR and 3′NR and in other genes. As is apparent in Fig. 3, any one of the 25 falciparum Csp sequences includes most of the RATs (at least eight of them). In fact every P. falciparum sequence contains one or more copies of each of the most common RATs of NANP (A, B,C, D, E, F, and G; accounting for 99.4% of all NANP motifs); and most sequences have all three common RATs of NVDP (M, N, and O; accounting for 98.9% of all NVDP motifs). Therefore, most of the RAT sequence variation likely could have been included in a single ancestral P. falciparum genome. Shuffling through intragenic recombination would rapidly generate the variation observed among the current falciparum sequences.
We propose that the Csp evidence favors the hypothesis that the population structure of P. falciparum is clonal (22) and that variation is generated by intragenic mitotic exchanges rather than by sexual recombination. Genetic variation can be created by intragenic recombination whenever diversified repeated motifs are present as is the case in the CR of P. falciparum Csp. Intragenic recombination events have been reported in the genes coding for the two major merozoite antigens, MSA-1 and MSA-2, and this result has been invoked as evidence against the hypothesis that P. falciparum has a clonal population structure (23, 24). However, merozoite surface antigens are subject to strong selection for diversity (25), which will drive even rare recombinant types to high frequency and maintain them in the population. The consequences of strong selection for antigenic diversity may well give the false impression that recombination is frequent when it may, in fact, be selection that enhances and preserves the outcomes of extremely rare recombination events. Moreover, the observation that recombination can generate diversity is not inconsistent with prevailing clonality. A clonal population structure implies that meiotic recombination is rare, not that it is totally lacking; but it impacts the distribution of genetic variation in populations and has long-term evolutionary consequences (22). It generally is accepted, for example, that Escherichia coli has a clonal population structure and that recombination between strains (clonal lineages) is extremely rare; yet the identification of recombinant segments within genes is not uncommon (26).
The evidence that we have elucidated in the case of Csp does not support the interpretation that intragenic exchanges result from sexual (meiotic) recombination. Intragenic recombination may occur by either one or two mechanisms: (i) interhelical exchanges associated with meiosis, which in the case of unequal crossing-over will increase or reduce the number of random repeated motifs; or (ii) intrahelical, by a mitotic slipped-strand exchange most frequently associated with simple repetitive DNA sequences, such as micro- and mini-satellites and variable numbers of tandem repeats (27). Interhelical recombination between dissimilar parasite strains will generate new variants, but so will asexual intrahelical exchanges whenever there is variation along the sequence.
Three items of Csp evidence favor mitotic, rather than meiotic, recombination as the mechanism that generates CR variation. First, the descent relationships show that the 5′NR and 3′NR types are mutually associated, but not with the RATs of the CR (Fig. 4). If we exclude singletons, there is only one case in which a particular 3′NR haplotype is associated with more than one 5′NR haplotype (indicated by arrows in Fig. 4), which might reflect a recombination event, but also could result from homoplasy (i.e., independent origin) (16). In all other cases there is a strong correlation between the 5′NR and 3′NR haplotypes, but not with the CR. For example, sequences M83156, M83167, and M83174 exhibit identical 5′NR and 3′NR haplotypes but have disparate RAT sequences (Table 2 and Fig. 3). Second, the incidence of recombination events does not increase with distance along the sequence, which would be expected if recombination came about by meiotic crossing-over. We detected four recombination events, one within the 5′NR, two within the 3′NR, and only one spanning across the CR. Third, the strength of linkage disequilibrium does not decrease with distance along the DNA sequence, contrary to what is expected with meiotic crossing-over (Fig. 2).
P. falciparum is haploid in the human host, but goes through fertilization and diploidy in the mosquito vector. The evidence just reviewed is not inconsistent with the possible occurrence of meiotic recombination and interhelical exchanges between identical DNA haplotypes, which might be the case if only a single genetic strain (haplotype) would be involved in the fertilization process. A clonal population structure is consistent with physiological sexuality, as it is required in Plasmodium to complete the life cycle. What it excludes is the prevalence of genetic sexuality, i.e., recombination between genetically heterogeneous haplotypes.
Repeat regions, such as the CR of the Csp, are common among the antigenic determinants of extracellular (sporozoite and merozoite schizonts) stages of the malaria parasite. Repeat regions are probably an adaptive mechanism that allows for “antigenic variation,” a phenomenon known in bacterial and protozoan parasites. In the strictest sense, it is a process by which parasites alter their antigenic determinants by switching the expression of particular allelic variants. The classical example is the variable surface glycoprotein system of Trypanosoma (28). In a broader sense, the phrase “antigenic variation” is applied to any specialized mechanism by which parasites generate diversity of antigenic determinants at a rate that is notably higher than observed in the rest of the genome. The RAT variation of the Csp is a case of adaptation for antigenic diversity by a process of slipped-strand mispairing of repetitive DNA sequences.
The CR of the Csp has a dual function in the parasite’s life cycle: (i) to increase the avidity of the sporozoite surface when interacting with the membrane of hepatocytes (29), and (ii) presumably to serve as a “smoke-screen” defense that induces an ineffectual immune response (30). The variable RATs allow for rapid CR evolution by intragenic recombination, which provides suitable opportunities for mispairing of repeating paralogues along the length of the gene sequence. The polymorphism preserved in the repeating units, as seen in the two sets of RATs that code for two distinct amino acid motifs, protects the parasites against stochastic reductions in variability such as result from demographic bottlenecks, to which parasites are particularly prone as a consequence of their adaptation to exploit small, discontinuous environments (hosts) (31); or as a consequence of strong selection that may drive one particular strain to a predominant role in natural populations (32). In a broad sense, the Csp’s CR, and perhaps several other antigenic repeat regions, are components of an antigenic diversity system. The structure of the genes for MSA-1 and MSA-2 is far more complex, which makes it more difficult to detect patterns of intragenic recombination, as we have uncovered in Csp. However, MSA-1 actually has a high degree of fidelity in its repeat structure. Pizzi et al. (33) have identified the latent periodicity of the repeating units by assigning certain simple “virtual” repeats, and have shown that the overwhelming repeat variability of MSA-1 can be explained by intragenic shuffling.
The genetic variation present in P. falciparum has been interpreted (i) as ancient on an evolutionary time scale, and (ii) as evidence of widespread sexual recombination between dissimilar strains of the parasite. We have presented evidence that suggests otherwise. First, our observation that silent site polymorphism is virtually absent in the NRs of Csp (as well as in nine other P. falciparum genes), provides strong evidence that the world populations of P. falciparum strains have recent common ancestry. Second, the linkage disequilibrium, patterns of recombination, and other evidence indicate that the genetic variation does not originate by sexual recombination. These observations are important for assessing the current levels of polymorphism in P. falciparum, as well as the potentiality for new variation, with significant bearing on public health efforts to control malaria.
Acknowledgments
We thank Allan Dickerman, Anthony James, Walter Fitch, Carlos Machado, and Michel Tibayrenc for helpful discussions. This work was supported by National Institutes of Health Grant GM42937 to F.J.A.
ABBREVIATIONS
- CR
central repeat region
- Csp
circumsporozoite gene
- NR
nonrepeat region
- RAT
repeat allotype
References
- 1.World Health Organization. Tropical Disease Report, Twelfth Programme Report. Geneva: World Health Organization; 1995. [Google Scholar]
- 2.Clyde D F, McCarthy V C, Miller R M, Hornick R B. Am J Med Sci. 1973;266:398–403. doi: 10.1097/00000441-197312000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Patarroyo M E, Amador L R, Clavijo P, Moreno A, Guzman F, Romero P, Tascon R, Franco A, Murillo L A, Ponton G, Trujillo G. Nature (London) 1988;332:158–161. doi: 10.1038/332158a0. [DOI] [PubMed] [Google Scholar]
- 4.Udhayakumar V, Shi Y-P, Kumar S, Jue D L, Wohlhueter R M, Lal A A. Infect Immun. 1994;62:1410–1413. doi: 10.1128/iai.62.4.1410-1413.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zevering Y, Khamboonruang C, Good M F. Eur J Immunol. 1994;24:1418–1425. doi: 10.1002/eji.1830240627. [DOI] [PubMed] [Google Scholar]
- 6.Babiker H, Walliker D. Parasitol Today. 1997;13:262–267. doi: 10.1016/s0169-4758(97)01075-2. [DOI] [PubMed] [Google Scholar]
- 7.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swofford D. PAUP: Phylogenetic Analysis Using Parsimony. Champaign, IL: Illinois Natural History Survey; 1993. [Google Scholar]
- 9.Hudson R R, Kaplan N L. Genetics. 1987;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rozas, J. & Rozas, R. (1997) Comput. Appl. Biosci., in press. [PubMed]
- 11.Lewontin R C, Kojima K. Evolution. 1960;14:458–472. [Google Scholar]
- 12.Qari S H, Collins W E, Lobel H O, Taylor F, Lal A A. Am J Trop Med Hyg. 1994;50:45–51. doi: 10.4269/ajtmh.1994.50.45. [DOI] [PubMed] [Google Scholar]
- 13.Jongwutiwes S, Tanabe K, Hughes M K, Kanbara H, Hughes A L. Am J Trop Med Hyg. 1994;51:659–668. doi: 10.4269/ajtmh.1994.51.659. [DOI] [PubMed] [Google Scholar]
- 14.Caspers P, Gentz R, Matile H, Pink J R, Sinigaglia F. Mol Biochem Parasitol. 1989;35:185–190. doi: 10.1016/0166-6851(89)90121-7. [DOI] [PubMed] [Google Scholar]
- 15.Arnot D. Parasitol Today. 1989;5:138–142. doi: 10.1016/0169-4758(89)90077-x. [DOI] [PubMed] [Google Scholar]
- 16.McCutchan T F, Lal A A, do Rosario V, Waters A P. Mol Biochem Parasitol. 1992;50:37–46. doi: 10.1016/0166-6851(92)90242-c. [DOI] [PubMed] [Google Scholar]
- 17.Conway D J. Parasitol Today. 1997;13:26–29. doi: 10.1016/s0169-4758(96)10077-6. [DOI] [PubMed] [Google Scholar]
- 18.Hughes A L. In: Mechanisms of Molecular Evolution. Takahata N, Clark A G, editors. Sunderland, MA: Sinauer; 1993. pp. 109–127. [Google Scholar]
- 19.McConkey G A, Waters A P, McCutchan T F. Annu Rev Microbiol. 1990;44:479–498. doi: 10.1146/annurev.mi.44.100190.002403. [DOI] [PubMed] [Google Scholar]
- 20.Qari S, Goldman I, F, Povoa M M, di Santi S, Alpers M P, Lal A A. Mol Biochem Parasitol. 1992;55:105–114. doi: 10.1016/0166-6851(92)90131-3. [DOI] [PubMed] [Google Scholar]
- 21.Escalante A A, Ayala F J. Proc Natl Acad Sci USA. 1994;91:11373–11377. doi: 10.1073/pnas.91.24.11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tibayrenc M, Keillberg F, Ayala F J. Proc Natl Acad Sci USA. 1990;87:2414–2418. doi: 10.1073/pnas.87.7.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kerr P J, Ranford-Cartwright L C, Walliker D. Mol Biochem Parasitol. 1994;66:241–248. doi: 10.1016/0166-6851(94)90151-1. [DOI] [PubMed] [Google Scholar]
- 24.Marshall V M, Coppel R L, Martin R K, Oduola A M J, Anders R F, Kemp D J. Mol Biochem Parasitol. 1991;45:349–352. doi: 10.1016/0166-6851(91)90104-e. [DOI] [PubMed] [Google Scholar]
- 25.Hughes A L, Hughes M K. Mol Biochem Parasitol. 1995;71:99–113. doi: 10.1016/0166-6851(95)00037-2. [DOI] [PubMed] [Google Scholar]
- 26.Hartl D L. Curr Opin Genet Dev. 1992;2:937–942. doi: 10.1016/s0959-437x(05)80119-4. [DOI] [PubMed] [Google Scholar]
- 27.Levinson G, Gutman G A. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 28.Borst P, Rudenko G. Science. 1994;264:1872–1874. doi: 10.1126/science.7516579. [DOI] [PubMed] [Google Scholar]
- 29.Aley S B, Bates M D, Tam J P, Hollingdale M R. J Exp Med. 1986;164:1915–1922. doi: 10.1084/jem.164.6.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nardin E H, Nussenzweig R S. Annu Rev Immunol. 1993;11:687–727. doi: 10.1146/annurev.iy.11.040193.003351. [DOI] [PubMed] [Google Scholar]
- 31.Price P W. Evolution. 1977;31:405–420. doi: 10.1111/j.1558-5646.1977.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 32.Maynard Smith J, Smith N H, O’Rourke M, Spratt B G. Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pizzi E, Liuni S, Frontali C. Nucleic Acids Res. 1990;18:3745–3752. doi: 10.1093/nar/18.13.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lockyer M J, Schwarz R T. Mol Biochem Parasitol. 1987;22:101–108. doi: 10.1016/0166-6851(87)90073-9. [DOI] [PubMed] [Google Scholar]
- 35.Dame J B, Williams J L, McCutchan T F, Weber J L, Wirtz R A, Hockmeyer W T, Maloy W L, Haynes J D, Schneider I, Roberts D, Sanders G S, Reddy P E, Diggs C L, Miller L H. Science. 1984;225:593–599. doi: 10.1126/science.6204383. [DOI] [PubMed] [Google Scholar]
- 36.del Portillo H A, Nussenzweig R S, Enea V. Mol Biochem Parasitol. 1987;24:289–294. doi: 10.1016/0166-6851(87)90161-7. [DOI] [PubMed] [Google Scholar]
- 37.Lockyer M J. Mol Biochem Parasitol. 1991;45:179–182. doi: 10.1016/0166-6851(91)90041-4. [DOI] [PubMed] [Google Scholar]
- 38.Davis J R, Cortese J F, Herrington D A, Murphy J R, Clyde D F, Thomas A W, Baqar S, Cochran M A, Thanassi J, Levine M M, Hackett C S. Exp Parasitol. 1992;74:159–168. doi: 10.1016/0014-4894(92)90043-a. [DOI] [PubMed] [Google Scholar]
- 39.Caspers P, Gentz R, Matile H, Pink J R, Sinigaglia F. Mol Biochem Parasitol. 1989;35:185–190. doi: 10.1016/0166-6851(89)90121-7. [DOI] [PubMed] [Google Scholar]
- 40.Campbell J R. Nucleic Acids Res. 1989;17:5854. doi: 10.1093/nar/17.14.5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lal A A, Goldman I F. J Biol Chem. 1991;266:6686–6689. [PubMed] [Google Scholar]