Abstract
Comparing experimental generalized N-H S2 order parameters to those calculated from molecular dynamics trajectories is increasingly used to judge force-field quality and completeness of sampling. Herein we demonstrate for the well-investigated system hen egg white lysozyme that different experimental starting structures can lead to significant differences in molecular-dynamics-derived S2 parameters that can be even larger than S2 parameter deviations due to different force fields. Caution should thus be taken in general when simulated S2 parameters are compared to experimental data with the aim of judging force-field quality. We show that adequately sampling flexible regions (∼100 ns) and only calculating S2 parameters averaged over short time windows proved necessary to obtain consistent results irrespective of the starting structure.
Molecular dynamics (MD) simulations and NMR spin relaxation spectroscopy are complementary tools to investigate the dynamics of biomolecules (1,2). Comparing experimental NMR spin relaxation data to those calculated from MD trajectories also allows assessing whether MD simulations accurately reproduce structural and dynamical properties of the system (3). A comparison of experimental and MD-derived N-H S2 order parameters (4) is increasingly used to judge force-field quality in this regard (5–8).
Aside from simulation conditions (5) and the approach used to extract S2 parameters (9), an influence of the MD starting structure on the computed S2 parameters ought to be expected too (10,11), considering that system dynamics and (local) structure are intimately related (12). This influence should become particularly pronounced in flexible parts of the molecule, where conformational variability in experimental structures is most likely to occur. Herein, we demonstrate for hen egg white lysozyme (HEWL) that different starting structures can lead to differences in MD-derived S2 parameters that can be larger than deviations due to different force fields. Accordingly, caution should be taken when simulated S2 parameters are compared to experimental data with the aim of judging force-field quality.
HEWL is a well-investigated model system that has become a standard for evaluating the quality of force fields in terms of internal dynamics (6–8,13). For investigating a starting structure dependence on calculated S2 values, we first clustered 92 unbound wild-type HEWL crystal structures with a resolution <2.0 Å and 50 conformations of an NMR ensemble (14) with respect to backbone torsion angles of residues Gly71 and Asn74 in the loop 2 region (residues 65–75). The conformations are characterized by the backbone N-H bonds of the two residues either pointing to the protein or to the solvent, revealing in total four major loop 2 conformations (cluster 1: 32 NMR structures, 37 crystal structures; cluster 2 and 4: 48 and 7 crystal structures; cluster 3: 18 NMR structures) (see Table S1 and Fig. S1 in Supplementary Material, Data S1). Of the three clusters containing crystals structures, we chose one crystal structure each as a starting structure (Fig. 1 a). All three structures have been crystallized in the same space group. Visual inspection of the crystal packings did not reveal any differences in stabilizing interactions by the crystal environment involving the loop 2 region. Hence, we believe that the conformational variance of loop 2 cannot be attributed to differences in crystal packing. For reasons of comparison, we also considered the first model of the NMR ensemble 1E8L (cluster 1).
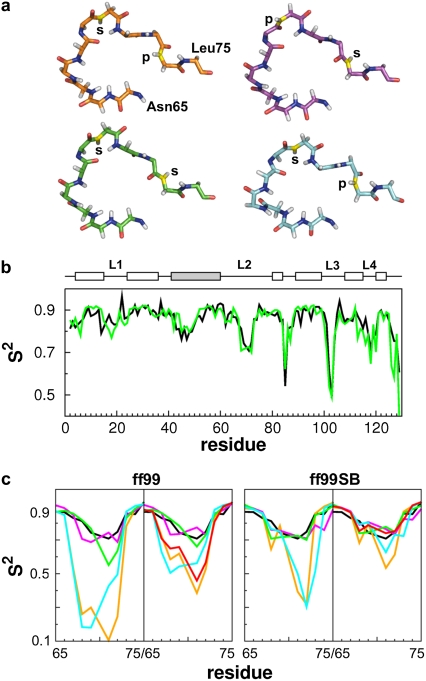
FIGURE 1.
(a) Residues 65–75 (loop 2) of the investigated HEWL structures (1HEL (orange, cluster 1); 1E8L model 1 (cyan, cluster 1); 1IEE (green, cluster 2); and 6LYT (magenta, cluster 4)). The N atoms of Gly71 and Asn74 are colored in yellow. Additionally, it is indicated if the N-H bond points to the protein (p) or to the solvent (s). (b) Experimental S2 values (18) (black) are compared to calculated values (green) from the 30 ns simulation of 1IEE with ff99SB. Secondary structure elements are indicated by boxes (white, helix; gray, β-sheet). (c) Comparison of experimental (18) (black) and calculated S2 values (colored as in a) of loop 2 using ff99 and ff99SB. The S2 values were either calculated over the whole 30 ns trajectory (left panels) or averaged over time windows of 1 ns length (right panels). The red line depicts 1 ns time window-averaged S2 values of 1HEL over 100 ns.
For each structure we performed MD simulations with periodic boundary conditions in the NVT ensemble using the TIP3P water model at 300 K for a minimum of 30 ns with AMBER9 (15) and applying the ff99 (16) and ff99SB (6) force fields. The MD simulations of 1HEL were extended to 100 ns. S2 parameters were calculated for backbone N-H bonds according to the isotropic reorientational eigenmode dynamics approach (17). Using an alternative approach applied by Hornak et al. (6) did not result in qualitatively different S2 values (see Fig. S8 in Data S1).
Initially, we analyzed the structural stability of loop 2 in terms of backbone dihedral angle changes for residues Gly71 and Asn74 (see Figs. S2–S5 in Data S1). The backbone dihedrals allow assigning each conformation from the MD simulations to one of the four clusters. With respect to loop 2, the trajectories originating from different starting structures revealed very different structural stabilities (Fig. S6 in Data S1). 6LYT and 1IEE almost exclusively exist in a loop 2 conformation similar to that of cluster 2, with 1IEE showing occasional transitions to the cluster 4 conformation in the case of ff99. In contrast, 1HEL and 1E8L display conformations that resemble those of cluster 1 and 3. Frequent exchanges between these two conformations occur in the case of ff99, but a more restricted loop 2 dynamics is observed in the case of ff99SB. In no case, transitions between cluster sets {1, 3} and {2, 4} are observed. As we believe that the observed clusters are not due to differences in the energetics of the crystal environments, this points to a poor sampling of the backbone dihedral angle transitions of the peptide bond between Arg73 and Asn74. From the observed differences in the structural stabilities, one can already anticipate computed S2 parameters of loop 2 to be different across the trajectories due to varying internal dynamics of that region.
Fig. 1 and Fig. S7 in Data S1 show calculated S2 parameters for the four different starting structures. In all cases, S2 parameters of ordered regions agree across the different simulations as well as with experiment (18) (RMSD < 0.085; Table S2), irrespective of the force field used (Fig. S7 in Data S1). This observation is in line with previously reported characteristics of the two force fields (5). In contrast, major differences occur in the loop regions of HEWL (residues 16–23, 65–75, 100–107, and 116–119) (Fig. 1, and Fig. S7 in Data S1) and are most pronounced for the loop 2 region. This is not unexpected (6) if different force fields are applied to generate trajectories originating from the same structure (as in the case of 1IEE, Fig. 1 c). However, it is disturbing that even larger S2 differences are observed if trajectories were generated from different structures, but using the same force field (Fig. 1 c). In our case, this holds true for both force fields tested. A better agreement between computed and experimental S2 for the loop 2 region of 6LYT has been found for ff99SB compared to ff99 by Hornak et al. (6), in agreement with similar findings on ubiquitin (5). In contrast, in our case, both ff99 and ff99SB result in S2 parameters for 6LYT that very well agree with experiment (RMSD = 0.047; 0.048). Similarly, a very good agreement is found for 1IEE with ff99 and ff99SB (RMSD = 0.064; 0.042). In contrast, 1HEL and 1E8L lead to too low S2 parameters irrespective of the force field used (RMSD > 0.175). These results demonstrate that depending on the initial loop conformation significant differences in computed S2 parameters are observed, if the whole trajectory is considered at once as done previously (5,6). Obviously, an agreement between computed and experimental S2 values should be considered fortuitous in this case.
If a simulation exceeds in length the overall tumbling correlation time, S2 parameters computed over the whole trajectory can include motions that would not be reflected in the experimental S2 values, leading to a bias in the computed S2 values (5,19). Thus, we also computed S2 parameters from the MD simulations after the isotropic reorientational eigenmode dynamics approach (17) for time windows of 1–5 ns in 1 ns steps. The S2 parameters were subsequently averaged over all available time windows. No qualitative change of S2 parameters was observed for 6LYT and 1IEE. However, in the case of 1HEL and 1E8L, increased S2 parameters were found for the loop 2 region for both the ff99 and ff99SB case, with the 1 ns S2 parameters providing the best results (RMSD = 0.058–0.153; Table S3 in Data S1), as also found recently (5,19). S2 parameters computed from the ff99SB trajectories now much better agree with experiment (RMSD = 0.085 (1HEL); 0.058 (1E8L)) than those from the ff99 trajectories (RMSD = 0.153 (1HEL); 0.143 (1E8L)). Encouragingly, S2 parameters computed from different starting structures are rather consistent in the ff99SB case. Overall, when calculating S2 parameters over short time windows, an improved description of internal dynamics by ff99SB over ff99 becomes obvious.
Longer MD simulations should in principle help to overcome a lack of adequate sampling and result in a better description of the dynamics of mobile protein regions (5). As an attempt to further improve the agreement between computed and experimental S2 values, we thus extended the trajectories of 1HEL to 100 ns simulation length. Assigning the loop 2 conformations again to one of the four clusters revealed no obvious change in the frequency of transitions between cluster 1 and 3 in the case of ff99 (Fig. S6 in Data S1). However, in the case of ff99SB, cluster 1 conformations almost exclusively prevail beyond a simulation time of 30 ns. The more restricted dynamics of the loop 2 region in the case of ff99SB compared to ff99 is also reflected in the S2 parameters. If calculated from time windows of 1 ns and subsequent averaging, the thus-obtained S2 parameters from the ff99SB trajectory agree well with those from 6LYT and 1IEE trajectories and show the best agreement with experiment (RMSD = 0.037). In contrast, ff99 simulation-derived S2 parameters are still at variance with experiment (RMSD = 0.122).
Our results demonstrate that caution should be taken in general when simulated S2 parameters are compared to experimental data with the aim of judging force-field quality. To assess the consistency of computed S2 parameters, trajectories started from conformationally varying structures should be evaluated in parallel whenever possible. Adequately sampling flexible regions (∼100 ns) with the aim to obtain an accurate representation of the structure and dynamics and only calculating S2 parameters over short time windows (∼1 ns) furthermore proved necessary to obtain consistent and accurate results irrespective of the starting coordinates.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Acknowledgments
This work was supported by startup funds from Goethe-University, Frankfurt am Main by the European Network-Nuclear Magnetic Resonance project (RII3 project/contract No.: 026145), and by the state of Hessen (Center for Biomolecular Magnetic Resonance). We are grateful for support by the Frankfurt Center for Scientific Computing.
Editor: Kathleen B. Hall.
References
- 1.Case, D. A. 2002. Molecular dynamics and NMR spin relaxation in proteins. Acc. Chem. Res. 35:325–331. [DOI] [PubMed] [Google Scholar]
- 2.Brüschweiler, R. 2003. New approaches to the dynamic interpretation and prediction of NMR relaxation data from proteins. Curr. Opin. Struct. Biol. 13:175–183. [DOI] [PubMed] [Google Scholar]
- 3.Smith, L. J., A. E. Mark, C. M. Dobson, and W. F. van Gunsteren. 1995. Comparison of MD simulations and NMR experiments for hen lysozyme. Analysis of local fluctuations, cooperative motions, and global changes. Biochemistry. 34:10918–10931. [DOI] [PubMed] [Google Scholar]
- 4.Lipari, G., and A. Szabo. 1982. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J. Am. Chem. Soc. 104:4546–4559. [Google Scholar]
- 5.Showalter, S. A., and R. Brüschweiler. 2007. Validation of molecular dynamics simulations of biomolecules using NMR spin relaxation as benchmarks: application to the AMBER99SB force field. J. Chem. Theory Comput. 3:961–975. [DOI] [PubMed] [Google Scholar]
- 6.Hornak, V., R. Abel, A. Okur, B. Strockbine, A. Roitberg, and C. Simmerling. 2006. Comparison of multiple AMBER force fields and development of improved protein backbone parameters. Proteins Struct. Funct. Bioinformat. 65:712–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buck, M., S. Bouguet-Bonnet, R. W. Pastor, and A. D. MacKerell. 2006. Importance of the CMAP correction to the CHARMM22 protein force field: dynamics of hen lysozyme. Biophys. J. 90:L36–L38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soares, T., X. Daura, C. Oostenbrink, L. Smith, and W. F. van Gunsteren. 2004. Validation of the GROMOS force-field parameter set 45A3 against nuclear magnetic resonance data of hen egg lysozyme. J. Biomol. NMR. 30:407–422. [DOI] [PubMed] [Google Scholar]
- 9.Clore, G. M., A. Szabo, A. Bax, L. E. Kay, P. C. Driscoll, and A. M. Gronenborn. 1990. Deviations from the simple two-parameter model-free approach to the interpretation of nitrogen-15 nuclear magnetic relaxation of proteins. J. Am. Chem. Soc. 112:4989–4991. [Google Scholar]
- 10.Horita, D. A., W. Zhang, T. E. Smithgall, W. H. Gmeiner, and R. A. Byrd. 2000. Dynamics of the Hck-SH3 domain: comparison of experiment with multiple molecular dynamics simulations. Protein Sci. 9:95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andrews, B. K., T. Romo, J. B. Clarage, B. M. Pettitt, and G. N. Phillips. 1998. Characterizing global substates of myoglobin. Structure. 6:587–594. [DOI] [PubMed] [Google Scholar]
- 12.Halle, B. 2002. Flexibility and packing in proteins. Proc. Natl. Acad. Sci. USA. 99:1274–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stocker, U., and W. F. van Gunsteren. 2000. Molecular dynamics simulation of hen egg white lysozyme: a test of the GROMOS96 force field against nuclear magnetic resonance data. Proteins. 40:145–153. [PubMed] [Google Scholar]
- 14.Schwalbe, H., S. B. Grimshaw, A. Spencer, M. Buck, J. Boyd, C. M. Dobson, C. Redfield, and L. J. Smith. 2001. A refined solution structure of hen lysozyme determined using residual dipolar coupling data. Protein Sci. 10:677–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Case, D. A., T. Cheatham, T. Darden, H. Gohlke, R. Luo, K. M. Merz, A. Onufriev, C. Simmerling, B. Wang, and R. Woods. 2005. The AMBER biomolecular simulation programs. J. Comput. Chem. 26:1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, J., P. Cieplak, and P. Kollman. 2000. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 21:1049–1074. [Google Scholar]
- 17.Prompers, J. J., and R. Brüschweiler. 2002. General framework for studying the dynamics of folded and nonfolded proteins by NMR relaxation spectroscopy and MD simulation. J. Am. Chem. Soc. 124:4522–4534. [DOI] [PubMed] [Google Scholar]
- 18.Buck, M., J. Boyd, C. Redfield, D. A. MacKenzie, D. J. Jeenes, D. B. Archer, and C. M. Dobson. 1995. Structural determinants of protein dynamics: analysis of 15N NMR relaxation measurements for main-chain and side-chain nuclei of hen egg white lysozyme. Biochemistry. 34:4041–4055. [DOI] [PubMed] [Google Scholar]
- 19.Nederveen, A. J., and A. Bonvin. 2005. NMR relaxation and internal dynamics of ubiquitin from a 0.2 microsec MD simulation. J. Chem. Theory Comput. 1:363–374. [DOI] [PubMed] [Google Scholar]