Abstract
Cac1p is a subunit of yeast chromatin assembly factor I (yCAF-I) that is thought to assemble nucleosomes containing diacetylated histones onto newly replicated DNA [Kaufman, P. D., Kobayashi, R. & Stillman, B. (1997) Genes Dev. 11, 345–357]. Although cac1Δ cells could establish and maintain transcriptional repression at telomeres, they displayed a reduced heritability of the repressed state. Single-cell analysis revealed that individual cac1Δ cells switch from transcriptionally “off” to transcriptionally “on” more often per cell cycle than wild-type cells. In addition, cac1Δ cells were defective for transcriptional silencing near internal tracts of C1–3A sequence, but they showed no defect in silencing at the silent mating type loci when analyzed by a reverse transcription–PCR assay. Despite the loss of transcriptional silencing at telomeres and internal C1–3A tracts, subtelomeric DNA was organized into nucleosomes that had all of the features characteristic of silent chromatin, such as hypoacetylation of histone H4 and protection from methylation by the Escherichia coli dam methylase. Thus, these features of silent chromatin are not sufficient for stable maintenance of a silent chromatin state. We propose that the inheritance of the transcriptionally repressed state requires the specific pattern of histone acetylation conferred by yCAF-I-mediated nucleosome assembly.
Telomeres, the protein–DNA structures at the ends of eukaryotic chromosomes, are essential for the stable maintenance of chromosomes (1). In some organisms, such as yeast and Drosophila, telomeres repress the transcription of nearby genes, a phenomenon called telomere position effect (TPE) (2–6). The transcriptional silencing of genes near telomeres is an epigenetic phenomenon, meaning that, in some cells, the telomeric gene is transcribed whereas in others the gene is repressed. Both the transcribed and the repressed states are stable for many generations, and both states are reversible (3). Genes near internal tracts of C1–3A⋅TG1–3 DNA are also transcriptionally repressed, a phenomenon called C1–3A-based silencing (CBS) (7).
Transcriptional silencing is thought to require a specialized chromatin structure. The C1–3A⋅TG1–3 repeats at the ends of yeast chromosomes are packaged into a nonnucleosomal chromatin structure, the telosome (8). Although the DNA next to yeast telomeres is organized into nucleosomes (8), these nucleosomes are distinguishable by two criteria from nucleosomes in most other regions of the genome. First, GATC sites in subtelomeric DNA are relatively inaccessible to methylation by the Escherichia coli dam methylase when this protein is expressed in yeast (9). Second, the histones in subtelomeric chromatin are hypoacetylated compared with histones in most other regions of the genome (10). These same chromatin features are also characteristic of the silent mating type genes or HM loci (10–12), as well as genes near internal tracts of telomeric sequence (D.d.B., W. H. Tham, and V.A.Z., unpublished results).
Because a repressed transcriptional state is inherited, cells must have a mechanism that promotes the assembly of the parental chromatin structure on both progeny DNA molecules. It has been suggested that histone acetylation plays a role in this process (13). Newly synthesized histones H3 and H4 are post-translationally modified by acetylation (14). Newly synthesized histone H4 is acetylated on lysines 5 and 12, and this pattern of acetylation, referred to as deposition-related acetylation, is absolutely conserved among Drosophila, humans, Tetrahymena (15–17), and probably yeast (18, 19). Although transcriptionally repressed chromatin is hypoacetylated in vivo relative to transcribed genes, lysine-12 of histone H4 is often acetylated at the HM loci (11), just as it is in Drosophila heterochromatin (20). Several lines of evidence indicate that the specific acetylation of histone H4 at lysine-5 or lysine-12 is critical for transcriptional repression in both yeast and Drosophila (11, 21–25).
In human cells, a three-subunit complex called human chromatin assembly factor I (hCAF-I) preferentially assembles newly synthesized histones with the deposition-related acetylation pattern onto newly replicated DNA (26–29). Yeast genes encoding proteins homologous to the three subunits of hCAF-I have recently been identified and are designated CAC1, CAC2, and CAC3 (chromatin assembly complex) (30). The CAC1 gene was also identified independently in a screen for mutants that stabilize plasmids carrying both centromeric and telomeric sequences (designated RLF2; rap1 localization factor 2) (31). Mutations in CAC1/RLF2 result in a reduction in TPE, increased UV sensitivity, and the mislocalization of the telomere-binding protein Rap1p as assayed by immunofluorescence with anti-Rap1p antibodies (30, 31). We will hereafter refer to this gene as CAC1 because this name describes the only known biochemical activity of the gene product.
In a two-hybrid screen, we identified Cac1p as a protein that interacts with the Pif1p DNA helicase, a protein that appears to inhibit telomere replication (32) (E.K.M. and V.A.Z., unpublished results). As previously reported (30, 31), we found that cac1Δ cells were defective in TPE. Here, we show that the defect in TPE resulted from an increased frequency with which cells switched from transcriptionally “off” to transcriptionally “on.” Furthermore, we demonstrate that cac1Δ cells were defective in the silencing of genes adjacent to internal tracts of C1–3A⋅TG1–3 sequence, whereas there was no defect in silencing at the silent mating type loci. In addition, cac1Δ cells assembled chromatin into nucleosomes at both subtelomeric and internal sites and showed no significant changes in chromatin structure as assayed by dam methylation protection or immunoprecipitation with antibodies to multiply acetylated histone H4. Thus, there can be significant alterations in TPE without changes in these aspects of chromatin structure. We propose that Cac1p is required for the inheritance of transcriptional silencing because the acetylation pattern of the histones assembled by yeast chromatin assembly factor I (yCAF-I) provides the signal that helps template formation of transcriptionally silent chromatin.
METHODS
All experiments were carried out in strains isogenic to YPH499 (33). The TPE assays, the dam methylation experiments, and the micromanipulation experiments (Figs. 1 and 4; Table 1) were done using strain YPH499UTAT and its derivatives, which have ADE2 at the chromosome V-R telomere (34) and URA3 at the chromosome VII-L telomere (3). The chromatin immunoprecipitations (Fig. 5) were done in DdB10UT and its derivatives, which is similar to YPH499UTAT except that it has ura3::LEU2 in place of ura3–52 and ADE2 is not at the chromosome V-R telomere. A PCR method (35) was used to delete the entire CAC1 ORF and replace it with TRP1 in YPH499UTAT. The cac1Δ::TRP1 allele was introduced into other strains by mating. E. Wiley made a sir2Δ derivative of YPH499UTAT by using the 2.7-kb SphI/EcoRV fragment of pJR531 (36). DdB10UTsir3Δ was made by transforming DdB10UT with EcoRI-digested pJR317 (36). Strains expressing the dam methylase were made by transforming YPH499UTAT or YPH499UTATsir2Δ with XhoI digested pDP6-dam (9). CBS assays were performed in YPH499CULT (made by J. Stavenhagen) and its derivatives, which have an ≈1-kb tract of C1–3A adjacent to URA3 within LYS2.
Figure 1.
TPE is reduced in cac1Δ cells. Strains with URA3 at the chromosome VII-L telomere were grown at 30°C and assayed for TPE by plating 10-fold serial dilutions of two independent colonies for each strain on FOA medium. Dilutions were also plated on YC–Ura medium to determine the total number of cells. Two independent platings are shown in Upper and Lower to illustrate the variation typically observed for cac1Δ cells. Photographs were taken after the indicated number of days. WT, wild type.
Figure 4.
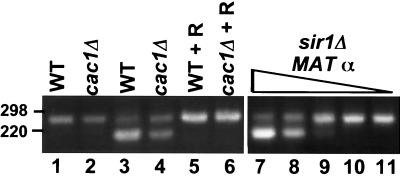
cac1Δ cells efficiently repress HMRa RT-PCR was done using total RNA prepared from MATα strains YPH500 and YPH500cac1Δ (lanes 1 and 2), MATa strains YPH499 and YPH499cac1Δ (lanes 3–6), and a MATα sir1Δ strain (lanes 7–11). Two micrograms of total RNA was used for reactions in lanes 1–6. Lanes 7–11 contain 10-fold serial dilutions of RNA from MATα sir1Δ cells starting with 0.1 μg in lane 7 down to 0.00001 μg in lane 11. RNA from strain YPH500, which does not express MATa1, was added to all reactions in lanes 7–11 so that there was 1 μg of total nucleic acid in each reaction mixture. Positions of the 220- and 298-bp markers are indicated. R indicates reactions where RNA was treated with RNase prior to reverse transcription.
Table 1.
Growth of individual cells after micromanipulation
| No. of divisions | FOA to FOA URA3-TEL
|
FOA to YC URA3-TEL
|
FOA to FOA ura3-52
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild type
|
cac1Δ
|
Wild type
|
cac1Δ
|
Wild type
|
cac1Δ
|
|||||||
| % | No. | % | No. | % | No. | % | No. | % | No. | % | No. | |
| 0 | 42 | 198 | 62 | 244 | 8 | 19 | 8 | 15 | 2 | 2 | 6 | 5 |
| 1–2 | 8.5 | 38 | 15 | 57 | 4 | 9 | 0.5 | 1 | 0 | 0 | 0 | 0 |
| 3 or more | 49 | 204 | 23 | 84 | 88 | 198 | 91 | 169 | 97 | 79 | 94 | 75 |
| Total cells | 100 | 440 | 100 | 385 | 100 | 226 | 100 | 185 | 100 | 81 | 100 | 80 |
| Stops* | 16% | 40% | 4% | 0.6% | 0% | 0% | ||||||
The percentage of cells in each growth category is presented; No. indicates the total number of cells analyzed.
The percentage of cells that stopped dividing after 1–2 divisions of the total cells that divided is presented. For example, in the first column, of the 242 (38 + 204) cells that started to divide, 38 or 16% of them stopped after 1–2 divisions. A χ2 test using the summed totals of the number of cells in each of the two categories (1–2 divisions and 3 or more divisions) for wild-type URA3-TEL and cac1Δ URA3-TEL, FOA to FOA, indicated that the difference between 16% and 40% is significant. χ2 = 16.864, P < 0.0001.
Figure 5.
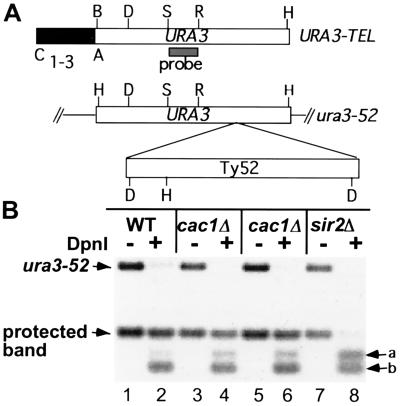
(A) Maps of the URA3 gene at the chromosome VII-L telomere (URA3-TEL) and at the internal ura3–52 locus (not to scale). Restriction endonuclease cleavage sites are BamHI (B), HindIII (H), StuI (S), and EcoRV (R). The shaded box indicates the StuI–EcoRV probe used for Southern hybridization. (B) The positions of the HindIII/BamHI fragments derived from the telomeric URA3 (“protected band”) and the ura3–52 locus are indicated on the left. The HindIII/BamHI/DpnI fragments derived from URA3-TEL (a) and ura3–52 fragments (b) are indicated on the right.
Methods for TPE and CBS (7), alpha factor confrontation (37), dam methylation protection (9), and chromatin immunoprecipitation (10) assays have been described. Antibodies raised against a histone H4 peptide acetylated at lysines 5, 8, 12, and 16 are described in ref. 38 and characterized for their activity against yeast histone H4 in ref. 10. For micromanipulations, cells were streaked on plates containing 5-fluoroorotic acid (FOA) and allowed to grow for 1–2 days at 30°C. Cells were resuspended in H2O and spread on FOA or yeast complete (YC) plates. Individual budding cells were micromanipulated to known locations on a plate by using a Singer MSM System micromanipulator. Plates were incubated at 30°C and cell divisions were monitored. Cell growth was scored after 2–3 hr and was monitored over a period of 24–48 hr. Reverse transcription of total RNA was done using Pharmacia Ready-to-go-You-Prime-First-Strand Beads according to instructions and using oligo(dT)16 to prime cDNA synthesis. Control samples were treated with 5 μg of RNase A for 15 min at 37°C prior to reverse transcription. The cDNAs were then PCR amplified by using Taq DNA polymerase and the 5′ and 3′ MATa1 primers described in ref. 39. PCR amplification was done for 40–45 cycles at 94°C for 1 min, 62°C for 30 sec, and 72°C for 1 min, followed by 7 min at 72°C for the final extension. PCR products were separated on a 3% agarose gel and stained with ethidium bromide.
RESULTS
TPE Is Reduced in cac1Δ Cells.
Because cells producing Ura3p cannot grow in the presence of FOA (40), TPE is often monitored by placing the URA3 gene near a telomere and determining the fraction of cells able to grow on plates containing FOA. TPE can also be monitored by placing the ADE2 gene next to a telomere (3). Whereas wild-type ADE2 cells produce white colonies and ade2 cells produce red colonies, cells with ADE2 next to a telomere generate largely red colonies with white sectors (ADE2 transcriptionally “off” with occasional switches to transcriptionally “on”) or largely white colonies with red sectors (ADE2 transcriptionally “on” with occasional switches to transcriptionally “off”). For the experiments reported here, TPE was determined in otherwise isogenic wild-type and mutant strains having URA3 near the left telomere of chromosome VII and ADE2 near the right telomere of chromosome V.
As monitored by the fraction of FOA-resistant (FOAR) cells, the cac1Δ strain had a variable 10- to 1,000-fold decrease in TPE relative to wild type, with a typical reduction of ≈100-fold (Fig. 1). Moreover, cac1Δ colonies were much smaller and more heterogeneous in size than wild-type cells on FOA medium, whereas there was no growth difference between cac1Δ and wild-type colonies on media without FOA. Thus, as reported previously (30, 31), cac1Δ cells had two phenotypes with respect to TPE, an absolute reduction in FOAR colonies and small colony size on FOA medium.
There are two possible explanations for the slow growth of cac1Δ colonies carrying a telomeric URA3 gene on FOA medium. First, the cac1Δ mutation might cause an increase in the level of basal transcription of URA3 in all or most cells, resulting in the growth inhibition of all cells on FOA medium. The second possibility is that the cac1Δ mutation increases the probability that a repressed URA3 gene switches to a transcriptionally active state, such that during colony growth, cells that die on FOA are continuously generated. To distinguish between these possibilities, the color of cac1Δ colonies with ADE2 located near the chromosome V-R telomere was determined. If the first possibility were correct, that all cells had increased basal transcription of telomeric genes, cac1Δ cells would produce colonies with a uniform color, without distinct sectors. If the second scenario were true, that a switch from a transcriptionally inactive to a transcriptionally active state occurred more frequently in a cac1Δ strain, cac1Δ cells would produce red/white sectored colonies.
Most (≈95%) colonies produced by wild-type cells with ADE2 at the chromosome V-R telomere were red with occasional white sectors (Fig. 2). ADE2 is repressed in a higher fraction of cells (≈95%) when it is located at the chromosome V-R telomere than when it is at the chromosome VII-L telomere (≈50% repressed) (3), confirming that chromosomal context is important for TPE (3). Most cac1Δ cells (≈95%) produced colonies that were white with numerous small red sectors (Fig. 2). A mostly red cac1Δ colony was rarely detected. As expected, sir3Δ colonies were completely white because of the complete loss of TPE (Fig. 2). The fact that cac1Δ cells produced mostly white colonies was consistent with the absolute reduction in TPE demonstrated by the reduced fraction of FOAR cells in the cac1Δ strain. The presence of small red sectors in virtually all cac1Δ colonies indicated that in a cac1Δ cell, the ADE2 gene was either “on” (white regions of the colony) or “off” (red sectors), and that cac1Δ cells can switch from a transcriptionally active to a transcriptionally repressed state. Thus, the appearance of cac1Δ colonies was consistent with a model in which a telomeric gene in a cac1Δ cell is either transcribed or repressed, rather than its being in an intermediate transcriptional state.
Figure 2.
A telomeric ADE2 gene is either “on” or “off” in cac1Δ cells. TPE was assayed using a color marker, the ADE2 gene, at the chromosome V-R telomere (34). Colonies were plated on YC medium with a low adenine concentration (10 mg/liter), allowed to grow at 30°C for 3 days, and then placed at 4°C for approximately 3 weeks to allow the red color to develop.
The Rate of Switching from a Transcriptionally Repressed to a Transcriptionally Active State Is Higher in a cac1Δ Strain.
To determine if cac1Δ cells switch transcriptional states more often than wild-type cells, the ability of individual cells to divide on FOA medium was assessed. Wild-type and cac1Δ cells were grown on FOA medium for 1–2 days to establish populations of cells in which URA3 transcription was repressed. The cells were then spread onto FOA or YC plates for micromanipulation. Individual budding cells were moved to gridded locations on a plate and their subsequent growth was observed. An FOA-grown cell that switches from a transcriptionally repressed state to a transcriptionally active state will stop dividing on FOA but will continue to divide on YC medium. Growth was classified into three categories: cells that did not divide at all, cells that underwent one or two divisions (most cells in this category underwent just one division—both the mother and daughter cells must have stopped dividing to be included in this category), and cells that divided three or more times (Table 1). For those cells that divided, the time between micromanipulation and the first division was noted.
Two viability controls were conducted. Mutant and wild-type cells with the URA3 gene at the chromosome VII-L telomere or without a telomeric URA3 gene (Ura−) were grown on FOA medium and then transferred as single cells to complete medium. In this experiment, 91% of the mutant and 88% of the wild-type cells carrying URA3 at the telomere formed colonies and >90% of the Ura− cells formed colonies (Table 1). These experiments established that the rate of killing from micromanipulation alone was ≈10% and that lack of Cac1p did not make cells supersensitive to FOA.
As observed previously for wild-type cells (3), a significant fraction of the micromanipulated cells from both the wild-type and cac1Δ strains failed to divide when transferred as individual cells to FOA medium (42% and 62%, respectively; Table 1). Because control experiments established that ≈90% of the mutant and wild-type cells grew after micromanipulation, the failure of some cells with URA3 near a telomere to continue to divide on FOA presumably reflects a change in the transcriptional status of the telomere-linked URA3 gene. However, it was difficult to determine if the failure of an individual cell to divide on FOA was due to a switch to a transcriptionally active state or if there was a low level of URA3 transcription in some of the repressed cells. Thus, only those cells that were able to divide at least once on FOA after micromanipulation were considered in determining switching rates.
Of the 141 cac1Δ cells that divided at least once on FOA, 40% (57/141) stopped dividing after one or two divisions. In contrast, of 242 wild-type cells that divided at least once on FOA, 16% (38/242) stopped dividing after one or two divisions (Table 1). We interpret the cessation of division to be the result of a switch from the URA3 “off” to the URA3 “on” transcriptional state. Therefore, cac1Δ mutants “switch” at a rate of roughly 0.4 per generation and wild-type cells “switch” at a rate of roughly 0.16 per generation, representing an ≈2.5-fold increase in the rate of switching in cac1Δ cells. Because a switch from “off” to “on” generates a cell that dies on FOA, an ≈2.5-fold increase in the switching rate would result in an ≈2.5-fold increase in the number of dead cells arising per generation and would account for the slow growth of cac1Δ colonies.
For those cells that divided on FOA, the first division on FOA occurred ≈3 hr after micromanipulation for both the cac1Δ and wild-type strains. Thus, the slow growth on FOA of cac1Δ cells with a telomeric URA3 gene was not due to cac1Δ cells having a longer cell cycle on FOA than wild-type cells. In addition, no growth rate differences were observed for Ura− wild-type and cac1Δ cells on FOA medium (data not shown). These data support the interpretation that the TPE defect in cac1Δ cells is due to an increased rate of switching from repressed to transcribed, rather than to an increase in the basal transcription of telomere-linked genes. The FOA assay cannot address the rate of switching from transcribed to repressed. However, the pattern of ADE2 sectoring observed in cac1Δ mutants is relevant to this issue. The rare mostly white colonies seen in the wild-type strain were very similar to cac1Δ cells in regard to both the number and size of their red sectors (data not shown), suggesting that cac1Δ cells did not switch from transcribed to repressed at an elevated rate.
CBS Is Virtually Eliminated but HM Silencing Is Not Altered in a cac1Δ Strain.
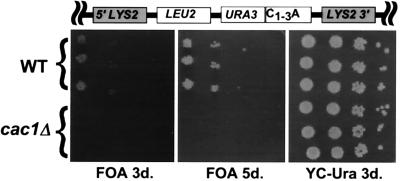
CBS, the transcriptional silencing of genes near internal tracts of telomeric sequence, requires many of the same gene products as TPE (7). To determine if Cac1p is important for silencing at internal chromosomal loci, CBS was assessed in otherwise isogenic wild-type and cac1Δ strains that had an ≈1-kb tract of C1–3A⋅TG1–3 DNA adjacent to a URA3 gene (J. Stavenhagen and V.A.Z., unpublished work) within the LYS2 locus, ≈400 kb from the chromosome II-R telomere. The fraction of FOAR cells was determined by plating cells on FOA and on YC medium. As expected from previous results with the 1-kb tract (J. Stavenhagen and V.A.Z., unpublished results), ≈2% of wild-type cells grew on FOA medium. In contrast, the frequency of FOAR Ura+ cells in the cac1Δ strain was ≈10−6, a reduction of ≈20,000 fold relative to wild-type (Fig. 3 and data not shown). Thus, Cac1p is critical for silencing both at telomeres and at internal tracts of C1–3A⋅TG1–3 DNA. It had not previously been demonstrated that Cac1p is involved in silencing at internal sites in the genome.
Figure 3.
CBS is virtually eliminated in cac1Δ cells. Wild-type and cac1Δ strains that contain a 1-kb tract of internal C1–3A sequence adjacent to a URA3 gene at the LYS2 locus (see map at the top) were assayed for CBS by plating 10-fold serial dilutions of cells on FOA and YC-Ura plates and incubating for 3 or 5 days at 30°C.
In contrast to TPE and CBS, transcriptional repression at the HM loci is very stable, with ≈10−6 cells expressing either of the HM loci. Derepression of an HM locus results in a nonmating phenotype. When quantitative mating was used as an assay, silencing at the HM loci was indistinguishable in cac1Δ and wild-type cells (data not shown), consistent with previously reported results (30). However, because mutations that cause partial loss of silencing at HML/HMR, such as sir1Δ mutations, result in no obvious defect in mating efficiency (41), we performed a reverse transcription (RT)-PCR assay (39) to assess whether HMRa RNA is expressed in cac1Δ MATα cells. By using primers that flank an intron in MATa1 (39), the RT-PCR assay distinguishes between a chromosomal DNA- and an RNA-derived MATa1 PCR product because the RNA-derived product is 53 bp smaller than the DNA-derived product. Contaminating DNA in the RNA preparations was not removed, hence both RNA- and DNA-derived products could be generated in the reaction. In wild-type MATα cells, the only copy of MATa1 is at the transcriptionally silent mating type locus, HMR. Since it is efficiently repressed, only the DNA-derived product was detected in the RT-PCR (Fig. 4, lane 1). Similarly, no MATa1 RNA was detected from cac1Δ MATα cells, demonstrating that the cac1Δ mutation does not cause detectable derepression of HMRa (Fig. 4, lane 2). As expected, both the RNA- and DNA-derived products were detected in wild-type and cac1Δ MATa cells (Fig. 4, lanes 3 and 4) and the RNA product was eliminated by pretreatment with RNase A (Fig. 4, lanes 5 and 6). HMRa1 RNA was detected in a sir1Δ MATα strain which is partially derepressed at HMRa (41). A dilution experiment showed that HMRa1 RNA was detectable in the sir1Δ MATα strain, even at a <10−3 dilution (0.001 μg of sir1Δ MATα RNA) (Fig. 4, lane 9). Thus, cac1Δ MATα cells express <10−3 of the amount of HMRa1 RNA present in a MATα sir1Δ strain. In addition, an alpha factor confrontation assay (37) revealed that transcription of HMLα occurred in fewer than 10−2 cells in a cac1Δ MATa strain. Taken together, these data suggest that the silencing defect in cac1Δ cells may be specific for genes next to tracts of C1–3A⋅TG1–3 DNA or that silencing at HML/HMR in cac1Δ cells is achieved by a function that is redundant with CAC1.
The Chromatin Structure of a Telomere-Linked Gene Is Similar in Wild-Type and cac1Δ Cells.
Micrococcal nuclease mapping experiments revealed that bulk DNA and subtelomeric DNA were assembled into nucleosomes in cac1Δ cells that appeared very similar in size and spacing to nucleosomes from wild-type cells (data not shown.) The cac1Δ or sir2Δ mutation was introduced into a strain expressing E. coli dam methylase and carrying a telomeric URA3 gene. DNA was isolated from otherwise isogenic wild-type, cac1Δ, and sir2Δ strains and digested with HindIII and BamHI in the presence or absence of DpnI (Fig. 5). DpnI cleaves its recognition sequence, GATC, only if the A is methylated. The DNA was transferred to membranes and probed with URA3. In the wild-type strain, the GATC site within the telomeric URA3 gene was partially protected from digestion by DpnI, indicating partial protection from methylation, consistent with previous results (9). As expected, this protection was eliminated in a sir2Δ strain. Although DNA from the cac1Δ strain showed a modest loss of protection of the GATC site from DpnI cleavage, >50% of fragment a was in the protected band. Because the subtelomeric URA3 gene was transcribed in >99% of cac1Δ cells as assayed from the fraction of FOAR cells (Fig. 1), >50% of these genes had a repressed chromatin structure as defined by accessibility to the dam methylase. Thus, the level of protection in cac1Δ DNA was much closer to that seen in the wild-type strain than it was to that of a sir2Δ strain.
To determine the acetylation state of histone H4, antibodies specific for acetylated H4 (38) were used to immunoprecipitate chromatin from otherwise isogenic wild-type, sir3Δ, and cac1Δ strains carrying a telomeric URA3 gene. This antiserum strongly recognizes yeast histone H4 only if it is acetylated at multiple positions (10, 11). The DNA in the immunoprecipitates was analyzed for URA3 sequences by slot-blot hybridization and quantitated by using phosphorimage analysis (10). The blots were then stripped and hybridized to a probe from the constitutively expressed, essential ACT1 (actin) gene or from the nonessential LEU2 gene. The fraction of total URA3 DNA immunoprecipitated with the H4 antibodies was normalized to the fraction of total ACT1 DNA or to the fraction of total LEU2 DNA immunoprecipitated with the H4 antibodies to control for possible differences in chromatin solubilization for the different genes and between different chromatin preparations (Fig. 6).
Figure 6.
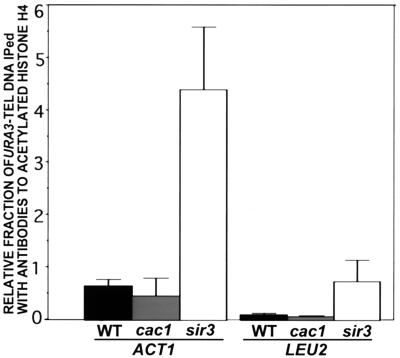
Relative fraction of URA3 DNA immunoprecipitated by antibodies to acetylated H4. Chromatin from strains DdB10UT, DdB10UTcac1Δ, and DdB10UTsir3Δ was immunoprecipitated with antibodies specific for multiply acetylated histone H4. The fraction of total URA3 DNA immunoprecipitated was divided by the fraction of total ACT1 DNA or the fraction of total LEU2 DNA immunoprecipitated by the antibodies to acetylated histone H4. The values represent the average of three independent immunoprecipitations for each strain. The standard deviation is indicated by the error bars.
When normalized to either ACT1 or LEU2, the transcribed telomeric URA3 gene in a sir3Δ background was immunoprecipitated at a higher level than the repressed, telomeric URA3 gene in a wild-type strain (Fig. 6). The relative fraction of URA3 chromatin immunoprecipitated from cac1Δ cells was very similar to that of wild-type cells, indicating that there was no major change in overall histone H4 acetylation at a telomere-adjacent gene in cac1Δ mutants. The relative fraction of telomeric URA3 DNA immunoprecipitated as compared with ACT1 was significantly higher than the relative fraction of URA3 DNA immunoprecipitated as compared with LEU2 in all strains tested. This result may reflect a difference in the densities of acetylated histone H4 between ACT1 and LEU2, or, more likely, differences in the relative accessibility of these genes to solubilization or immunoprecipitation. Nevertheless, normalization of the URA3 data to either the ACT1 or LEU2 probes showed that a telomeric URA3 gene in a sir3Δ background was immunoprecipitated at a substantially higher level than in either a wild-type or a cac1Δ strain, and that there was no significant difference in URA3 immunoprecipitation between wild-type and cac1Δ cells.
DISCUSSION
Two lines of evidence indicate that the reduction in TPE in cac1Δ cells was the result of an increased rate of switching from transcriptionally “off” to transcriptionally “on” of the telomeric gene. First, colonies of a strain carrying a telomeric ADE2 gene produced mostly white colonies with red sectors, indicating that there were two populations of cells within the colony, those with the ADE2 gene “on” (white) and those with the ADE2 gene “off” (red). Second, single-cell analysis showed that cac1Δ cells “switch” from a transcriptionally repressed state (FOAR) to a transcriptionally active state (FOA-sensitive) ≈2.5 times more often per generation than do wild-type cells. The increased switching in a cac1Δ mutant was not accompanied by substantial changes in chromatin structure as assayed by dam methylation protection (Fig. 5), overall histone H4 acetylation (Fig. 6), or nucleosome mapping (data not shown).
The fact that no major changes in these aspects of chromatin structure were seen suggests two possibilities. One is that the fraction of cells subject to TPE in a population of cac1Δ cells does not differ significantly from that of wild-type cells, but a TPE defect is detected because of the increased frequency with which cells switch from transcriptionally “off” to “on.” This model requires that cac1Δ cells also have an increased rate of switching from the transcribed to the repressed state, which is not consistent with the appearance of cells with ADE2 at the telomere. Although only ≈5% of wild-type cells produced mostly white colonies, the number and size of the red sectors in these largely white colonies were indistinguishable from those in cac1Δ cells (data not shown), suggesting that there is not a significant change in the rate of switching from transcribed to repressed. Therefore, cac1Δ cells probably have an alteration(s) in chromatin structure that is too subtle to be detected with antibodies to fully acetylated histone H4 or by dam methylation protection.
Because both the repressed and the active transcriptional states are relatively stable, there must be a mechanism that acts during or after each S phase that usually causes the parental chromatin to be reassembled on both daughter molecules in wild-type cells. We propose that transcriptionally silenced genes near telomeres or internal tracts of C1–3A sequence are usually assembled into chromatin by yCAF-I, which, like human CAF-I (28, 29), preferentially uses histones with a deposition pattern of acetylation. This pattern is best characterized for newly made histone H4, which is acetylated on lysines 5 and 12 (17). We propose that appropriately acetylated histones are not only essential for transcriptional silencing but also provide a signal that helps perpetuate a repressed chromatin state. In the absence of Cac1p, an alternate pathway can assemble telomeric chromatin, but according to our model this pathway does not discriminate between deposition-acetylated and other forms of histones H3 and H4. This model requires that cac1Δ cells have a pool of free histones that lack a deposition acetylation pattern, a point that has not been addressed experimentally in either wild-type or cac1Δ yeast. Another possibility is that the alternative chromatin assembly pathway utilizes histones with a deposition pattern of acetylation, but assembly by this pathway results in chromatin that is more accessible to histone acetylases/deacetylases. In either case, chromatin assembled in the absence of yCAF-I would be relatively deficient in histones with a deposition pattern of acetylation, a situation that would not affect its ability to be immunoprecipitated by antibodies to multiply acetylated histone H4 (Fig. 6). That is, histone H4 in chromatin synthesized by an alternative assembly pathway would not necessarily be acetylated at lysines 5 and 12. Although the alternative pathway could result in assembly of histones with the appropriate acetylation state some of the time, thereby explaining why some cac1Δ cells exhibit TPE, assembly by yCAF-I increases the probability that histones will have the appropriate acetylation pattern. We propose that HM chromatin is also normally synthesized by yCAF-I, but that there is a redundant mechanism that assures chromatin is silenced at the HM loci in a cac1Δ strain.
This model also requires a mechanism for targeting yCAF-I specifically to telomeres. Mammalian CAF-I must discriminate between deposition pattern histones and other histones because it is preferentially associated with the former (29). Thus, yCAF-I may have a higher affinity for chromatin containing histones with a deposition pattern of acetylation and thereby be targeted preferentially to chromatin with these modifications. Alternatively, yCAF-I may be targeted to telomeres by a specific interaction with a telomere replication complex, perhaps by its ability to interact with the Pif1p DNA helicase (E.K.M. and V.A.Z., unpublished results) or by its being targeted to the same subnuclear compartment, the nuclear periphery, as telomeres (42).
Yeast Cac1p is dispensable for assembling DNA into nucleosomes but is critical for propagating the repressed chromatin state both at telomeres and at internal sites on the chromosome. The proteins that constitute CAF-I are conserved between humans and yeast, suggesting that this complex serves an important function. TPE is only one of many examples where a repressed transcriptional state is inherited. Perhaps, in higher cells, CAF-I not only is involved in maintaining stable transcriptional repression, such as position effect variegation in Drosophila and X chromosome inactivation in mammals, but also functions to maintain the transcriptional repression of developmentally regulated genes.
Acknowledgments
We thank C. D. Allis for antibodies to acetylated histone H4, J. Stavenhagen for help with the CBS experiment, and all the members of the Zakian laboratory for input throughout the course of this work. This work was supported by National Institutes of Health Grants GM26938 and GM43265 to V.A.Z., a National Institutes of Health Postdoctoral Fellowship to E.K.M. (GM16593), and an American Cancer Society Postdoctoral Fellowship to D.d.B.
ABBREVIATIONS
- TPE
telomere position effect
- CBS
C1–3A-based silencing, FOA, 5-fluoroorotic acid
- FOAR
FOA-resistant
- yCAF-I
yeast chromatin assembly factor I
- RT-PCR
reverse transcription–PCR
References
- 1.Sandell L L, Zakian V A. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 2.Levis R, Hazelrigg T, Rubin G M. Science. 1985;229:558–561. doi: 10.1126/science.2992080. [DOI] [PubMed] [Google Scholar]
- 3.Gottschling D E, Aparicio O M, Billington B L, Zakian V A. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 4.Nimmo E R, Cranston G, Allshire R C. EMBO J. 1994;13:3801–3811. doi: 10.1002/j.1460-2075.1994.tb06691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rudenko G, Blundell P A, Dirks-Mulder A, Kieft R, Borst P. Cell. 1995;83:547–553. doi: 10.1016/0092-8674(95)90094-2. [DOI] [PubMed] [Google Scholar]
- 6.Horn D, Cross G A. Cell. 1995;83:555–561. doi: 10.1016/0092-8674(95)90095-0. [DOI] [PubMed] [Google Scholar]
- 7.Stavenhagen J B, Zakian V A. Genes Dev. 1994;8:1411–1422. doi: 10.1101/gad.8.12.1411. [DOI] [PubMed] [Google Scholar]
- 8.Wright J H, Gottschling D E, Zakian V A. Genes Dev. 1992;6:197–210. doi: 10.1101/gad.6.2.197. [DOI] [PubMed] [Google Scholar]
- 9.Gottschling D E. Proc Natl Acad Sci USA. 1992;89:4062–4065. doi: 10.1073/pnas.89.9.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 11.Braunstein M, Sobel R E, Allis C D, Turner B M, Broach J R. Mol Cell Biol. 1996;16:4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh J, Klar A J S. Genes Dev. 1992;6:186–196. doi: 10.1101/gad.6.2.186. [DOI] [PubMed] [Google Scholar]
- 13.Wolffe A P. Dev Genet. 1994;15:463–470. doi: 10.1002/dvg.1020150604. [DOI] [PubMed] [Google Scholar]
- 14.Ruiz-Carrillo A, Wangh L J, Allfrey V G. Science. 1975;190:117–128. doi: 10.1126/science.1166303. [DOI] [PubMed] [Google Scholar]
- 15.Chicoine L G, Schulman I G, Richman R, Cook R G, Allis C D. J Biol Chem. 1986;261:1071–1076. [PubMed] [Google Scholar]
- 16.Sobel R E, Cook R G, Allis C D. J Biol Chem. 1994;269:18576–18582. [PubMed] [Google Scholar]
- 17.Sobel R E, Cook R G, Perry C A, Annunziato A T, Allis C D. Proc Natl Acad Sci USA. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleff S, Andrulis E D, Anderson C W, Sternglanz R. J Biol Chem. 1995;270:24674–24677. doi: 10.1074/jbc.270.42.24674. [DOI] [PubMed] [Google Scholar]
- 19.Parthun M R, Widom J, Gottschling D E. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 20.Turner B M, Birley A J, Lavender J. Cell. 1992;69:375–384. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]
- 21.Park E-C, Szostak J W. Mol Cell Biol. 1990;10:4932–4934. doi: 10.1128/mcb.10.9.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Megee P C, Morgan B A, Mittman B A, Smith M M. Science. 1990;247:841–845. doi: 10.1126/science.2106160. [DOI] [PubMed] [Google Scholar]
- 23.Thompson J S, Ling X, Grunstein M. Nature (London) 1994;369:245–247. doi: 10.1038/369245a0. [DOI] [PubMed] [Google Scholar]
- 24.de Rubertis F, Kadosh D, Henchoz S, Pauli D, Reuter G, Struhl K, Spierer P. Nature (London) 1996;384:589–591. doi: 10.1038/384589a0. [DOI] [PubMed] [Google Scholar]
- 25.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith S, Stillman B. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 27.Smith S, Stillman B. EMBO J. 1991;10:971–980. doi: 10.1002/j.1460-2075.1991.tb08031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaufman P D, Kobayashi R, Kessler N, Stillman B. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- 29.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 30.Kaufman P D, Kobayashi R, Stillman B. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 31.Enomoto S, McCune-Zierath P D, Gerami-Nejad M, Sanders M A, Berman J. Genes Dev. 1997;11:358–370. doi: 10.1101/gad.11.3.358. [DOI] [PubMed] [Google Scholar]
- 32.Schulz V P, Zakian V A. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 33.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiley E, Zakian V A. Genetics. 1995;139:67–79. doi: 10.1093/genetics/139.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimmerly W J, Rine J. Mol Cell Biol. 1987;7:4225–4237. doi: 10.1128/mcb.7.12.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahoney D J, Marquardt R, Shei G-J, Rose A B, Broach J R. Genes Dev. 1991;5:605–615. doi: 10.1101/gad.5.4.605. [DOI] [PubMed] [Google Scholar]
- 38.Lin R, Leone J W, Cook R G, Allis C D. J Cell Biol. 1989;108:1577–1588. doi: 10.1083/jcb.108.5.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smeal T, Claus J, Kennedy B, Cole F, Guarente L. Cell. 1996;84:633–642. doi: 10.1016/s0092-8674(00)81038-7. [DOI] [PubMed] [Google Scholar]
- 40.Boeke J D, LaCroute F, Fink G R. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 41.Pillus L, Rine J. Cell. 1989;59:637–647. doi: 10.1016/0092-8674(89)90009-3. [DOI] [PubMed] [Google Scholar]
- 42.Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser S M. J Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]