Abstract
Apolipoprotein B (apoB) mRNA editing catalyzed by apoB mRNA editing catalytic subunit 1 (APOBEC-1) has been proposed to be a nuclear process. To test this hypothesis, the subcellular distribution of hemagglutinin-(HA) tagged APOBEC-1 expressed in transiently transfected hepatoma cells was determined by indirect immunofluorescence microscopy. HA-APOBEC-1 was detected in both the nucleus and cytoplasm of rat and human hepatoma cells. Mutagenesis of APOBEC-1 demonstrated that the N-terminal 56 amino acids (1–56) were necessary for the nuclear distribution of APOBEC-1, but this region did not contain a functional nuclear localization signal (NLS). However, we identified a 24-amino acid domain in the C terminus of APOBEC-1 with characteristics of a cytoplasmic retention signal (CRS) or a nuclear export signal (NES). These data suggest, therefore, that the nuclear import of APOBEC-1 may not be mediated by a positive NLS; rather, it may be achieved by overcoming the effect of a CRS/NES. We also demonstrated that the nuclear distribution of APOBEC-1 occurred only in cell lines that were capable of editing apoB RNA. We propose that the cellular distribution of APOBEC-1 is determined by multiple domains within this protein, and a nuclear localization of the enzyme may be regulated by cell type-specific factors that render these cells uniquely editing competent.
Apolipoprotein B (apoB) mRNA undergoes posttranscriptional deamination editing of a cytidine at nucleotide 6666, converting a CAA glutamine codon to a UAA in-frame translation stop codon (1, 2). Translation of unedited and edited variants of apoB mRNA generates two isoforms of apoB proteins, apoB100 and apoB48, which play distinct roles in lipid metabolism pathways (3). A tripartite RNA sequence motif consisting of a mooring sequence, a spacer, and a regulatory element is required for site-specific RNA editing (4–8).
apoB mRNA editing catalytic subunit 1 (APOBEC-1) is the catalytic subunit of the editosome, a multiprotein complex assembled to edit apoB RNA (9–11). It is a cytidine deaminase and shares amino acid homology in its catalytic domain with a variety of cytidine and adenosine deaminases (12–14). In the absence of editosomal proteins, APOBEC-1 cannot edit apoB RNA (11, 12, 15). Proteins that complement APOBEC-1 in apoB mRNA editing activity (auxiliary proteins) have been identified in diverse tissues and cell types (11, 12, 16–18) and seem to play important but uncharacterized roles in regulating editing activity during tissue development as well as in response to nutritional stress and hormone stimulation (17–19).
Compelling evidence for a nuclear localization of the editing activity has been provided through the demonstration that a portion of cellular apoB RNA molecules is edited before polyadenylation and splicing (20, 21). APOBEC-1 has two short stretches of basic amino acids separated by 12 residues in its N terminus, which closely resemble the consensus bipartite nuclear localization signal (NLS) (18, 22).
Indirect immunofluorescence microscopy was used to study the intracellular distribution of hemagglutinin (HA)-tagged APOBEC-1 in transiently transfected cells. The data demonstrate that APOBEC-1 can distribute in the nucleus and the cytoplasm, but a nuclear distribution is unique to editing-competent cells. Moreover, these studies revealed a presumptive cytoplasmic localization signal in the C terminus of APOBEC-1 that plays a prominent role in determining the enzyme’s intracellular distribution.
MATERIALS AND METHODS
Plasmids.
The vector expressing HA fusion proteins (6his-HA-pcDNA3) was derived from pcDNA3 (Invitrogen, San Diego) by inserting a 60-bp fragment containing the translational initiation ATG and sequence encoding six histidine residues and the HA epitope YPYDVPDYA at the BamHI/EcoRV sites.
Wild-type apobec-1 cDNA was PCR amplified by pfu DNA polymerase (Stratagene) from pPROEX-apobec-1 (23) by using primers YY5′/YY3′. The PCR products were digested and subcloned into 6his-HA-pcDNA3 at the EcoRV/XbaI sites (6his-HA-apobec-1/pcDNA3). Plasmids encoding the N- or C-terminal deletion mutants [APOBEC delNT(1–56) and APOBEC delCT(173–229)] were constructed analogously by cloning PCR fragments generated by primers N1/SP6 and T7/C1, respectively, into 6his-HA-pcDNA3 at the EcoRV/XbaI sites.
A chicken muscle pyruvate kinase (CMPK) fragment obtained by SmaI/XbaI digestion of myc-CMPK/pcDNA3 (24) was subcloned in 6his-HA-pcDNA3 at the EcoRV/XbaI sites to create 6his-HA-CMPK/pcDNA3. A 27-bp fragment encoding the simian virus 40 (SV40) NLS (PKKKRKV) was inserted in-frame with CMPK into an EcoRI site immediately upstream of the CMPK cDNA to create the NLS-CMPK construct.
Plasmids encoding APOBEC-1-CMPK were constructed by subcloning PCR products amplified from 6his-HA-apobec-1/pcDNA3 using primers T7/7–14-a in 6his-HA-CMPK/pcDNA3 at HindIII/EcoRI sites. Plasmids encoding N- or C-terminal deletion variants of APOBEC-1 fused to the N terminus of CMPK were generated by an analogous PCR strategy. Individual constructs and the primers and templates used for PCR in their constructions are as follows: APOBEC delNT(1–56)-CMPK, primers T7/7–14-a and plasmid encoding APOBEC delNT(1–56); APOBEC delCT(173–229)-CMPK, primers T7/7–14-b and plasmid encoding APOBEC delCT(173–229); and APOBEC NT(1–47)-CMPK, primers T7/N27 and plasmid 6his-HA-apobec-1/pcDNA3. All PCR products were subcloned into 6his-HA-CMPK/pcDNA3 at the HindIII/EcoRV sites.
Plasmids encoding CMPK-APOBEC-1 chimeric proteins were constructed by cloning PCR products generated from 6his-HA-CMPK/pcDNA3 using primers T7/ck3′ into 6his-HA-apobec-1/pcDNA3 at HindIII/EcoRV sites. This HindIII/EcoRV fragment was also cloned into plasmids encoding APOBEC delNT(1–56) and APOBEC delCT(173–229) to make constructs expressing CMPK-APOBEC delNT(1–56) and CMPK-APOBEC delCT(173–229), respectively.
NLS-CMPK and APOBEC-1 fusions were generated as described above except using plasmid encoding NLS-CMPK as PCR template, and the PCR products were cloned into corresponding plasmids encoding APOBEC-1 variants at HindIII/EcoRV sites. Plasmid for expressing NLS-CMPK-APOBEC CT(173–229) was constructed by fusing EcoRV/XbaI-digested PCR fragments encoding the C-terminal 57 amino acids of APOBEC-1 using primers 27l/SP6 to the 3′ end of NLS-CMPK cDNA. This plasmid was used as a template to generate PCR fragments by using primers T7/27–196, which were cloned into 6his-HA-pcDNA3 at HindIII/XbaI sites to make plasmids encoding NLS-CMPK-APOBEC CT(173–196). A 60-bp fragment encoding the leucine-rich (LR) motif (180–196) was fused to the 3′ end of NLS-CMPK cDNA to express NLS-CMPK-APOBEC-CT(180–196).
Plasmids for expression of recombinant proteins in Escherichia coli were constructed by cloning PCR fragments generated from constructs encoding corresponding APOBEC-1 variants by using primers HT/SP6 in pPROEX-1 (GIBCO/BRL) at NdeI/XbaI sites. Plasmid for expression APOBEC delLR(180–196) was constructed by cloning PCR fragments generated from 6his-HA-apobec-1/pcDNA3 by using primers DL/SP6 in HA-apobec-1/pPROEX-1 at SnaBI/XbaI sites.
Cell Culture and Transfection.
Rat hepatoma McArdle RH7777, Chinese hamster ovary (CHO), monkey kidney COS-7, and human hepatoma HepG2 cells were obtained from the American Type Culture Collection and maintained under the recommended culture conditions. Cells were seeded on two-well Lab-Tek Chamber Slide (Nunc) 18 h before transfection. McArdle, HepG2, and CHO cells were transfected with 4–5 μg of DNA per 105 cells by the calcium phosphate precipitation method as described previously (25). COS-7 cells were transfected with 1 μg of DNA per 105 cells by Lipofectamine Reagent (GIBCO/BRL).
Immunofluoresence Microscopy.
Cells were fixed 48 h after transfection in 2% paraformaldehyde for 15 min on ice and permeabilized for 15 min with ice-cold 0.4% Triton X-100 in PBS. Immunostaining was performed by incubating fixed cells for 1 h at room temperature with anti-HA mAb (Babco, Berekely, CA; 1:500 in PBS containing 3% BSA), followed by incubation for 1 h at room temperature with fluorescein-conjugated goat anti-mouse antibody (Organon Teknika–Cappel; 1:25 in 3% BSA/PBS). Coverslips were mounted on the glass slides with PBS containing 90% glycerol, 4% n-propyl gallate, and 1.5 μg of 4′,6-diamidino-2-phenylindole (DAPI)/ml. Slides were observed under an Olympus BH-2 fluorescence microscope using a 40× Olympus objective or as otherwise stated. Proteins expressed in transfected cells were determined to be full-length by Western blotting analysis.
Preparation of Cellular Extracts and Recombinant Proteins.
Fifty 150-mm dishes of 80% confluent cells were harvested by scraping into cold buffer A (50 mM Tris⋅HCl, pH 8.0/5 mM MgCl2/2 mM EGTA/150 mM NaCl/0.25 mM DTT) with 1 mM phenylmethylsulfonyl fluoride (Sigma), 5 mg/ml aprotinin (Sigma), and 5 mg/ml leupeptin (Boehringer Mannheim). Cells were collected by centrifugation (800 × g, 3 min), washed once in buffer A, and processed into whole cell S100 extracts as described previously (9, 10).
Recombinant proteins were expressed in E. coli and purified by Ni-NTA (Qiagen, Chatsworth, CA) metal-chelating chromatography as described previously (23).
Western Blot Analysis.
Cell extracts were resolved by SDS/10.5% PAGE, transferred to nitrocellulose membrane (Schleicher & Schuell), probed with anti-HA mAb and a secondary peroxidase-conjugated goat anti-mouse IgG (Zymed Laboratories), and visualized with the ECL kit (Amersham).
Editing Assay.
In vitro apoB RNA editing assay was performed, and the editing efficiency was evaluated by poisoned primer extension as previously described (23). The primer extension products were resolved on denaturing 10% polyacrylamide gels and quantified by laser densitometric scanning (PhosphoImager model 425E, Molecular Dynamics). The percentage of editing activity was calculated as the number of densitometric units corresponding to edited RNA (UAA) divided by the sum of this number plus that corresponding to unedited RNA (CAA).
Primers.
Primers used were as follows, with their 5′ nucleotide in parentheses. YY5′, GGGGCCGATATCGTGAGTTCCGAGAC (nucleotide 14 of apobec-1 cDNA); YY3′, GCTCTAGAGCTCATTTCAACCCTGTG (nucleotide 675 of apobec-1 cDNA); N1, CGGATATCAACACCAACAAACACG (nucleotide 184 of apobec-1 cDNA); SP6, GCTCTAGCATTTAGGTGACACTATAG; T7, TAATACGACTCACTATAGGG; C1, GCTCTAGATCAGAATTCATGGGGGTACCTTGG (nucleotide 501 of apobec-1 cDNA); 7-14-a, CCGGAATTCCCTTTCAACCCTGTGGC (nucleotide 673 of apobec-1 cDNA); 7-14-b, CCGGAATTCAGATGGGGGTACCTTGG (nucleotide 501 of apobec-1 cDNA); N27, CCGGAATTCCTTCCTCCCCAG (nucleotide 170 of apobec-1 cDNA); ck3′, GGCGATATCTGGCACGGGCACCAC (nucleotide 1573 of CMPK cDNA); 27l, GGCGATATCCTGTGGGTGAGGCTG (nucleotide 531 of apobec-1 cDNA); 27-196, GGCTCTAGATCATAAAATATTTAAACAGGGTGG (nucleotide 568 of apobec-1 cDNA); HT, GGGAATTCCATATGTACCCCTACGACGTGC; and DL, CCGTACGTAAGAAGAAAACAACCTCAACTC (nucleotide 619 of apobec-1 cDNA).
RESULTS
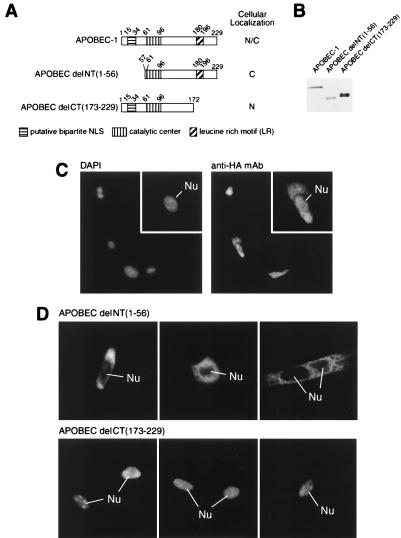
The expression level of APOBEC-1 in cells and tissues is typically below the detection limit of most immunoassays. We have therefore evaluated the intracellular distribution of the enzyme by engineering an HA-tag to its N terminus (Fig. 1A) and expressing this construct in McArdle cells (Fig. 1 B and C). Indirect immunofluorescence microscopy revealed that HA-APOBEC-1 distributed in both the nucleus and the cytoplasm of transiently transfected McArdle cells (Fig. 1C).
Figure 1.
Structure and subcellular localization of APOBEC-1 and deletion mutants. (A) The diagram of each construct and the corresponding domains are drawn in scale with a summary of their intracellular distribution. (B) An equivalent number of cells from the corresponding transfections were analyzed by Western blotting. Note that the C-terminal deletion mutant migrated slower than expected for its predicted molecular mass. (C) DAPI-stained nucleus and anti-HA mAb-stained HA-APOBEC-1 in transiently transfected McArdle cells. Insets show a high magnification of one of the transfected cells. Nu, nucleus. (D) The N- or C-terminal deletion mutants were localized in McArdle cells by indirect immunofluorescence microscopy. Three representative fields are shown for each transfected construct.
The molecular mass of HA-APOBEC-1 is approximately 30 kDa, well below the molecular cut-off size of the nuclear pore complex for diffusion (22). To evaluate whether the nuclear distribution of APOBEC-1 was due to diffusion, the N-terminal first 56 amino acids containing a presumptive NLS were deleted, and the mutant construct [APOBEC delNT(1–56)] was transfected into McArdle cells (Fig. 1 B and D). Despite its smaller size (Fig. 1B), the mutant protein was excluded from the nucleus (Fig. 1D). As a control for the effect of protein size on the cellular distribution, the C-terminal 57 amino acids of APOBEC-1 were removed, and the mutant construct was transfected into McArdle cells [APOBEC delCT(173–229)] (Fig. 1, B and D). This construct was entirely localized in the nucleus (Fig. 1D). The data suggest that the N-terminal 56 amino acids are required for the nuclear distribution of APOBEC-1. They also demonstrate that despite slightly higher expression of APOBEC delCT(173–229) compared with expression of APOBEC-1 and APOBEC delNT(1–56) (Fig. 1B), this protein was capable of virtually quantitative nuclear localization. We interpret these data as suggesting that under our conditions, the intracellular distribution of protein is a consequence of its amino acid sequence rather than being the result of overexpression of protein in the cells.
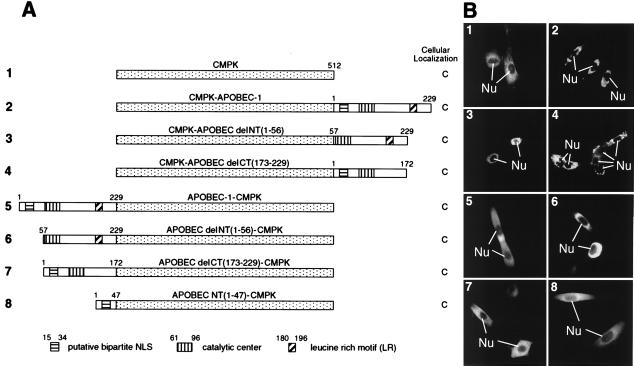
To determine the sequence responsible for APOBEC-1 nuclear distribution, we adopted the scenario most frequently used to define an NLS, which is to test the ability of a specific sequence of interest to target a heterologous nonnuclear protein to the nucleus. CMPK is a well characterized reporter protein for this purpose (24, 26). CMPK was localized in the cytoplasm when expressed in McArdle cells (Fig. 2, B1), but it could be targeted to the nucleus as an SV40 NLS-CMPK fusion protein (Fig. 3 A and B1). To evaluate the possibility of an NLS in APOBEC-1, a series of chimeric proteins containing CMPK and either full-length or truncated APOBEC-1 were created and expressed in McArdle cells (Fig. 2A). All of these fusion proteins were localized in the cytoplasm regardless of the relative orientation of CMPK and APOBEC in the chimeras (Fig. 2B2–7). In addition, the N-terminal 47 amino acids of APOBEC-1, including the putative bipartite-like NLS, failed to target CMPK to the nucleus (Fig. 2B8). Taken together, the data suggest that the N terminus of APOBEC-1 is necessary for nuclear distribution of the enzyme, but it is not sufficient to function as an NLS. The data also suggest the possibility that the C terminus of APOBEC-1 may have a cytoplasmic retention signal (CRS) or a nuclear export signal (NES).
Figure 2.
Subcellular localization of CMPK and APOBEC fusions. (A) The diagram of each construct and the corresponding domains are drawn in scale with a summary of their intracellular distribution. (B) Subcellular localization of the various CMPK-APOBEC chimeric proteins. Numbering of the panels indicates the constructs used in transfection and corresponds to that in A.
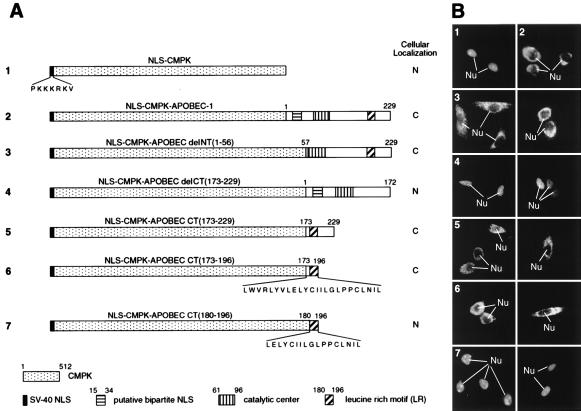
Figure 3.
Delineation of the putative CRS of APOBEC-1. (A) The diagram of each construct and the corresponding domains are drawn in scale with a summary of their intracellular distribution. (B) Subcellular localization of the various fusion proteins. Numbering of the panels indicates the constructs used in transfection and corresponds to that in A.
To evaluate these possibilities, a fusion protein in which the well characterized SV40 NLS (26) was fused to the N terminus of CMPK was constructed (NLS-CMPK, Fig. 3A). As predicted, this protein was localized in the nucleus (Fig. 3B1). However, fusion of APOBEC-1 to the C terminus of NLS-CMPK resulted in a cytoplasmic localization (Fig. 3B2). This is unlikely because of the size of the fusion protein, as SV40 NLS has been reported to be able to target much larger proteins into the nucleus (27, 28).
To further delineate the region of APOBEC-1 capable of overriding the SV40 NLS, we fused NLS-CMPK with portions of APOBEC-1 and localized these proteins in McArdle cells after transient transfection (Fig. 3A). Proteins fused with the N-terminal deletion of APOBEC-1 [NLS-CMPK-APOBEC delNT(1–56)] were localized in the cytoplasm (Fig. 3B3), whereas those fused with the C-terminal deletion of APOBEC-1 [NLS-CMPK-APOBEC delCT(173–229)] were localized in the nucleus (Fig. 3B4). Fusion of the C-terminal 57 amino acids of APOBEC-1 to NLS-CMPK [NLS-CMPK-APOBEC CT(173–229)] was sufficient for localizing the chimeric protein to the cytoplasm (Fig. 3B5). NLS-CMPK chimeras were localized to the cytoplasm with as little as 24 amino acids from the C terminus of APOBEC-1 [NLS-CMPK-APOBEC CT(173–196); Fig. 3, B6], but further deletions to just the 17 amino acid LR motif [NLS-CMPK-APOBEC CT(180–196)] seemed to abolish the signal as this chimeric protein was localized within the nucleus (Fig. 3, B7). These results, taken together with those from the localization of just the N- and C-terminal deletions of APOBEC-1, strongly support the possibility that the C terminus of APOBEC-1 contains either a CRS or an NES.
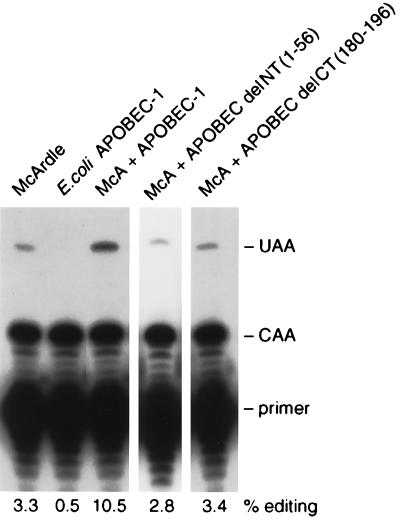
The intracellular distributions of the N- and C-terminal deletion constructs of APOBEC-1 may have been related to their editing activity. To evaluate this, purified E. coli-expressed recombinant proteins were assayed for in vitro editing activity in the presence of auxiliary proteins from McArdle cell extracts. Both N- and C-terminal LR motif deletion constructs were unable to carry out apoB RNA editing (Fig. 4). The essential role of the C-terminal LR motif for APOBEC-1 editing activity has been demonstrated (11, 15, 18, 29, 30), but the significance of the N-terminal 56 amino acids in RNA editing is a novel finding.
Figure 4.
N-terminal 56 amino acids and the LR motif are required for editing activity of APOBEC-1. Primer extension analysis of editing activity of 5 μg of recombinant protein in the presence of 34 μg of McArdle cell extracts is shown. The positions of primer and the extension products generated from unedited (CAA) and edited (UAA) RNAs are indicated. Similar results were obtained from two independent preparations of the recombinant proteins.
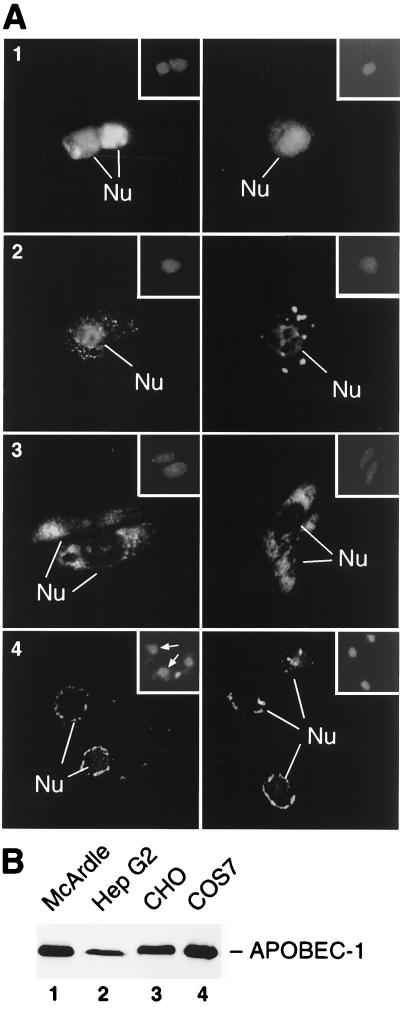
To evaluate whether the subcellular distribution of APOBEC-1 was related to the expression of auxiliary factors, the enzyme’s localization was determined in several cell types. HepG2 cells express apoB mRNA but do not synthesize APOBEC-1 (31). These cells express auxiliary factors and will edit apoB mRNA when transfected with apobec-1 cDNA (31). HA-APOBEC-1 displayed prominent nuclear staining in HepG2 cells with large cytoplasmic speckles (Fig. 5A2). Despite the difference in transfection efficiency and the total expression level of APOBEC-1 in HepG2 comparing with that in McArdle cells (Fig. 5, A1, B, and legend), HepG2 still displayed a cytoplasmic component, suggesting once again that the observed distributions were a function of cell signaling.
Figure 5.
Subcellular localization of APOBEC-1 in apoB RNA-editing-competent versus incompetent cell lines. (A) HA-APOBEC-1 was expressed and localized in McArdle (A1), HepG2 (A2), CHO (A3), and COS-7 (A4) cells by immunofluorescence analysis. Insets correspond to DAPI-stained fields indicating the location of the cell nucleus. The transfection efficiencies were as follows: McArdle, 15% ± 6%, n = 4; HepG2, 4% ± 2%, n = 5; CHO 7% ± 2%, n = 4; COS-7 23% ± 7%, n = 4. Two representative fields for each transfection are shown. (B) An equal amount of extracts from 105 cells of each cell line was used to evaluate the expression level of APOBEC-1 by immunoblotting analysis.
The subcellular distribution of HA-APOBEC-1 in cell lines known to lack the auxiliary factors was strikingly different from that seen in the editing competent cell lines. CHO and COS-7 were selected for these studies, because these cell lines do not contain either APOBEC-1 or auxiliary factors (refs. 16, 18, 32, and 33, and data not shown). APOBEC-1 demonstrated an intense general cytoplasmic distribution in CHO cells and cytoplasmic but perinuclear distribution in COS-7 cells (Fig. 5 A3 and 4). Neither cell line demonstrated significant nuclear distribution of APOBEC-1 despite the fact that the expression of APOBEC-1 was comparable with that seen in McArdle and HepG2 cells (Fig. 5B). The data suggest that nuclear localization of APOBEC-1 is cell type specific and that this capability and the expression of complementing auxiliary factors are coincident and unique characteristics of cells capable of editing apoB mRNA.
DISCUSSION
We have evaluated the intracellular distribution of APOBEC-1 through the expression of HA-tagged protein in transfected cells. The data revealed four novel characteristics of APOBEC-1. The enzyme was distributed in both the nucleus and cytoplasm of transfected human and rat hepatoma cells. This is an important finding, as it provides direct evidence that APOBEC-1 can reside within the nucleus, where it might have access to and edit pre-mRNA.
Second, the N terminus of APOBEC-1 was required for nuclear localization but was not sufficient in localizing a reporter protein to the nucleus and therefore did not meet the criteria of an NLS as previously proposed (18). The finding that the C-terminal deletion construct of APOBEC-1 quantitatively localized in the nucleus suggested that the remaining portion of APOBEC-1 contained a nuclear distribution determinant. This signal was not sufficient for nuclear import of the larger CMPK fusion proteins, suggesting that it may be a weak signal or highly dependent on conformational status. Whatever the mechanism for APOBEC-1 nuclear distribution is, the data suggest that it must involve coregulation of signaling from its C terminus.
In this regard, a 24-amino acid domain within the C terminus of the enzyme has been identified as a powerful determinant for cytoplasmic distribution in all cell types examined, a characteristic consistent with a putative CRS. Interestingly, this domain shared no homology with the CRS defined in other proteins. The domain’s leucine rich (LR) characteristic is similar to an NES found in cAMP-dependent protein kinase inhibitor and HIV Rev protein (34, 35). The LR motif has been shown to be required for APOBEC-1 homodimerization and RNA editing activity (29, 30). It shares high homology with the repeat unit of the leucine-rich repeats, which have been proposed to provide a general structure for diverse molecular interactions (36). However, the APOBEC-1 LR motif was not sufficient as a putative CRS/NES, and therefore additional information in the form of primary amino acid sequence or conformation must be involved. Moreover, the NES of protein kinase inhibitor is a slightly weaker signal than SV40 NLS, and a reporter protein bearing both signals had a relatively even distribution in the nucleus and the cytoplasm (34). This is in contrast to what was observed for the C terminus of APOBEC-1, which was dominant over the SV40 NLS. Despite these differences, the possibility must be considered that the C terminus of APOBEC-1 may serve as an NES and that the cytoplasmic distribution of the enzyme could be the result of nuclear export. Because of these uncertainties, the 24-amino acid domain will be referred to as a CRS/NES.
Finally, our data suggest that the nuclear and cytoplasmic targeting signals in APOBEC-1 are biologically significant. The coincidence of a nuclear distribution of APOBEC-1 and RNA editing competence suggests that a factor(s) that affects the subcellular distribution of APOBEC-1 might be expressed in a cell type-specific manner. Our preliminary data indicated that in contrast to the wild-type protein, APOBEC delCT(173–229) was able to migrate into the nucleus when expressed in CHO or COS-7 cells, but APOBEC delNT(1–56) was localized exclusively in the cytoplasm in these two cell types (data not shown). These data suggest that the difference in subcellular distribution of APOBEC-1 in editing competent cells versus editing incompetent cells is the result of a cell type-specific neutralization or masking of an otherwise generic putative CRS/NES. The ability of APOBEC delCT(173–229) to localize in the nucleus of CHO or COS-7 cells suggests that cellular factors necessary for nuclear import of APOBEC-1 may be commonly expressed among different cell types.
Selective nuclear uptake of proteins, which are otherwise restricted to cytoplasm, is mediated by two major themes: masking/unmasking of NLS and anchoring/releasing from the cytoplasm (37–39). NF-κB is one of the best documented examples for the first mechanism. NF-κB is sequestered in the cytoplasm through interaction with the inhibitor I-κB, whose binding masks its NLS. Upon cellular stimulation, I-κB is phosphorylated and proteolyzed, thereby unmasking the NLS of NF-κB and enabling it to redistribute to the nucleus (40, 41). Our data suggest that the intracellular distribution of APOBEC-1 may be regulated differently than that of NF-κB.
Xenopus xnf 7 is an example of the second mechanism for nuclear distribution in that it has both a bipartite NLS and a cytoplasmic retention domain. xnf 7 is a maternally expressed, putative transcription factor that is retained inactive in the cytoplasm of oocytes until the midblastula transition, when it enters the nucleus and presumably assumes its function (42). The cytoplasmic retention of xnf 7 arises from a novel anchoring mechanism in which the cytoplasmic retention domain interacts with an anchor protein in the cytoplasm. Anchoring and releasing of xnf 7 from the cytoplasm has been proposed to be regulated by phosphorylation and dephosphorylation (42).
A similar mechanism has been implicated in modulating nuclear import of the Xenopus myogenic factor XMyo D. In most cells, XMyo D is anchored in the cytoplasm through its C terminus, but in embryonic myogenic progenitor cells, XMyo D resides in the nucleus (43). The cytoplasmic retention functions of the CRS in xnf 7 or XMyo D cannot be bypassed by a consensus NLS (42, 43). This is strikingly similar to what we have observed on the CRS/NES of APOBEC-1, suggesting that the cytoplasmic distribution of APOBEC-1 is the default pattern and that, like xnf 7 and XMyo D, nuclear distribution of APOBEC-1 requires cell type-specific factors.
Cyclin B1 is an example of a protein that has a CRS but no recognizable NLS, a characteristic similar to that of APOBEC-1. Cyclin B1 is a cytoplasmic protein in interphase but rapidly enters the nucleus at the start of mitosis. Cytoplasmic retention of cyclin B1 is conferred by a CRS that differs from that of xnf 7, XMyo D, and APOBEC-1 in that it can be overridden by a consensus NLS (44). Phosphorylation of serine residues within the CRS disrupts interactions in the cytoplasm and enables nuclear transport of cyclin B1, presumably mediated through proteins associated with cyclin-CDK complexes (45).
Some proteins enter the nucleus by passive diffusion after dissociation from the cytoplasmic anchoring subunit. Type II cAMP-dependent protein kinase consists of two catalytic and two cAMP-binding regulatory subunits associated with the Golgi apparatus. In response to increased levels of cAMP, the regulatory subunits bind to cAMP and release catalytic subunits, which are then free to migrate into the nucleus by passive diffusion (46). If APOBEC-1 does not have a nuclear localization determinant or chaperone binding site, then the nuclear distribution of the C-terminal deletion of APOBEC-1 could be taken as evidence that the protein can enter the nucleus by passive diffusion.
The cellular distribution of APOBEC-1 seems therefore to be the result of multiple determinants that have been observed individually in other proteins but have not been described previously in the combination as appeared in APOBEC-1. The characteristics of APOBEC-1 are similar to CRS function in xnf 7 and XMyo D but, like cyclin B1, without a conical NLS, suggesting that a chaperone may be required or that entry into the nucleus occurs by means of diffusion subsequent to the neutralization of the putative CRS/NES.
In conclusion, we have demonstrated that the HA-APOBEC-1 distributes both in the nucleus and the cytoplasm in transiently transfected McArdle cells, giving evidence that APOBEC-1 is a nuclear protein. Although the N terminus of APOBEC-1 does not contain a consensus NLS, it is required for nuclear uptake of APOBEC-1. A 24-amino acid CRS/NES at the C terminus of APOBEC-1 has been identified and may play a role in regulating the nuclear to cytoplasmic distribution ratio of APOBEC-1. Our data suggest a hypothesis wherein editing competence in cells requires factors that regulate the nuclear distribution of APOBEC-1 as well as factors that interact with and regulate the enzyme in the context of editosomes. It is possible that some or all of the auxiliary factors function at both levels of regulation. We propose that nuclear distribution of APOBEC-1 may be regulated by orchestrating different domains through a pathway expressed only in apoB RNA editing-competent cells.
Acknowledgments
We thank Dr. David L. Spector and Dr. David S. Goldfarb for helpful suggestions, Dr. Haruhiko Siomi for myc-CMPK/pcDNA3 plasmid, Katherine Kovalski for excellent technical assistance, and Jenny M. L. Smith for the preparation of the figures.This work was supported in part by U.S. Public Health Service Grant DK 43739 and grants from The Council for Tobacco Research and the Rochester Area Pepper Center for Research on Aging (to H.C.S.).
ABBREVIATIONS
- apoB
apolipoprotein B
- APOBEC-1
apoB mRNA editing catalytic subunit 1
- HA
hemagglutinin
- NLS
nuclear localization signal
- CRS
cytoplasmic retention signal
- NES
nuclear export signal
- CMPK
chicken muscle pyruvate kinase
- LR
leucine-rich
- CHO
Chinese hamster ovary
- DAPI
4′,6-diamidino-2-phenylindole
- SV40
simian virus 40
References
- 1.Chen S-H, Habib G, Yang C-Y, Gu Z-W, Lee B R, Weng S-A, Silberman S R, Cai S-J, Deslypere J P, Rosseneu M, Gotto A M, Jr, Li W-H, Chan L. Science. 1987;328:363–366. doi: 10.1126/science.3659919. [DOI] [PubMed] [Google Scholar]
- 2.Powell L M, Wallis S C, Pease R J, Edwards Y H, Knott T J, Scott J. Cell. 1987;50:831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- 3.Davidson N O. Ann Med. 1993;25:539–543. [PubMed] [Google Scholar]
- 4.Shah R R, Knott T J, Legros J E, Navaratnam N, Greeve J C, Scott J. J Biol Chem. 1991;266:16301–16304. [PubMed] [Google Scholar]
- 5.Backus J W, Smith H C. Nucleic Acids Res. 1992;20:6007–6014. doi: 10.1093/nar/20.22.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Backus J W, Smith H C. Biochim Biophys Acta. 1994;1217:65–73. [PubMed] [Google Scholar]
- 7.Backus J W, Schock D, Smith H C. Biochim Biophys Acta. 1994;1219:1–14. doi: 10.1016/0167-4781(94)90240-2. [DOI] [PubMed] [Google Scholar]
- 8.Smith H C. In: Seminars in Cell Biology. Stuart K, editor. Vol. 4. London: Saunders Science Publications/Academic; 1993. pp. 267–278. [Google Scholar]
- 9.Smith H C, Kuo S R, Backus J W, Harris S G, Sparks C E, Sparks J D. Proc Natl Acad Sci USA. 1991;88:1489–1493. doi: 10.1073/pnas.88.4.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris S G, Sabio I, Mayer E, Steinberg M F, Backus J W, Sparks J D, Sparks C E, Smith H C. J Biol Chem. 1993;268:7382–7392. [PubMed] [Google Scholar]
- 11.Teng B-B, Burant C F, Davidson N O. Science. 1993;260:1816–1819. doi: 10.1126/science.8511591. [DOI] [PubMed] [Google Scholar]
- 12.Navaratnam N, Morrison J R, Bhattacharya S, Patel D, Funahashi T, Giannoni F, Teng B-B, Davidson N O, Scott J. J Biol Chem. 1993;268:20709–20712. [PubMed] [Google Scholar]
- 13.Navaratnam N, Bhattacharya S, Fujino T, Patel D, Jarmuz A L, Scott J. Cell. 1995;81:187–195. doi: 10.1016/0092-8674(95)90328-3. [DOI] [PubMed] [Google Scholar]
- 14.Smith H C, Sowden M P. Trends Genet. 1996;12:418–424. doi: 10.1016/0168-9525(96)10042-1. [DOI] [PubMed] [Google Scholar]
- 15.MacGinnitie A J, Anant S, Davidson N O. J Biol Chem. 1995;270:14768–14775. [PubMed] [Google Scholar]
- 16.Driscoll D M, Zhang Q. J Biol Chem. 1994;269:19843–19847. [PubMed] [Google Scholar]
- 17.Inui Y, Giannoni F, Funahashi T, Davidson N O. J Lipid Res. 1994;35:1477–1489. [PubMed] [Google Scholar]
- 18.Yamanaka S, Poksay K S, Balestra M E, Zeng G-Q, Innerarity T L. J Biol Chem. 1994;269:21725–21734. [PubMed] [Google Scholar]
- 19.Funahashi T, Giannoni F, DePaoli A M, Skarosi S F, Davidson N O. J Lipid Res. 1995;36:414–428. [PubMed] [Google Scholar]
- 20.Lau P P, Xiong W, Zhu H-T, Chen S-H, Chan L. J Biol Chem. 1991;266:20550–20554. [PubMed] [Google Scholar]
- 21.Sowden M, Hamm J K, Spinelli S, Smith H C. RNA. 1996;2:274–288. [PMC free article] [PubMed] [Google Scholar]
- 22.Dingwall C, Laskey R A. Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y, Smith H C. Biochem Biophys Res Commun. 1996;218:797–801. doi: 10.1006/bbrc.1996.0142. [DOI] [PubMed] [Google Scholar]
- 24.Siomi H, Dreyfuss G. J Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sowden M, Hamm J K, Smith H C. J Biol Chem. 1996;271:3011–3017. doi: 10.1074/jbc.271.6.3011. [DOI] [PubMed] [Google Scholar]
- 26.Kalderon D, Roberts B L, Richardson W D, Smith A E. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 27.Goldfarb D S, Gariepy J, Schoolnik G, Kornberg R D. Nature (London) 1986;322:641–644. doi: 10.1038/322641a0. [DOI] [PubMed] [Google Scholar]
- 28.Lanford R E, Kanda P, Kennedy R C. Cell. 1986;46:575–582. doi: 10.1016/0092-8674(86)90883-4. [DOI] [PubMed] [Google Scholar]
- 29.Lau P P, Zhu H-J, Baldini A, Charnsangavej C, Chan L. Proc Natl Acad Sci USA. 1994;91:8522–8526. doi: 10.1073/pnas.91.18.8522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oka K, Kobayashi K, Sullivan M, Martinez J, Teng B-B, Ishimura-Oka K, Chan L. J Biol Chem. 1997;272:1456–1460. doi: 10.1074/jbc.272.3.1456. [DOI] [PubMed] [Google Scholar]
- 31.Giannoni F, Bonen D K, Funahashi T, Hadjiagapion C, Burant C F, Davidson N O. J Biol Chem. 1994;269:5932–5936. [PubMed] [Google Scholar]
- 32.Boström K, Garcia Z, Poksay K S, Johnson D F, Lusis A J, Innerarity T L. J Biol Chem. 1990;265:22446–22452. [PubMed] [Google Scholar]
- 33.Teng B-B, Davidson N O. J Biol Chem. 1992;267:21265–21272. [PubMed] [Google Scholar]
- 34.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 35.Fisher U, Huber J, Boelens W C, Mattaj I W, Lührmann R. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 36.Kobe B, Deisenhofer J. Trends Biochem Sci. 1994;19:415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 37.Nigg E A. Nature (London) 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 38.Vandromme M, Gauthier-Rouvière C, Lamb N, Fernandez A. Trends Biochem Sci. 1996;21:59–64. [PubMed] [Google Scholar]
- 39.Whiteside S T, Goodbourn S. J Cell Sci. 1993;104:949–955. doi: 10.1242/jcs.104.4.949. [DOI] [PubMed] [Google Scholar]
- 40.Beg A A, Ruben S M, Scheinman R I, Haskill S, Rosen C A, Baldwin A S., Jr Genes Dev. 1992;6:1899–1913. doi: 10.1101/gad.6.10.1899. [DOI] [PubMed] [Google Scholar]
- 41.Imbert V, Rupec R A, Livolsi A, Pahl H L, Traenckner B M, Mueller-Dieckmann C, Farahifar D, Rossi B, Auberger P, Baeuerle P A, Peyron J-F. Cell. 1996;86:787–798. doi: 10.1016/s0092-8674(00)80153-1. [DOI] [PubMed] [Google Scholar]
- 42.Li X, Shou W, Kloc M, Reddy B A, Etkin L D. J Cell Biol. 1994;124:7–17. doi: 10.1083/jcb.124.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rupp R A W, Snider L, Weintraub H. Genes Dev. 1994;8:1311–1323. doi: 10.1101/gad.8.11.1311. [DOI] [PubMed] [Google Scholar]
- 44.Pines J, Hunter T. EMBO J. 1994;13:3772–3781. doi: 10.1002/j.1460-2075.1994.tb06688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J, Meyer A N, Donoghue D J. Proc Natl Acad Sci USA. 1997;94:502–507. doi: 10.1073/pnas.94.2.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nigg E A, Schäfer G, Hilz H, Eppenberger H M. Cell. 1985;41:1039–1051. doi: 10.1016/s0092-8674(85)80084-2. [DOI] [PubMed] [Google Scholar]