Abstract
Fanconi anemia (FA) is a genetically heterogeneous autosomal recessive syndrome associated with chromosomal instability, hypersensitivity to DNA crosslinking agents, and predisposition to malignancy. The gene for FA complementation group A (FAA) recently has been cloned. The cDNA is predicted to encode a polypeptide of 1,455 amino acids, with no homologies to any known protein that might suggest a function for FAA. We have used single-strand conformational polymorphism analysis to screen genomic DNA from a panel of 97 racially and ethnically diverse FA patients from the International Fanconi Anemia Registry for mutations in the FAA gene. A total of 85 variant bands were detected. Forty-five of the variants are probably benign polymorphisms, of which nine are common and can be used for various applications, including mapping studies for other genes in this region of chromosome 16q. Amplification refractory mutation system assays were developed to simplify their detection. Forty variants are likely to be pathogenic mutations. Seventeen of these are microdeletions/microinsertions associated with short direct repeats or homonucleotide tracts, a type of mutation thought to be generated by a mechanism of slipped-strand mispairing during DNA replication. A screening of 350 FA probands from the International Fanconi Anemia Registry for two of these deletions (1115–1118del and 3788–3790del) revealed that they are carried on about 2% and 5% of the FA alleles, respectively. 3788–3790del appears in a variety of ethnic groups and is found on at least two different haplotypes. We suggest that FAA is hypermutable, and that slipped-strand mispairing, a mutational mechanism recognized as important for the generation of germ-line and somatic mutations in a variety of cancer-related genes, including p53, APC, RB1, WT1, and BRCA1, may be a major mechanism for FAA mutagenesis.
Fanconi anemia (FA) is an autosomal recessive disorder characterized by congenital abnormalities, bone marrow failure, and predisposition to acute myelogenous leukemia and other malignancies (1–3). FA cells are hypersensitive to DNA crosslinking agents such as diepoxybutane and mitomycin C (4). Eight complementation groups (FA-A through FA-H) have been described, with FA-A accounting for approximately two-thirds of FA families (5–7). The gene responsible for the defect in FA-C (FAC) was isolated by functional complementation and mapped to chromosome 9q22.3 (5, 8). The FAD gene was mapped to chromosome 3p22–26, but has not yet been isolated (9). Recently, FAA was mapped to 16q24.3 (10, 11), and the cDNA was isolated by two independent approaches: positional and expression cloning (12, 13). Both FAA and FAC encode unique proteins, which do not exhibit any homology to known proteins that might suggest a function.
A few mutations in FAA were described in the initial cloning reports (12, 13), including two mutations involving base substitutions that result in utilization of a cryptic splice site leading to insertion of 30 bp and six intragenic deletions (4–879 bp). The consequence of the majority of these deletions is predicted to be a truncated protein. Some of the deletions were identified only in cDNA, and the detailed breakpoints in genomic DNA were not described.
To identify additional mutations in FAA, we screened a panel of 97 FA patients from the International Fanconi Anemia Registry (IFAR). The complete coding sequence (43 exons) was amplified from genomic DNA and screened by single-strand conformational polymorphism (SSCP) analysis. Forty mutations and 45 polymorphisms were identified; 17 mutations as well as one polymorphism are short deletions that are flanked by short direct repeats.
MATERIALS AND METHODS
Family Resource and Screening Panels Selection.
Genomic DNA samples from unrelated FA patients enrolled in the IFAR at The Rockefeller University were studied. The clinical diagnosis of FA was confirmed by a positive diepoxybutane test (14). Three families previously were classified as FA-A by somatic cell hybridization. The first screening panel included 50 FA patients that were selected based on the following criteria: consanguinity, ethnicity, or positive logarithm of odds score at 16q24.3. The two largest groups with a single origin were from Brazil (30 patients) and Turkey (6 patients). Based on the data found in the first screening, the total IFAR population was screened for some mutations, and a second screening panel of 47 patients was constructed in which the majority of the patients carried one known mutation. When our screen of FAA exons indicated the presence of a mutation, other family members were analyzed.
PCR-SSCP Analysis.
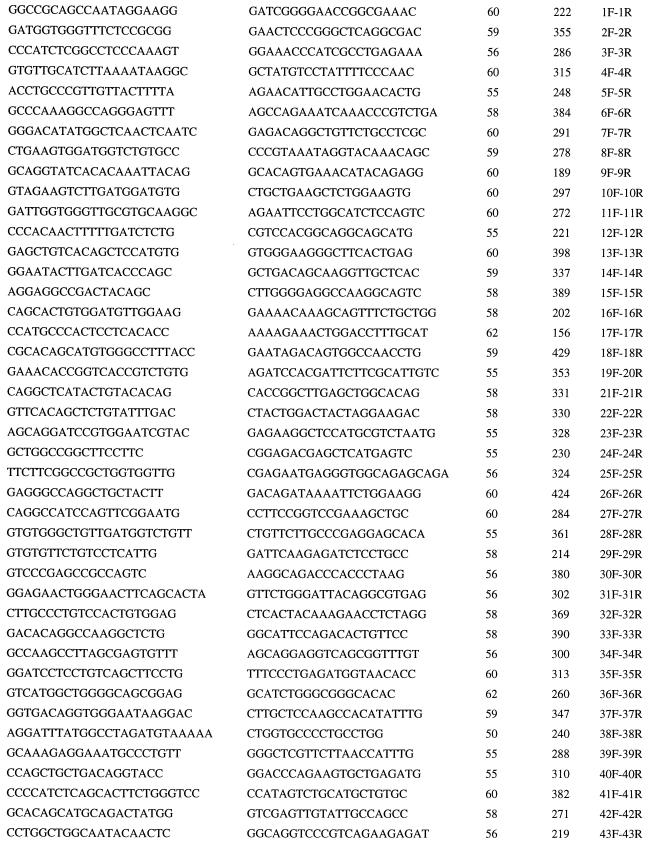
All FAA exonic sequences were amplified from genomic DNA by PCR and analyzed by SSCP. Primer pairs were generated from intron sequences (15) in collaboration with C. Mathew (Guy’s Hospital, London) to amplify 43 exons in 42 PCR reactions, with products ranging from 156 to 429 bp (Table 1). PCRs contained 10 ng of genomic DNA, 10 mM Tris⋅HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.25 mM spermidine, 0.2 mM dNTP, 0.4 μM unlabeled forward primer, 0.12 μM 33P-labeled reverse primer, and 0.3 units of Taq DNA polymerase (Boehringer Mannheim), in a total volume of 10 μl. Ten percent dimethyl sulfoxide and 1% formamide were added to amplify exon 1 and exon 2, respectively. Exons 3, 30, 31, and 43 were amplified in 20 mM (NH4)2SO4/20 mM NaCl/80 mM Tris, pH 9.0/2 mM MgCl2. PCR conditions were: 3 min at 95°C; then 30 cycles of 30 sec at 94°C, 30 sec at 55–62°C, and 30 sec at 72°C; and then a final 5 min at 72°C. The amplification of exon 1 started with 5 min at 95°C and 5 min at 58°C before the regular cycles. Two microliters of 6× loading dye (Perkin–Elmer) were added to the 10-μl PCR, and the samples were denatured at 95°C and cooled on ice, and 5 μl were loaded onto a 7.5% GeneAmp Detection Gel (Perkin–Elmer). Electrophoresis was performed in 0.5× TBE (0.089 M tris borate/0.089 M boric acid/0.002 M EDTA) at 12 W at room temperature for 3–4 hr with a fan directed at the gel. Gels were dried and exposed to film overnight at −70°C. The patterns of both single strands and heteroduplexes were detected on a single gel.
Table 1.
PCR primers for FAA
| Forward primer | Reverse primer | Annealing temperature (°C) | Size (bp) | Names |
|---|---|---|---|---|
All the primers are indicated in the 5′–3′ direction.
Sequence Analysis and Family Segregation Analysis.
Upon detection of an abnormal SSCP and/or heteroduplex pattern, genomic DNA was amplified in three parallel PCRs from the patient and a control. The 50-μl reactions were essentially similar to the PCR-SSCP reactions but contained unlabeled primers. After amplification, the combined products were purified on QIAquick spin columns (Qiagen) and both strands were sequenced directly with fluorescently labeled primers on an Applied Biosystems 377 DNA sequencer. When a sequence variant was identified, additional family members as well as normal chromosomes were screened by SSCP and/or restriction analysis to determine if the variant was associated with the FA phenotype.
Restriction Analysis.
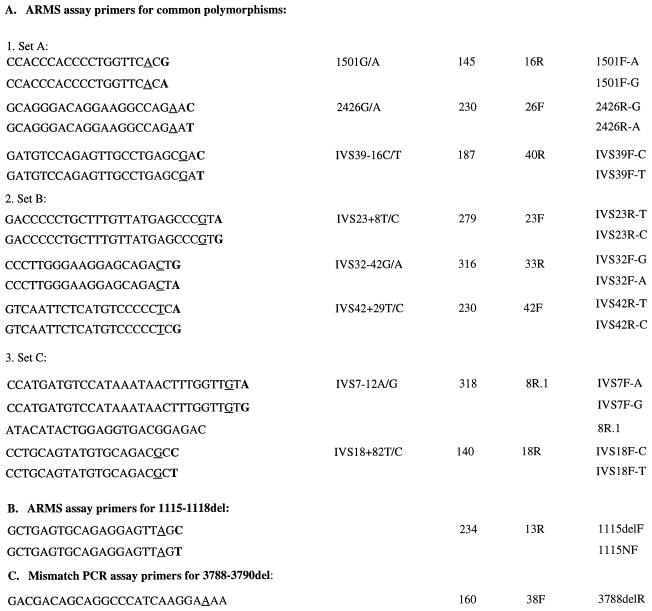
Restriction site assays were developed for the following variants: 401–402insC and 2426G/A, which eliminate DraIII site, 3091C>T and IVS11–1delG, which eliminate PstI site, 3715–3729del, which eliminates MboII site, and 4249C>G, which creates DdeI site. A modified mismatch PCR assay was developed for the 3788–3790del, as follows (Table 2C): An artificial 26-mer (3788delR) in the reverse orientation was designed to abolish the single MboII site in the mutant allele and to leave intact one of the two MboII sites in the normal allele. After a 15-μl PCR amplification with the flanking intronic forward primer (38F), 0.6 μl of MboII (3 units) was added directly to the PCR together with 1.4 μl of 100 mM MgCl2 (final concentration 10 mM) and incubated for 1 hr at 37°C. The products were separated on 2.5% agarose gels.
Table 2.
Mutation-specific PCR primers
| Polymorphism | Product size (bp) | Second primer | Name | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
All the primers are indicated in the 5′–3′ direction. Boldface type indicates a base that matches a variant. Underline indicates a base that mismatches the wild-type sequence.
Amplification Refractory Mutation System Assays.
Amplification refractory mutation system assays were developed to screen for eight of the common polymorphisms, as described in Table 2A. Six multiplex reactions were designed such that each detects two or three variants (Set A: 1501G/A, 2426G/A, and IVS39 −16C/T; Set B: IVS 23 +8T/C, IVS 32 −42G/A, and IVS42 +29T/C; Set C: IVS7 −12A/G and IVS18 +82T/C). Exon 6 of the FAA gene was amplified simultaneously in all of the reactions as an internal control. The haplotype was determined in two stages where the first PCR contained primers that specifically amplified one variant allele, and the second reaction contained primers that specifically amplify the second variant. The 15-μl PCR amplification contained 10 ng of genomic DNA, 10 mM Tris⋅HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.25 mM spermidine, 0.2 mM dNTP, 0.6 μM of each primer, and 0.5 units of Taq DNA polymerase (Boehringer Mannheim). PCR conditions were as described above for the PCR-SSCP analysis with an annealing temperature of 58°C. The PCR products were separated on 2.5% agarose gels.
Amplification refractory mutation system assays also were developed for the 1115–1118del mutation (Table 2B). The mutation-specific primer 1115delF was used together with the normal intronic primer 13R to amplify a PCR product of 234 bp. Exon 1 of the FAC gene (16) was included as an internal control. When a mutant product was amplified, a second PCR was performed with a normal primer (1115NF) to check for the heterozygosity status of the mutation. PCR conditions were as described above, with an annealing temperature of 54°C.
RESULTS
PCR-SSCP Analysis.
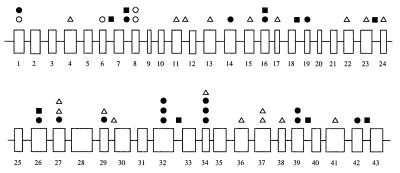
The complete coding sequence of the FAA gene was screened for mutations in 97 FA patients and four controls. Eighty-five abnormal SSCP patterns were found; 40 of these cosegregate with the FAA phenotype and do not appear in the normal alleles screened and therefore are likely to be pathogenic mutations (Fig. 1; Table 3). Mutation types include frameshift, splicing, nonsense, and missense mutations. Thirty-two of the mutations were found in a single patient, six were found in two unrelated patients, and two were found in multiple patients. Two classes of mutations, base substitutions and microdeletions/microinsertions, were identified in this study.
Figure 1.
Schematic representation of the distribution of sequence variants in the FAA gene, including point mutations (•), point mutations that are part of a complex allele (○), microdeletions/microinsertions (▵), and common polymorphisms (▪).
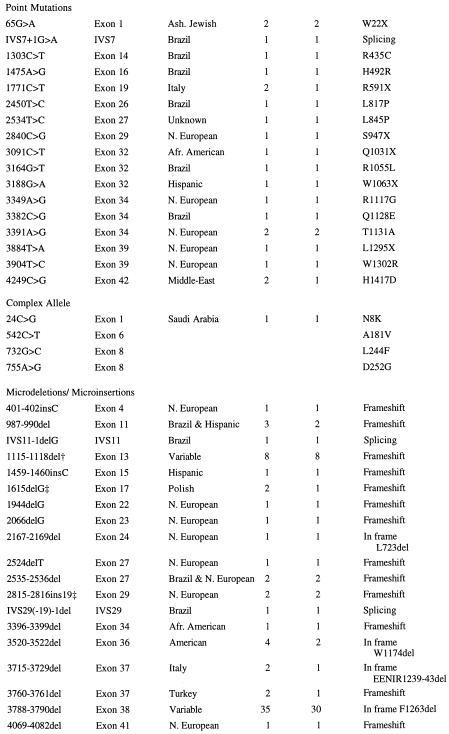
Table 3.
Mutations in the FAA gene
| Nucleotide change* | Location | Ethnicity | No. of alleles | No. of patients | Mutation type |
|---|---|---|---|---|---|
Variants are designated as mutations based on inheritance pattern and screening of normal alleles. Pathogenicity has not been proven.
Ref. 12.
These mutations also were described by J. Pronk and A. Savoia, respectively (Fanconi Anemia Mutation Database).
Base Substitutions.
Ten missense mutations (1303C>T, 1475A>G, 2450T>C, 2534T>C, 3164G>T, 3349A>G, 3382C>G, 3391A>G, 3904T>C, and 4249C>G), six nonsense mutations (65G>A, 1771C>T, 2840C>G, 3091C>T, 3188G>A, and 3884T>A), and one potential splicing mutation (IVS7+1G>A) were identified. In addition, a complex allele of four nucleotide changes (24C>G, 542C>T, 732G>C, and 755A>G) was identified; phase was determined by analysis of segregation of these mutations in a large pedigree.
Microdeletions/Microinsertions.
All but two (IVS11 −1delG and 3396–3399del) of the small deletions/insertions occurred at repeats of 1–5 nucleotides, either in tandem or separated by intervening nucleotides (Table 4). Four of these (2167–2169del, 3520–3522del, 3715–3729del, and 3788–3790del) cause in-frame deletions of 1–5 amino acids. Thirteen deletions/insertions (401–402insC, 987–990del, 1115–1118del, 1459–1460insC, 1615delG, 1944delG, 2066delG, 2524delT, 2535–2536del, 2815–2816ins19, 3396–3399del, 3760–3761del, and 4069–4082del) result in a frameshift and premature stop codon that predicts a truncated protein, and two deletions (IVS29(-19)-1del and IVS11–1delG) cause potential splicing mutations. 1115–1118del previously was described in a patient with German ancestry (12). 1615delG and 2815–2816ins19 previously were described by J. Pronk and A. Savoia, respectively (Fanconi Anemia Mutation Database). 3788–3790del was found in 10 of 30 Brazilian patients in the SSCP screen.
Table 4.
Microdeletions/microinsertions and short direct repeats in the FAA gene
| Microdeletion | Sequence | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
Lowercase indicates intronic sequence. Boldface type indicates repeats. The deletion hot spot consensus sequence (CCTG/CAGG) is underlined; MboII restriction site is indicated. The deleted/inserted sequence is boxed (for deletions in short direct repeats, the most 3′ nucleotide is arbitrarily assigned).
This deletion is denoted by the 5′ intronic direct repeat for simplicity.
Nonpathogenic variant.
Deletion Hot Spot Consensus Sequences.
The tetranucleotide CCTG (CAGG) motif, previously identified to be a mutation hot spot consensus sequence (17), was identified near approximately half of the deletions/insertions (Table 4). The CCTG motif was found in close proximity to some point mutations as well, including 65G>A, 1303C>T, 1475A>G, 2450T>C, 3164G>T, 3382C>G, 3904T>C, and the polymorphism 2426G/A. The TTC (GAA) repeat (MboII restriction site) mutation hot spot (18) was identified as part of the deletion in 764–766del, 3715–3729del, and 3788–3790del. It also was found downstream of 3396–3399del and 2535–2536del (Table 4).
IFAR Population Screening.
After the completion of the first panel, 300 additional FA patients without any known FAC mutations (16) were screened for 3788–3790del and 1115–1118del. In the total IFAR population 3788–3790del was found in 33 patients (5% allele frequency) from different ethnic groups; this mutation was associated with at least two different haplotypes. 1115–1118del was carried by 12 patients (2% allele frequency) from diverse ethnic groups; this mutation was associated with a single haplotype for the common FAA polymorphisms.
Polymorphisms.
The FAA gene is highly polymorphic; 45 variants that are likely to represent polymorphisms were identified and are described in the Fanconi Anemia Mutation Database (http://www.rockefeller.edu/fanconi/mutate). Some of these are rare intronic variants that segregate with the disease phenotype and require further study at the RNA level to rule out their pathogenicity. Most of the variants were found in only one or few chromosomes and are likely to represent rare polymorphisms, because they segregate independently of the FA phenotype and/or were found in normal controls. Interestingly, one of these variants, 764–766del, is an in-frame microdeletion of a direct repeat (R255del; Table 4) that did not segregate with the disease phenotype as was also the case for the missense variant 17T>A (V6D). Another phenomena that needs further investigation is that a number of individuals, including normal controls, appear to carry multiple nonpathogenic variants that are arranged in complexes. Three missense mutations, 2216C>T (P739L), 3859G>A (V1287I), and 3982A>G (T1328A), probably are nonpathogenic because they each were found in a patient also carrying two frameshift mutations, but we cannot exclude the possibility of a complex pathogenic allele. Nine variants (IVS6+74G/A, IVS7–12A/G, 1501G/A, IVS18+82T/C, IVS23+8T/C, 2426G/A, IVS32–42G/A, IVS39–16C/T, and IVS42+29T/C) were found to be common biallelic polymorphisms in which the frequency of the more common allele ranges from 55% to 74% in the FA population. Two of these polymorphisms (1501G/A and 2426G/A) cause amino acid substitutions (501S/G and 809G/D, respectively). The 1501G/A polymorphism was identified in the original cDNA clones (12), and the 2426G/A and the IVS7–12A/G were independently described by A. Savoia (Fanconi Anemia Mutation Database). Preliminary haplotype analysis of FA-A families revealed the existence of at least six large genomic deletions, as shown by loss of heterozygosity for some of the variants.
DISCUSSION
We have screened a panel of 97 DNA samples from ethnically diverse FA patients by SSCP to identify the spectrum of mutations in the FAA gene. The majority of the patients were likely to belong to complementation group A, based on the relatively high frequency of this complementation group and on linkage analysis with chromosome16q24.3 markers in a subset of these families. We have identified 40 mutations and 45 polymorphisms; nine of the polymorphisms are common and can be used for various applications including mapping studies for other genes in this region of chromosome 16q. At least 67 of the 97 patients screened by SSCP belong to FA-A based on the presence of at least one mutation in the FAA gene that segregates with the FA phenotype. Of the remaining 30 patients, some may belong to other complementation groups, whereas some may carry mutations that do not result in a detectable shift on SSCP gels. Two major types of mutations were identified in the present study: base substitutions and microdeletions/microinsertions. These mutations were spread throughout the gene (Fig. 1).
A total of 350 non-FA-C IFAR patients were screened for 3788–3790del, the most common mutation found by SSCP, and for 1115–1118del, which previously was described in a patient of German ancestry (12). 3788–3790del was found in 10% of the patients and was especially common among the Brazilian patients. 1115–1118del was found in 3.4% of the total 350 patients analyzed for these microdeletions. We propose that 3788–3790del had at least two independent founders; haplotype analysis showed two haplotypes that were unlikely to arise from a common mutant chromosome (Table 2, Set A: GGC vs. AAT). Because the haplotype of one of these founders (GGC) is common in the general population, it is not possible to predict whether there were more than two founders for this mutation. We currently are extending our haplotype analysis for this purpose. A preliminary screen of the total IFAR panel by using mutation-specific assays for several of the other mutations detected by SSCP indicates that the other mutations are likely to be rare (data not shown).
In the original cloning studies of FAA, reverse transcription–PCR products were used for mutation detection (12, 13). Five of the eight mutations described in these studies were large deletions that would not be detected by SSCP analysis of genomic DNA, unless the patient was homozygous for a deleted exon. Data from several patients in the SSCP screen, as well as results from haplotype analysis that used eight of the nine common polymorphisms described above, suggest the existence of six large genomic deletions in patients in our study. We currently are using other methods to characterize these deletions.
The close association of short direct repeats and homonucleotide tracts with microdeletions/microinsertions as seen in FAA has been reported to be common in human genes (19), including cystic fibrosis transmembrane conductance regulator, β-globin, factor IX (20), hypoxanthine phosphoribosyltransferase (21), and many cancer-related genes including p53 (22, 23), retinoblastoma (RB1) (24), adenomatous polyposis coli (APC) (25), Wilms tumor (WT1) (17), and breast cancer (BRCA1) (26). Such deletions also are induced by mutagens (21). The repeat units that are associated with microdeletions usually are between 2 and 8 bp (19). These deletions/insertions can be explained by a slipped-strand mispairing mechanism initially proposed by Streisinger et al. (27), in which one DNA strand of a repeat can be misaligned by chance with the downstream repeat of the complementary strand (28, 29). The resulting loop subsequently is excised, fixing the deletion before the next round of replication. This slippage-misalignment mechanism has been proposed to be a ubiquitous mechanism of mutagenesis and is responsible for a significant proportion of mutations in mammalian cells (21). The FAA coding sequence and flanking intronic sequences contain many such hypermutable repetitive sequences.
Our results show that the single bp substitutions identified in FAA are associated with three major motifs: CpG motifs (20%), homonucleotide tracts (20%), and short direct repeats (33%); the remainder of the point mutations do not appear to fit into any of these categories. DNA methylation is considered responsible for bp substitutions at CpG sites, and it was shown that CpG mutation hot spots in BRCA1 (26), NF1 (30), and p53 (31), for example, are methylated. Further study is needed to learn if CpG sites associated with mutations in FAA also are methylated. We propose that misalignment-mediated errors during DNA synthesis and methylation-induced mutagenesis account for the majority of the mutations described in this analysis.
A variety of mutation hot spot consensus sequences have been reported in the literature. The sequence CCTG (CAGG), first identified as a homologous recombination hot spot in the murine major histocompatibility complex (32), has been observed to be a mutation hot spot in a large number of human genes (19), especially when it occurs near direct repeat sequences (17). We have found this motif in the vicinity of many of the mutations, both microdeletions/microinsertions and point mutations, described in this study. The TTC repeat (MboII restriction site) motif, described in the Chinese hamster APRT gene as a hot spot for spontaneous deletions (18) also was identified near several of the microdeletions/microinsertions in FAA.
One important question that remains to be answered is the relevancy of FAA mutations to the pathogenesis of cancer in FA patients. We speculate that FAA is a member of the “caretaker” gene family that includes xeroderma pigmentosum (XP), hereditary non-polyposis colorectal cancer (HNPCC) genes, ataxia-telangiectasia (ATM), and probably BRCA1 and BRCA2, as recently was suggested by Kinzler and Vogelstein (33). A mutator phenotype with strong specificity for deletions was also implied for Werner syndrome (WRN) (34) and Bloom syndrome (BLM) (35). Each of these genes is responsible in a unique way for the integrity of the genome and when mutated causes predisposition to cancer. According to the caretaker-gatekeeper model, inactivation of a caretaker gene results in a higher mutation rate in all genes, including gatekeeper genes that directly regulate tumor growth. This model is consistent with the high proportion of deletions at the hypoxanthine phosphoribosyltransferase locus (36), and the loss of heterozygosity at the glycophorin A (GPA) locus in FA cells (ref. 37 and A.D.A., unpublished results), and could explain the cancer predisposition of Fanconi patients. The potential hypermutability of the FAA gene also could result in an increased risk of cancer in heterozygous carriers of FAA mutations. We hypothesize that carriers of a germ-line mutation in FAA are at high risk for a somatic mutation in the second FAA allele as a result of sequence-specific hypermutable regions; this recently has been shown to be the mechanism for converting a benign variant (I1307K) into pathogenic mutation in APC (38). The resulting FA cellular phenotype of genomic instability would predispose cells to the accumulation of two additional mutations in a gatekeeper gene, causing cancer to develop. The epidemiological and molecular implications of this hypothesis currently are being tested.
In conclusion, the large number of different variants found in this mutation screen indicates that the FAA gene is highly polymorphic and may be hypermutable. Based on our sample population there are probably many private or semiprivate mutations as well as ethnic specific mutations, making large-scale mutation screening difficult. An analysis of genotype-phenotype correlation is in progress and may provide insight into protein structure-function relationships.
Acknowledgments
We gratefully acknowledge the contribution of the many physicians who referred patients to the IFAR. We thank Dr. R. Pasquini for the blood samples from the Brazilian families and the technical assistance of Y. Flit and C. Koenigsberg. This work was supported by National Institutes of Health Grant HL32987 (A.D.A.), the Fanconi Anemia Research Fund, Inc. (A.D.A.), and Associação Alirio Pfiffer - Curitiba, Brazil (N.M.).
ABBREVIATIONS
- FA
Fanconi anemia
- SSCP
single-strand conformational polymorphism
- IFAR
International Fanconi Anemia Registry
References
- 1.Butturini A, Gale R P, Verlander P C, Adler-Brecher B, Gillio A P, Auerbach A D. Blood. 1994;84:1650–1655. [PubMed] [Google Scholar]
- 2.Giampietro P F, Adler-Brecher B, Verlander P C, Pavlakis S G, Davis J G, Auerbach A D. Pediatrics. 1993;91:1116–1120. [PubMed] [Google Scholar]
- 3.Giampietro P F, Verlander P C, Davis J G, Auerbach A D. Am J Med Genet. 1996;68:58–61. [PubMed] [Google Scholar]
- 4.Auerbach A D. Exp Hematol. 1993;21:731–733. [PubMed] [Google Scholar]
- 5.Strathdee C A, Duncan A M V, Buchwald M. Nat Genet. 1992;1:196–198. doi: 10.1038/ng0692-196. [DOI] [PubMed] [Google Scholar]
- 6.Joenje H, Lo Ten Foe J R, Oostra A B, van Berkel C G M, Rooimans M A, Schroeder-Kurth T M, Wegner R-D, Gille J J P, Buchwald M, Arwert F, Joenje H, Lo Ten Foe J R, Oostra A B, van Berkel C G M, Rooimans M A, Schroeder-Kurth T M, Wegner R-D. Blood. 1995;86:2156–2160. [PubMed] [Google Scholar]
- 7.Joenje H, Oostra A B, Wijker M, di Summa F M, van Berkel C G M, Rooimans M A, Ebell W, van Weel M, Pronk J C, Buchwald M, Arwert F. Am J Hum Genet. 1997;61:940–944. doi: 10.1086/514881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strathdee C A, Gavish H, Shannon W R, Buchwald M. Nature (London) 1992;356:736–767. doi: 10.1038/356763a0. [DOI] [PubMed] [Google Scholar]
- 9.Whitney M, Thayer M, Reifsteck C, Olson S, Smith L, Jakobs P M, Leach R, Naylor S, Joenje H, Grompe M. Nat Genet. 1995;11:341–343. doi: 10.1038/ng1195-341. [DOI] [PubMed] [Google Scholar]
- 10.Pronk J C, Gibson R A, Savoia A, Wijker M, Morgan N V, Melchionda S, Ford D, Temtamy S, Ortega J J, Jansen S, Havenga C, Cohn R I, de Ravel T J, Roberts I, Westerveld A, Easton D F, Joenje H, Mathew C G, Arwert F. Nat Genet. 1995;11:338–340. doi: 10.1038/ng1195-338. [DOI] [PubMed] [Google Scholar]
- 11.Gschwend M, Levran O, Kruglyak L, Ranade K, Verlander P C, Shen S, Faure S, Weissenbach J, Altay C, Lander E S, Auerbach A D, Botstein D. Am J Hum Genet. 1996;59:377–384. [PMC free article] [PubMed] [Google Scholar]
- 12.The Fanconi Anaemia/Breast Cancer Consortium. Nat Genet. 1996;14:324–328. doi: 10.1038/ng1196-324. [DOI] [PubMed] [Google Scholar]
- 13.Lo Ten Foe J R, Rooimans M A, Bosnoyan-Collins L, Alon N, Wijker M, Parker L, Lightfoot J, Cheng N C, van Berkel C G M, Strunk M H P, Gille J J P, Kruyt F A E, Pronk J C, Arwert F, Buchwald M, Joenje H. Nat Genet. 1996;14:320–323. doi: 10.1038/ng1196-320. [DOI] [PubMed] [Google Scholar]
- 14.Auerbach A D, Rogatko A, Schroeder-Kurth T M. Blood. 1989;73:391–396. [PubMed] [Google Scholar]
- 15.Ianzano L, d’ Apolitol M, Centra M, Savino M, Levran O, Auerbach A D, Clenton-Jansen A-M, Doggett N A, Pronk J C, Tipping A J, Gibson R A, Mathew C G, Whitmore S A, Apostotou D F, Callen D F, Zelante L, Savoia A. Genomics. 1997;41:309–314. doi: 10.1006/geno.1997.4675. [DOI] [PubMed] [Google Scholar]
- 16.Verlander P C, Lin J, Udono M U, Zhang Q, Gibson R, Mathew C G, Auerbach A D. Am J Hum Genet. 1994;54:595–601. [PMC free article] [PubMed] [Google Scholar]
- 17.Huff V, Jaffe N, Saunders G F, Strong L C, Villalba F, Ruteshouser E C. Am J Hum Genet. 1995;56:84–90. [PMC free article] [PubMed] [Google Scholar]
- 18.Smith D G, Adair G M. Mut Res. 1996;352:87–96. doi: 10.1016/0027-5107(96)00007-3. [DOI] [PubMed] [Google Scholar]
- 19.Krawczak M, Cooper D N. Hum Genet. 1991;86:425–441. doi: 10.1007/BF00194629. [DOI] [PubMed] [Google Scholar]
- 20.Darvasi A, Kerem B. Eur J Hum Genet. 1995;3:14–20. doi: 10.1159/000472269. [DOI] [PubMed] [Google Scholar]
- 21.Kimura H, Iyehara-Ogawa H, Kato T. Mutagenesis. 1994;9:395–400. doi: 10.1093/mutage/9.5.395. [DOI] [PubMed] [Google Scholar]
- 22.Jego N, Thomas G, Hamelin R. Oncogene. 1993;8:209–213. [PubMed] [Google Scholar]
- 23.Greenbalt M S, Grollman A P, Harris C C. Cancer Res. 1996;56:2130–2136. [PubMed] [Google Scholar]
- 24.Canning S, Dryja T P. Proc Natl Acad Sci USA. 1989;86:5044–5048. doi: 10.1073/pnas.86.13.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyoshi Y, Ando H, Nagase H, Nishisho I, Horii A, Miki Y, Mori T, Utsunomiya J, Baba S, Petersen G, Hamilton S R, Kinzler K W, Vogelstein B, Nakamura Y. Proc Natl Acad Sci USA. 1992;89:4452–4456. doi: 10.1073/pnas.89.10.4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodenhiser D, Chakraborty P, Andrews J, Ainsworth P, Mancini D, Lopes E, Singh S. Oncogene. 1996;12:2623–2629. [PubMed] [Google Scholar]
- 27.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Cold Spring Harbor Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 28.Kunkel T A, Soni A. J Biol Chem. 1988;263:14784–14789. [PubMed] [Google Scholar]
- 29.Kunkel T A. Biochemistry. 1990;29:8003–8011. doi: 10.1021/bi00487a001. [DOI] [PubMed] [Google Scholar]
- 30.Rodenhiser D I, Coulter-Mackie M B, Singh S M. Hum Mol Genet. 1993;2:439–444. doi: 10.1093/hmg/2.4.439. [DOI] [PubMed] [Google Scholar]
- 31.Denissenko M F, Chen J X, Tang M-S, Pfeifer G P. Proc Natl Acad Sci USA. 1997;94:3893–3898. doi: 10.1073/pnas.94.8.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steinmetz M, Ueumatsu Y, Lindahl K F. Trends Genet. 1987;3:7–10. [Google Scholar]
- 33.Kinzler K W, Vogelstein B. Nature (London) 1997;386:761–763. doi: 10.1038/386761a0. [DOI] [PubMed] [Google Scholar]
- 34.Fukuci K-I, Martin G M, Monnat R J., Jr Proc Natl Acad Sci USA. 1989;86:5893–5897. doi: 10.1073/pnas.86.15.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langlois R G, Bigbee W L, Jensen R H, German J. Proc Natl Acad Sci USA. 1989;86:670–674. doi: 10.1073/pnas.86.2.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laquerbe A, Moustacchi E, Fuscoe J C, Papadopoulo D. Proc Natl Acad Sci USA. 1995;92:831–835. doi: 10.1073/pnas.92.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sala-Trepat M, Boyse J, Richard P, Papadopoulo D, Moustacchi E. Mutat Res. 1993;289:115–126. doi: 10.1016/0027-5107(93)90137-5. [DOI] [PubMed] [Google Scholar]
- 38.Laken S J, Petersen G M, Gruber S G, Oddoux C, Ostrer H, Giardiello F M, Hamilton S R, Hampel H, Markowitz A, Klimstra D, Jhanwar S, Winawer S, Offit K, Luce M C, Kinzler W K, Vogelstein B. Nat Genet. 1997;17:79–83. doi: 10.1038/ng0997-79. [DOI] [PubMed] [Google Scholar]