Abstract
We have developed high-density DNA microarrays of yeast ORFs. These microarrays can monitor hybridization to ORFs for applications such as quantitative differential gene expression analysis and screening for sequence polymorphisms. Automated scripts retrieved sequence information from public databases to locate predicted ORFs and select appropriate primers for amplification. The primers were used to amplify yeast ORFs in 96-well plates, and the resulting products were arrayed using an automated micro arraying device. Arrays containing up to 2,479 yeast ORFs were printed on a single slide. The hybridization of fluorescently labeled samples to the array were detected and quantitated with a laser confocal scanning microscope. Applications of the microarrays are shown for genetic and gene expression analysis at the whole genome level.
The genome sequencing projects have generated and will continue to generate enormous amounts of sequence data. The genomes of Saccharomyces cerevisiae, Haemophilus influenzae (1), Mycoplasma genitalium (2), and Methanococcus jannischii (3) have been completely sequenced. Other model organisms have had substantial portions of their genomes sequenced as well including the nematode Caenorhabditis elegans (4) and the small flowering plant Arabidopsis thaliana (5). Given this ever-increasing amount of sequence information, new strategies are necessary to efficiently pursue the next phase of the genome projects—the elucidation of gene expression patterns and gene product function on a whole genome scale.
One important use of genome sequence data is to attempt to identify the functions of predicted ORFs within the genome. Many of the ORFs identified in the yeast genome sequence were not identified in decades of genetic studies and have no significant homology to previously identified sequences in the database. In addition, even in cases where ORFs have significant homology to sequences in the database, or have known sequence motifs (e.g., protein kinase), this is not sufficient to determine the actual biological role of the gene product. Experimental analysis must be performed to thoroughly understand the biological function of a given ORF’s product. Model organisms, such as S. cerevisiae, will be extremely important in improving our understanding of other more complex and less manipulable organisms.
To examine in detail the functional role of individual ORFs and relationships between genes at the expression level, this work describes the use of genome sequence information to study large numbers of genes efficiently and systematically. The procedure was as follows. (i) Software scripts scanned annotated sequence information from public databases for predicted ORFs. (ii) The start and stop position of each identified ORF was extracted automatically, along with the sequence data of the ORF and 200 bases flanking either side. (iii) These data were used to automatically select PCR primers that would amplify the ORF. (iv) The primer sequences were automatically input into the automated multiplex oligonucleotide synthesizer (6). (v) The oligonucleotides were synthesized in 96-well format, and (vi) used in 96-well format to amplify the desired ORFs from a genomic DNA template. (vii) The products were arrayed using a high-density DNA arrayer (7–10). The gene arrays can be used for hybridization with a variety of labeled products such as cDNA for gene expression analysis or genomic DNA for strain comparisons, and genomic mismatch scanning purified DNA for genotyping (11).
METHODS
Script Design.
All scripts were written in UNIX Tool Command Language. Annotated sequence information from GenBank was extracted into one file containing the complete nucleotide sequence of a single chromosome. A second file contained the assigned ORF name followed by the start and stop positions of that ORF. The actual sequence contained within the specified range, along with 200 bases of sequence flanking both sides, was extracted and input into the primer selection program primer 0.5 (Whitehead Institute, Boston). Primers were designed so as to allow amplification of entire ORFs. The selected primer sequences were read by the 96-well automated multiplex oligonucleotide synthesizer instrument for primer synthesis. The forward and reverse primers were synthesized in two separate 96-well plates in corresponding wells. All primers were synthesized on a 20-nmol scale.
ORF Amplification and Purification.
Genomic DNA was isolated as described (12) and used as template for the amplification reactions. Each PCR was done in a total volume of 100 μl. A total of 0.2 μM each of forward and reverse primers were aliquoted into a 96-well PCR plate (Robbins Scientific, Sunnyvale, CA); a master mix containing 0.24 mM each dNTP, 10 mM Tris (pH 8.5), 50 mM MgCl2, 2.5 units Taq polymerase, and 10 ng of template was added to the primers, and the entire mix was thermal cycled for 30 cycles as follows: 15 min at 94°C, 15 min at 54°C, and 30 min at 72°C. Products were ethanol precipitated in polystyrene v-bottom 96-well plates (Costar). All samples were dried and stored at −20°C.
Arraying Procedure and Processing.
Microarrays were made as described (8).
A custom built arraying robot was used to print batches of 48 slides. The robot utilizes four printing tips which simultaneously pick up ≈1 μl of solution from 96-well microtiter plates. After printing, the microarrays were rehydrated for 30 sec in a humid chamber and then snap dried for 2 sec on a hot plate (100°C). The DNA was then UV crosslinked to the surface by subjecting the slides to 60 millijoules of energy. The rest of the poly-l-lysine surface was blocked by a 15-min incubation in a solution of 70 mM succinic anhydride dissolved in a solution consisting of 315 ml of 1-methyl-2-pyrrolidinone (Aldrich) and 35 ml of 1 M boric acid (pH 8.0). Directly after the blocking reaction, the bound DNA was denatured by a 2-min incubation in distilled water at ≈95°C. The slides were then transferred into a bath of 100% ethanol at room temperature.
Probe Preparation: cDNA.
Yeast cultures (100 ml) were grown to ≈1 ODA600 and total RNA was isolated as described (13). Up to 500 μg total RNA was used to isolate mRNA (Qiagen, Chatsworth, CA). Oligo(dT)20 (5 μg) was added and annealed to 2 μg of mRNA by heating the reaction to 70°C for 10 min and quick chilling on ice, plus 2 μl SuperScript II (200 units/μl) (Life Technologies, Gaithersburg, MD), 0.6 μl 50× dNTP mix (final concentrations were 500 μM dATP, dCTP, dGTP, and 200 μM dTTP), 6 μl 5× reaction buffer, and 60 μM Cy3-dUTP or Cy5-dUTP (Amersham). Reactions were carried out at 42°C for 2 h, after which the mRNA was degraded by the addition of 0.3 μl 5 M NaOH and 0.3 μl 100 mM EDTA and heating to 65°C for 10 min. The sample was then diluted to 500 μl with TE and concentrated using a Microcon-30 (Amicon) to 10 μl.
Probe Preparation: Genomic DNA.
Fluorescent DNA was prepared from total genomic DNA as follows: 1 μg of random nonamer oligonucleotides was added to 2.5 μg of genomic DNA. This mixture was boiled for 2 min and then chilled on ice. A reaction mixture containing dNTPs (25 μM dATP, dCTP, dGTP, 10 μM dTTP, and 40 μM Cy3-dUTP or Cy5-dUTP) reaction buffer (New England Biolabs), and 20 units exonuclease free Klenow enzyme (United States Biochemical) was added, and the reaction was incubated at 37°C for 2 h. The sample was then diluted to 500 μl with TE and concentrated using a Microcon-30 (Amicon) to 10 μl.
Hybridization.
Purified, labeled probe was resuspended in 11 μl of 3.5× SSC containing 10 μg Escherichia coli tRNA, and 0.3% SDS. The sample was then heated for 2 min in boiling water, cooled rapidly to room temperature, and applied to the array. The array was placed in a sealed, humidified, hybridization chamber. Hybridization was carried out for 10 h in a 62°C water bath, after which the arrays were washed immediately in 2× SSC/0.2% SDS. A second wash was performed in 0.1× SSC.
Analysis and Quantitation.
Arrays were scanned on a scanning laser fluorescence microscope developed by Steve Smith with software written by Noam Ziv (Stanford University). A separate scan was done for each of the two fluorophores used. The images were then combined for analysis. A bounding box, fitted to the size of the DNA spots, was placed over each array element. The average fluorescent intensity was calculated by summing the intensities of each pixel present in a bounding box and then dividing by the total number of pixels. Local area background was calculated for each array element by determining the average fluorescent intensity at the edge of the bounding box. To normalize for fluorophore-specific variation, control spots containing yeast genomic DNA were applied to each quadrant during the arraying process. These elements were quantitated and the ratios of the signals were determined. These ratios were then used to normalize the photomultiplier sensitivity settings such that the ratios of the fluorescence of the genomic DNA spots were close to a value of 1.0. The average signal intensity at any given spot was regarded as significant if it was at least two standard deviations above background. Each experiment was conducted in duplicate, with the fluorophores representing each channel reversed. The ratios presented here are the average of the two experiments, except in the case in which the signal for the element in question was below the reliability threshold. The reliability threshold also determined the dynamic range of the experiment. For all of the experiments presented, the average dynamic range was ≈1 to 100. In the case where the fluorescence from a very bright spot saturates the detector, differential ratios will, in general, be underestimated. This can be compensated for by scanning at a lower overall sensitivity.
RESULTS
The accumulation of sequence information from model organisms presents an enormous opportunity and challenge to understand the biological function of many previously uncharacterized genes. To do this accurately and efficiently, a directed strategy was developed that enables the monitoring of multiple genes simultaneously. Microarraying technology provides a method by which DNA can be attached to a glass surface in a high-density format (8). In practice, it is possible to array over 6,000 elements in an area less than 1.8 cm2. Given that the yeast genome consists of ≈6,100 ORFs, the entire set of yeast genes can be spotted onto a single glass slide.
With this capability and the availability of the entire sequence of the yeast genome, our strategy was to use a directed approach for generating the complete genome array. This procedure involved synthesizing a pair of oligonucleotide primers to amplify each ORF. The PCR product containing each gene of interest was arrayed onto glass and used, for example, as probe for monitoring gene expression levels by hybridizing to the array labeled cDNA generated from isolated mRNA of a culture grown under any experimental condition.
Primer Selection and Synthesis.
The primer selection was fully automated using Tool Command Language scripts and primer 0.5. (Whitehead). Primer pairs were automatically selected successfully for >99% of the ORFs tested. Primer sequences can thus be selected rapidly with minimal manual processing. A complete set of forward and reverse primers were selected initially for each ORF on chromosomes I, II, III, V, VI, VIII, IX, X, and XI. Primers for a representative set of ORFs (15% coverage) were chosen for the remaining chromosomes. With the release of the entire yeast genome sequence, the complete set of primers has now been selected.
Because each ORF requires a unique pair of synthetic primers, a total of approximately 12,200 oligonucleotides will be required to individually amplify each target. This costly component was addressed with the automated multiplex oligonucleotide synthesizer (6) which efficiently synthesizes primers in a 96-well format. Each primer, synthesized on a 20-nmol scale, provides enough material for 100 amplification reactions, whereas a given PCR product provides enough material to generate an element on 500–1,000 arrays. Thus, a single primer pair provides enough starting material for up to ≈50,000 arrays.
Primers were synthesized to amplify yeast ORFs. Primer synthesis had a failure rate of <1% in over 18 plates of synthesis as determined by standard trityl analysis (6). The success rate of the PCR amplifications using the primer pairs was 94% based on agarose gel analysis of each PCR. The purified PCR products were used to generate arrays. Two versions of the arrays were created for the experimental results presented here. The first array contained 2,287 elements and the second array batch contained 2,479 elements.
Genome Arrays.
The amplified ORFs were arrayed onto glass at a spacing of 345 microns (Fig. 1). The high-density spacing of DNA samples allows the hybridization volumes to be minimized—volumes are a maximum of 10 μl. The labeled probe can thus be maintained at relatively high concentrations, making 1–2 μg of mRNA sufficient for analysis. This also obviates the need for a subsequent amplification step and thus avoids the risk of altering the relative ratios of different cDNA species in the sample.
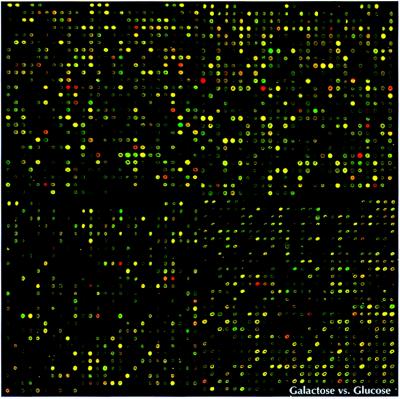
Figure 1.
Two-color fluorescent scan of a yeast microarray containing 2,479 elements (ORFs). The center-to-center distance between elements is 345 μm. A probe mixture consisting of cDNA from yeast extract/peptone (YEP) galactose (green pseudocolor) and YEP glucose (red pseudocolor) grown yeast cultures was hybridized to the array. Intensity per element corresponds to ORF expression, and pseudocolor per element corresponds to relative ORF expression between the two cultures.
Genetic Analysis: Genomic Comparison of Unrelated Strains.
Microarrays allow efficient comparison of the genomes of different strains. Genomic DNA from Y55, an S. cerevisiae strain divergent from the reference strain S288c, was randomly labeled with Cy3-dUTP and hybridized simultaneously with the S288c DNA labeled with Cy5-dUTP. When a comparison between the hybridization of the DNA from the two strains was done, several elements gave relatively little or no signal above background from the Cy3 channel (data not shown). These include SGE1, ASP3A-D, YLR156, YLR159, YLR161, ENA2 (YDR039 is ENA2), and YCR105. These results imply that the regions containing these genes are extremely divergent, or all together deleted from the strain. Subsequent attempts to generate PCR products from SGE1, ENA2, and ASP3A using Y55 DNA failed. This result supports the conclusion that these genes are likely to be missing from the Y55 genome. It is interesting to note that at least two of the regions absent in the Y55 genome have been previously shown or suggested to be deleted in mutant laboratory strains (14–16). In particular, the Asp-3 region appears to be highly prone to being deleted (15, 16).
These results indicate that gene arrays can be used to efficiently screen different strains of an organism for large deletion polymorphisms. A single hybridization and scan will reveal differences based on differential hybridization to particular elements. It is reasonable to suppose that an equivalent number of genes are present in the Y55 genome and absent in the S288c genome. This result should be viewed as a minimum estimate of the deletion polymorphisms that exist between these two unrelated strains as intergenic deletions or small intragenic deletions would not be detected because considerable hybridizing material would be remain. Sequence polymorphisms, such as deletions, are present in populations of every species and must at some level affect phenotype. One of the challenges of the genome era will be to critically examine sequence polymorphisms that exist in the natural gene pool relative to the reference genome sequence.
Gene Expression Analysis.
The arrays were used to examine gene expression in yeast grown under a variety of different conditions. Expression analysis is an ideal application of these arrays because a single hybridization provides quantitative expression data for thousands of genes. To better understand results for genes of known function, ORFs were placed in biologically relevant categories on the basis of function (e.g., amino acid catabolic genes) and/or pathways (e.g., the histidine biosynthesis pathway).
Heat Shock Results.
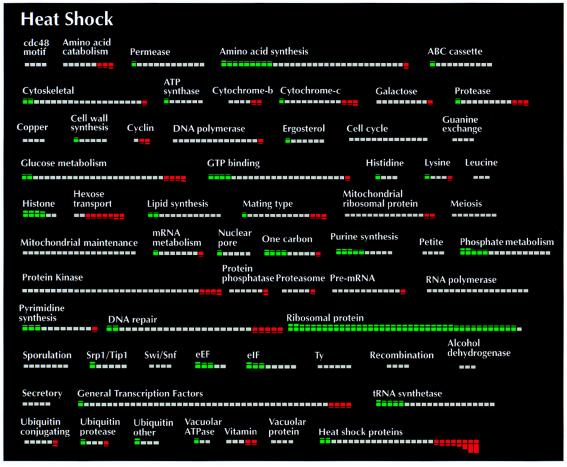
A log phase culture growing in YEP/dextrose medium at 25°C was split in half. One half of the culture remained at 25°C whereas the other half of the culture was shifted to 39°C. mRNA was isolated from both cultures 1 h after heat shock for comparison on microarrays and, although this time point is not optimal for measuring induction of heat shock mRNAs (17), many known heat shock genes exhibited considerable induction at this time point (Table 1; Fig. 2). Down-regulation of genes in the ribosomal protein and histone gene categories was also observed. Differential expression between the heat-shocked culture and the control was also observed for many other genes. Genes in many categories, such as amino acid catabolism and amino acid synthesis, exhibited a mixed response with some genes showing little or no differential expression and other genes showing a significant increase or decrease in gene expression in response to heat shock (Table 1; Fig. 2).
Table 1.
Heat shock vs. control expression data
| Ratio of gene expression
|
ORF | Gene | Description | |
|---|---|---|---|---|
| Control | Heat | |||
| 2.2 | YLR142 | PUT1 | Proline oxidase | |
| 2.0 | YOL140 | ARG8 | Acetylornithine aminotransferase | |
| 2.3 | YGL148 | ARO2 | Chorismate synthase | |
| 36.0 | YFL014 | HSP12 | Heat shock protein | |
| 27.4 | YBR072 | HSP26 | Heat shock protein | |
| 6.7 | YBR054 | YRO2 | Similarity to HSP30 heat shock protein Yrolp | |
| 3.4 | YCR021 | HSP30 | Heat shock protein | |
| 2.6 | YER103 | SSA4 | Heat shock protein | |
| 2.5 | YLR259 | HSP60 | Mitochondrial heat shock protein HSP60 | |
| 2.1 | YBR169 | SSE2 | Heat shock protein of the HSP70 family | |
| 1.7 | YBL075 | SSA3 | Cytoplasmic heat shock protein | |
| 1.4 | YPL240 | HSP82 | Heat shock protein | |
| 1.4 | YDR258 | HSP78 | Mitochondrial heat shock protein of clpb family of ATP-dependent proteases | |
| 1.0 | YNL007 | SIS1 | Heat shock protein | |
| 1.1 | YEL030 | 70-kDa heat shock protein | ||
| 1.9 | YHR064 | Heat shock protein | ||
| 1.3 | YBL008 | HIR1 | Histone transcription regulator | |
| 2.6 | YBL002 | HTB2 | Histone H2B.2 | |
| 3.3 | YBL003 | HTA2 | Histone H2A.2 | |
| 3.3 | YBR010 | HHT1 | Histone H3 | |
| 3.9 | YBR009 | HHF1 | Histone H4 | |
| 2.4 | YDR343 | HXT6 | High-affinity hexose transporter | |
| 2.1 | YHR092 | HXT4 | Moderate- to low-affinity glucose transporter | |
| 3.6 | YAR071 | PHO11 | Secreted acid phosphatase, 56 kDa isozyme | |
| 2.3 | YLR096 | KIN2 | Ser/Thr protein kinase | |
| 2.5 | YER102 | RPS8B | Ribosomal protein S8.e | |
| 2.6 | YBR181 | RPS101 | Ribosomal protein S6.e | |
| 2.6 | YCR031 | CRY1 | 40S ribosomal protein S14.e | |
| 2.7 | YLR441 | RP10A | Ribosomal protein S3.a.e | |
| 2.8 | YHR141 | RPL41B | Ribosomal protein L36a.e | |
| 2.8 | YBL072 | RPS8A | Ribosomal protein S8.e | |
| 2.8 | YHL015 | URP2 | Ribosomal protein | |
| 2.8 | YBR191 | URP1 | Ribosomal protein L21.e | |
| 3.1 | YLR340 | RPLA0 | Acidic Ribosomal protein L10.e | |
| 3.3 | YGL123 | SUP44 | Ribosomal protein | |
| 5.8 | YLR194 | Hypothetical protein | ||
Figure 2.
ORF categories displaying differential expression between heat shocked and untreated cultures. Bars within categories correspond to individual ORFs. Green shaded bars correspond to relative increases in ORF expression under 25°C growth conditions. Red shaded bars correspond to relative increases in ORF expression under 39°C growth conditions.
Cold Shock Results.
A log phase culture growing in YEP/dextrose medium at 37°C was split in half. One half of the culture remained at 37°C while the other half of the culture was shifted to 18°C. mRNA was isolated from both cultures 1 h after cold shock for comparison on microarrays. As expected, two known cold shock genes (TIP1, TIR1) were expressed at a significantly higher level in the cold-shocked culture. Genes in other functional categories, such as glucose metabolism and heat shock displayed a mixed response with expression of some genes being unaffected and other genes exhibiting significant up- or down-regulation in response to cold shock (Table 2).
Table 2.
Cold shock vs. control expression data
| Ratio of gene expression
|
ORF | Gene | Description | |
|---|---|---|---|---|
| Control | Cold | |||
| 3.3 | YOR153 | PDR5 | Pleiotropic drug resistance protein | |
| 2.4 | YCR012 | PGK1 | Phosphoglycerate kinase | |
| 2.9 | YCL040 | GLK1 | Aldohexose specific glucokinase | |
| 1.4 | YHR064 | Heat shock protein | ||
| 2.0 | YJL034 | KAR2 | Nuclear fusion protein | |
| 2.1 | YDR258 | HSP78 | Mitochondrial heat shock protein of clpb family of ATP-dependent proteases | |
| 2.2 | YLL039 | UBI4 | Ubiquitin precursor | |
| 2.7 | YLL026 | HSP104 | Heat shock protein | |
| 3.1 | YER103 | SSA4 | Heat shock protein | |
| 3.3 | YBR126 | TPS1 | α, α-Trehalose-phosphate synthase (UDP-forming) | |
| 3.8 | YPL240 | HSP82 | Heat shock protein | |
| 7.9 | YBR054 | YRO2 | Similarity to HSP30 heat shock protein Yrolp | |
| 7.9 | YBR072 | HSP26 | Heat shock protein | |
| 16.5 | YCR021 | HSP30 | Heat shock protein | |
| 1.8 | YDR343 | HXT6 | High-affinity hexose transporter | |
| 2.1 | YHR096 | HXT5 | Putative hexose transporter | |
| 2.4 | YFR053 | HXK1 | Hexokinase I | |
| 2.8 | YHR092 | HXT4 | Moderate- to low-affinity glucose transporter | |
| 3.4 | YHR094 | HXT1 | Low-affinity hexose (glucose) transporter | |
| 2.3 | YHR089 | GAR1 | Nucleolar rRNA processing protein | |
| 1.7 | YLR048 | NAB1B | 40S ribosomal protein p40 homolog b | |
| 1.7 | YLR441 | RP10A | Ribosomal protein S3a.e | |
| 1.7 | YLL045 | RPL4B | Ribosomal protein L7a.e.B | |
| 1.6 | YLR029 | RPL13A | Ribosomal protein L15.e | |
| 1.6 | YGL123 | SUP44 | Ribosomal protein | |
| 3.1 | YBR067 | TIP1 | Cold- and heat-shock-induced protein of the Srp1/Tip1p family | |
| 2.2 | YER011 | TIR1 | Cold-shock-induced protein of the Tir1p, Tip1p family | |
| 2.0 | YCR058 | Hypothetical protein | ||
| 4.2 | YKL102 | Hypothetical protein | ||
Steady-State Galactose vs. Glucose Results.
mRNA was isolated from steady-state log phase YEP galactose and YEP glucose grown cultures for comparison on the microarrays. As expected, the GAL genes were expressed at a much higher level in the galactose culture. Many genes were differentially expressed in these cultures that were not a priori expected to exhibit differential expression. For example, some genes in the amino acid catabolic category were up-regulated in the galactose culture whereas genes in the one-carbon metabolism and purine categories were largely or entirely down-regulated in the galactose culture (Table 3). Genes in other categories, such as amino acid synthesis, abc transporter, cytochrome c, and cytochrome b, exhibited mixed responses; some genes in a category showed little or no obvious differential expression whereas other genes in the same category showed significant differential expression in the galactose and glucose cultures.
Table 3.
Glucose vs. galactose expression data
| Ratio of gene expression
|
ORF | Gene | Description | |
|---|---|---|---|---|
| Glucose | Galactose | |||
| 2.1 | YHR018 | ARG4 | Arginosuccinate lyase | |
| 3.5 | YPR035 | GLN1 | Glutamate–ammonia ligase | |
| 2.8 | YML116 | ATR1 | Aminotriazole and 4-nitroquinoline resistance protein | |
| 2.0 | YMR303 | ADH2 | Alcohol dehydrogenase II | |
| 3.7 | YBR145 | ADH5 | Alcohol dehydrogenase V | |
| 3.2 | YBL030 | AAC2 | ADP, ATP carrier protein 2 | |
| 2.9 | YBR085 | AAC3 | ADP, ATP carrier protein | |
| 2.7 | YDR298 | ATP5 | H+-transporting ATP synthase δ chain precursor | |
| 2.5 | YBR039 | ATP3 | H+-transporting ATP synthase γ chain precursor | |
| 5.5 | YML054 | CYB2 | Lactate dehydrogenase cytochrome b2 | |
| 3.4 | YML054 | CYB2 | Lactate dehydrogenase cytochrome b2 | |
| 2.3 | YKL150 | MCR1 | Cytochrome-b5 reductase | |
| 4.2 | YBL045 | COR1 | Ubiquinol–cytochrome c reductase 44K core protein | |
| 3.5 | YDL067 | COX9 | Cytochrome c oxidase chain VIIA | |
| 2.7 | YLR038 | COX12 | Cytochrome c oxidase, subunit VIB | |
| 2.6 | YHR051 | COX6 | Cytochrome c oxidase subunit VI | |
| 2.4 | YLR395 | COX8 | Cytochrome c oxidase chain VIII | |
| 2.3 | YFR033 | QCR6 | Ubiquinol–cytochrome c reductase 17K protein | |
| 23.7 | YLR081 | GAL2 | Galactose (and glucose) permease | |
| 21.9 | YBR018 | GAL7 | UDP-glucose-hexose-1-phosphate uridylyltransferase | |
| 21.8 | YBR020 | GAL1 | Galactokinase | |
| 19.5 | YBR019 | GAL10 | UDP-glucose 4-epimerase | |
| 14.7 | YLR081 | GAL2 | Galactose (and glucose) permease | |
| 8.6 | YDR009 | GAL3 | Galactokinase | |
| 3.0 | YML051 | GAL80(1) | Negative regulator for expression of galactose-induced genes | |
| 2.8 | YML051 | GAL80(2) | Negative regulator for expression of galactose-induced genes | |
| 2.7 | YER055 | HIS1 | ATP phosphoribosyltransferase | |
| 3.4 | YBR248 | HIS7 | Glutamine amidotransferase/cyclase | |
| 7.4 | YCL030 | HIS4 | Phosphoribosyl-AMP cyclohydrolase/phosphoribosyl-ATP pyrophosphatase/histidinol dehydrogenase | |
| 5.8 | YKR080 | MTD1 | Methylenetetrahydrofolate dehydrogenase (NAD+) | |
| 6.0 | YDR019 | GCV1 | Glycine decarboxylase T subunit | |
| 6.1 | YLR058 | SHM2 | Serine hydroxymethyltransferase | |
| 8.1 | YML123 | PHO84 | High-affinity inorganic phosphate/H+ symporter | |
| 3.5 | YDR408 | ADE8 | Phosphoribosylglycinamide formyltransferase (GART) | |
| 3.6 | YDR408 | ADE8 | Phosphoribosylglycinamide formyltransferase (GART) | |
| 4.4 | YAR015 | ADE1 | Phosphoribosylamidoimidazole-succinocarboxamide synthase | |
| 5.6 | YMR300 | ADE4 | Amidophosphoribosyltransferase | |
| 5.6 | YOR128 | ADE2 | Phosphoribosylaminoimidazole carboxylase | |
| 6.0 | YGL234 | ADE5,7 | Phosphoribosylamine-glycine ligase and phosphoribosylformylglycinamidine cyclo-ligase | |
| 6.3 | YBL015 | ACH1 | Acetyl-CoA hydrolase | |
DISCUSSION
The results of these experiments show that many genes are differentially expressed under the three environmental conditions described here. The expected and predicted changes in gene expression, such as HSP12 in the heat-shocked culture, TIP1 in the cold-shocked culture, and GAL2 in the steady-state galactose culture, were observed in every case. However, in addition to the expected changes in gene expression, significant differential expression was also observed for many other genes that would not, a priori, be expected to be differentially expressed. For example, expression of PHO11 decreased and expression of YLR194, KIN2, and HXT6 increased in the heat shocked culture. Expression of MST1 and APE3 decreased and expression of PDR5 and GAR1 increased in the cold-shocked culture. In addition, ADE4 and SER2 were expressed at reduced levels whereas PHO84 and ACH1 were expressed at higher levels in cells grown in galactose compared with cells grown in glucose. Differential expression of these and many other genes was specific to one of these three environmental conditions.
Many other genes were found to be differentially expressed under more than one condition. When differentially expressed genes in cold- and heat-shocked cultures were compared, 30 genes were found in common. Of these 30 genes, 28 showed inverse expression (i.e., increased expression under one condition and decreased expression under the other condition). Two genes, YCR058 and YKL102, showed elevated expression in response to both cold and heat shock. Fifteen genes were found to be differentially expressed in both the heat-shocked and steady-state galactose cultures: 9 genes showed increased expression and 5 showed decreased expression under both conditions. Twenty genes were differentially expressed in both the cold-shocked and steady-state galactose cultures: 8 genes showed decreased expression and 5 genes showed increased expression under both conditions. Six genes showed increased expression in the galactose culture and decreased expression in the cold shocked culture. One gene (ODP1) showed increased expression in both the cold-shocked and steady-state galactose cultures.
Gene expression is affected in a global fashion when environmental conditions are changed and both expected and unexpected genes are affected. There is also overlap in the genes that are differentially expressed under quite different environmental conditions. These results can be rationalized by considering the high degree of cross-pathway regulation in yeast. For example, there is evidence for cross-pathway regulation between (i) carbon and nitrogen metabolism (18), (ii) phosphate and sulfate metabolism (19), and (iii) purine, phosphate, and amino acid metabolism (20–24). There are also examples of the interaction of general and specific transcription factors (25, 26). Finally, within the broad class of amino acid biosynthetic genes, there is evidence for amino acid specific regulation of some genes, regulation via general control for other genes, and regulation via both specific and general control for other genes (22, 27–30).
Cross-pathway regulation arises from the complex structure of promoters. Virtually all promoters contain sites for multiple transcription factors and, therefore, virtually all genes are subject to combinatorial regulation. For example, the HIS4 promoter contains binding sites for GCN4 (the general amino acid control transcription factor), PHO2/BAS2 (a transcriptional regulator of phosphatase and purine biosynthetic genes), and BAS1 (a transcriptional regulator of purine biosynthetic genes) (31). It is likely that the complex effects on gene expression described in this work are a direct consequence of the combinatorial regulation of gene expression.
These findings illustrate the power of the highly parallel whole genome approach when examining gene expression. The global effects of environmental change on gene expression can now be directly visualized. It is clear that determining the mechanism(s) and the functional role of the dramatic global effects on gene expression in different environments will be a significant challenge. The era of whole genome analysis will, ultimately, allow researchers to switch from the very focused single gene/promoter view of gene expression and instead view the cell more as a large complex network of gene regulatory pathways.
With the entire sequence of this model organism known, new approaches have been developed that allow for genome wide analyses (32, 33) of gene function. The genome microarrays represent a novel tool for genetic and expression analysis of the yeast genome. This pilot study uses arrays containing >35% of the yeast ORFs and it is clear that the entire set of ORFs from the yeast genome can be arrayed using the directed primer based strategy detailed here. Recent advances in arraying technology will allow all 6,100 ORFs to be arrayed in an area of less than 1.8 cm2. Furthermore, as the technology improves, detection limits will allow less than 500 ng of starting mRNA material to be used for making probe.
The genome arrays provide for a robust, fully automated approach toward examining genome structure and gene function. They allow for comparisons between different genomes as well as a detailed study of gene expression at the global level. This research will help to elucidate relationships between genes and allow the researcher to understand gene function by understanding expression patterns across the yeast genome.
Acknowledgments
Support was provided by National Institutes of Health Grant P0/HG00205.
ABBREVIATION
- YEP
yeast extract/peptone
References
- 1.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 2.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, et al. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 3.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 4.Sulston J, Du Z, Thomas K, Wilson R, Hillier L, et al. Nature (London) 1992;356:37. doi: 10.1038/356037a0. [DOI] [PubMed] [Google Scholar]
- 5.Newman T, de Bruijn F J, Green P, Keegstra K, Kende H, et al. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lashkari D A, Hunicke-Smith S P, Norgren R M, Davis R W, Brennan T. Proc Natl Acad Sci USA. 1995;92:7912–7915. doi: 10.1073/pnas.92.17.7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 8.Shalon D, Smith S, Brown P O. Genome Res. 1996;6:639–645. doi: 10.1101/gr.6.7.639. [DOI] [PubMed] [Google Scholar]
- 9.Heller R A, Schena M, Chai A, Shalon D, Bedilion T, Gilmore J, Woolley D E, Davis R W. Proc Natl Acad Sci USA. 1997;94:2150–2155. doi: 10.1073/pnas.94.6.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeRisi J, Penland L, brown P O, Bittner M L, Meltzer P S, Ray M, Chen Y, Su Ya, Trent J M. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 11.Nelson S F, McCusker J H, Sander M A, Kee Y, Modrich P, Brown P O. Nat Genet. 1993;4:11–18. doi: 10.1038/ng0593-11. [DOI] [PubMed] [Google Scholar]
- 12.Hoffman C S, Winston F. Gene. 1989;84:473–479. doi: 10.1016/0378-1119(89)90523-4. [DOI] [PubMed] [Google Scholar]
- 13.Schmitt M, Brown T, Trumpower B. Nucleic Acids Res. 1990;18:3091. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehrenhofer-Murray A E, Wurgler F E, Sengstag C. Mol Gen Genet. 1994;244:287–294. doi: 10.1007/BF00285456. [DOI] [PubMed] [Google Scholar]
- 15.Kim K-W, Kamerud J Q, Livingston D M, Roon R J. J Biol Chem. 1988;263:11948–11953. [PubMed] [Google Scholar]
- 16.Kim K-W, Roon R J. J Bacteriol. 1984;157:958–961. doi: 10.1128/jb.157.3.958-961.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Craig E A. In: The Molecular Biology of the Yeast Saccharomyces: Gene Expression. Jones E W, Pringle J R, Broach J R, editors. Vol. 2. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 501–537. [Google Scholar]
- 18.Dang V D, Bohn C, Bolotin-Fukuhara M, Daignan-Fornier B. J Bacteriol. 1996;178:1842–1849. doi: 10.1128/jb.178.7.1842-1849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Connell K F, Baker R E. Genetics. 1992;132:63–73. doi: 10.1093/genetics/132.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braus G, Mosch H U, Vogel K, Hinnen A, Hutter R. EMBO J. 1989;8:939–945. doi: 10.1002/j.1460-2075.1989.tb03455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosch H U, Scheier B, Lahti R, Mantsala P, Braus G H. J Biol Chem. 1991;266:20453–20456. [PubMed] [Google Scholar]
- 22.Mitchell A P, Magasanik B. Mol Cell Biol. 1984;4:2767–2773. doi: 10.1128/mcb.4.12.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daignan-Fornier B, Fink G R. Proc Natl Acad Sci USA. 1992;89:6746–6750. doi: 10.1073/pnas.89.15.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tice-Baldwin K, Fink G R, Arndt K T. Science. 1989;246:931–935. doi: 10.1126/science.2683089. [DOI] [PubMed] [Google Scholar]
- 25.Messenguy F, Dubois E. Mol Cell Biol. 1993;13:2586–2592. doi: 10.1128/mcb.13.4.2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Devlin C, Tice-Baldwin K, Shore D, Arndt K T. Mol Cell Biol. 1991;11:3642–3651. doi: 10.1128/mcb.11.7.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magasanik B. In: The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression. Jones E W, Pringle J R, Broach J R, editors. Vol. 2. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 283–317. [Google Scholar]
- 28.Hinnebusch A G. In: The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression. Jones E W, Pringle J R, Broach J R, editors. Vol. 2. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 319–414. [Google Scholar]
- 29.Brisco P R, Kohlhaw G B. J Biol Chem. 1990;265:11667–11675. [PubMed] [Google Scholar]
- 30.O’Connell K F, Surdin-Kerjan Y, Baker R E. Mol Cell Biol. 1995;15:1879–1888. doi: 10.1128/mcb.15.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arndt K T, Styles C, Fink G R. Science. 1987;237:874–880. doi: 10.1126/science.3303332. [DOI] [PubMed] [Google Scholar]
- 32.Smith V, Chou K N, Lashkari D, Botstein D, Brown P O. Science. 1996;274:2069–2074. doi: 10.1126/science.274.5295.2069. [DOI] [PubMed] [Google Scholar]
- 33.Shoemaker D D, Lashkari D A, Morris D, Mittman M, Davis R W. Nat Genet. 1996;14:450–456. doi: 10.1038/ng1296-450. [DOI] [PubMed] [Google Scholar]