Abstract
3-methyladenine (3MeA) DNA glycosylases remove 3MeAs from alkylated DNA to initiate the base excision repair pathway. Here we report the generation of mice deficient in the 3MeA DNA glycosylase encoded by the Aag (Mpg) gene. Alkyladenine DNA glycosylase turns out to be the major DNA glycosylase not only for the cytotoxic 3MeA DNA lesion, but also for the mutagenic 1,N6-ethenoadenine (ɛA) and hypoxanthine lesions. Aag appears to be the only 3MeA and hypoxanthine DNA glycosylase in liver, testes, kidney, and lung, and the only ɛA DNA glycosylase in liver, testes, and kidney; another ɛA DNA glycosylase may be expressed in lung. Although alkyladenine DNA glycosylase has the capacity to remove 8-oxoguanine DNA lesions, it does not appear to be the major glycosylase for 8-oxoguanine repair. Fibroblasts derived from Aag −/− mice are alkylation sensitive, indicating that Aag −/− mice may be similarly sensitive.
In the face of inescapable DNA-damaging agents and inevitable spontaneous DNA degradation, the constant challenge to preserve genomic integrity has been met by the evolution of numerous pathways that protect against the genotoxic effects of DNA-damaging agents. Unless processed properly, DNA damage can be mutagenic, carcinogenic, and teratogenic, and DNA damage also may contribute to aging (1).
Alkylating agents are found in our environment, in our food, inside all cells as natural metabolites, and in the clinic as cancer chemotherapeutic agents. Base excision repair (BER) is one of the major pathways for the repair of damaged DNA bases and proceeds through a sequence of reactions requiring several different enzymes. The first step involves excision of the damaged base (for which most cells are known to have several different DNA glycosylases). Base excision by glycosylases is followed by strand cleavage in the vicinity of the abasic site (by AP endonuclease or AP lyase), and preparation of the DNA ends for gap filling and ligation. DNA polymerase fills the gap, and DNA ligase seals the remaining nick, thus completing the BER process (1).
Human and rat 3-methyladenine (3MeA) DNA glycosylases, so named because 3MeA was the first substrate identified for this class of enzymes (2), actually display an unexpectedly broad substrate range, including guanines methylated at the N3 or the N7 position (3–6), deaminated adenine [i.e., hypoxanthine (Hx)] (7), oxidized guanine 8-oxoguanine (8oxoG) (8), cyclic etheno adducts on both adenine and guanine (9, 10), and haloethylated purines (unpublished observations). The mouse alkyladenine DNA glycosylase (Aag) has not been assayed for release of all of these substrates, though it has been shown to act on N3 and N7 methylpurines and on 8oxoG (8, 11). The precise biological effects of all of the DNA lesions repaired by mammalian 3MeA DNA glycosylases are not yet known for mammals, though there is strong evidence that 3MeA is cytotoxic (12), and other lesions may be mutagenic, namely Hx (13), 8oxoG (14), 1,N6-ethenoadenine (ɛA), and N2,3-ethenoguanine (15, 16). It is important to note that each of these DNA lesions can arise spontaneously in the DNA of untreated mammalian cells (17–21), that Hx and 8oxoG are induced by cigarette smoke (22, 23), and that ethenopurines are induced by the carcinogenic industrial chemical vinyl chloride (24). These findings raise the possibility that the mammalian 3MeA DNA glycosylase may influence spontaneous mutation as well as protect against environmental genotoxic agents. Here we describe the generation of Aag null animals, a BER-deficient animal model. Surprisingly, the Aag gene product seems to be the only measurable DNA glycosylase, in several mouse tissues, that is active against 3MeA, Hx, and ɛA.
MATERIALS AND METHODS
Reagents.
Enzymes were purchased from New England Biolabs and Boehringer Mannheim. DNA probes were synthesized with NEBlot (New England Biolabs) and labeled with [32P]α-dCTP (NEN/DuPont). E14 embryonic stem (ES) cells were provided by A. Berns and H. te Riele, Netherlands Cancer Institute, Amsterdam.
Partial Purification of Mouse Aag.
The Aag cDNA was subcloned into pRSETA to express polyhistidine-tagged Aag (His-Aag) and purified according to manufacturer’s instructions (Invitrogen). The identical purification procedure was performed on control JM109 cells lacking any expression vector to create negative control extracts. Purification of His-Aag away from the endogenous 3MeA DNA glycosylase activity of JM109 was confirmed by showing that control JM109 extracts have no residual 3MeA DNA glycosylase activity (data not shown).
Creation of Aag Heterozygous ES Cells by Targeted Homologous Recombination.
E14 ES cells were cultured on 0.1% gelatin-coated dishes in DMEM/60% Buffalo rat liver-conditioned medium (25), supplemented with 10% fetal calf serum/1% nonessential amino acids (GIBCO/BRL)/0.1 mM 2-mercaptoethanol/50 units/ml of penicillin/50 μg/ml of streptomycin/1,000 units/ml of leukemia inhibitory factor (GIBCO/BRL). Ten to 15 micrograms of Aag-targeting construct was prepared as described (12) and electroporated into 107 129/Ola-derived E14 ES cells in PBS at 385 V/cm, 1,200 μF, for 10 msec by using a Progenitor II PG200, Hoefer Gene-pulser. G418 (200 μg/ml) and 1-(2′-deoxy,2′-fluoro-β-d-arabinofuranosyl)-5 iodouracil (0.2 μM; Bristol Myers Squibb) resistant clones were expanded into duplicate 24-well dishes without feeders. Half of the cultures were frozen at −80°C in ES medium/10% dimethyl sulfoxide, and genomic DNA was isolated from the other half as described (26). Homologous recombinants were identified by Southern analysis (12) and reconfirmed by probing with neo and herpes simplex virus-tk specific probes (data not shown).
Genotyping Mouse Cells and Tissues.
Ear punches were prepared for PCR as described (27). DNA was isolated from spleen by using a QIAmp Tissue Kit (Qiagen). Southern analysis was performed as described (12). Three primers were used in multiplex PCR analysis of DNA. PI (5′GCAGCGACTGGCAGATTC) and PIII (5′GAAATGCACGGGCTAGGG) detect the wild-type allele (164 bp), and primers PII (which hybridizes the neo cassette; 5′AGAAGGGTGAGAACAGAG) and PIII detect the targeted allele (523 bp). The faint band above the 523-bp product appearing in Fig. 1B disappeared under optimal reaction conditions. The 2,160-bp PI/PIII product is not observed under these PCR conditions. Cycling was done at: 95°C 5 min/55°C 2 min/72°C 1 min (1 cycle); 95°C 1 min/55°C 1 min/72°C 1 min (30 cycles); 95°C 1 min/55°C 4 min/72°C 10 min (1 cycle).
Figure 1.
Genotyping Aag +/+, +/−, and −/− mice. (A) Schematic representation of a portion of the wild-type and targeted Aag alleles indicating the location of three primers and HindIII (H) sites. Note that Aag exon II is disrupted by insertion of a neo expression cassette to create an Aag null allele. Primers PI and PIII yield a 164-bp fragment in the presence of the wild-type allele. In the presence of the targeted allele, primers PII and PIII yield a 523-bp fragment. HindIII sites are loosely indicated to show that disruption of Aag by neo insertion increases the size of the HindIII fragment (details have been presented elsewhere) (12). (B) (Upper) PCR genotyping of MEFs (lanes 1–5). PCR genotyping of DNA from spleens of mice used in subsequent biochemical analysis (lanes 6–7). (Lower) Southern blot analysis of the DNA used for PCR analysis above. HindIII-digested DNA was probed with a fragment of the Aag gene that lies outside of the targeting construct (12). The Aag null allele (8.4 kb) and the wild-type Aag allele (6.3 kb) are indicated.
Generation of Aag +/+, +/−, and −/− Mice.
Ten to fifteen karyotypically normal ES clones were injected into C57BL/6 blastocysts and transferred into pseudopregnant female BCBA mice. Male chimeras were crossed with C57BL/6 females, and germ-line transmission was determined by the transmission of the agouti coat color.
Preparation of Protein Extracts.
Aag +/+ and Aag −/− ES cell protein extracts were prepared as described (12). Mouse tissues were thawed on ice in 2.5 vol of glycosylase buffer (20 mM Tris⋅HCl, pH 7.8/100 mM KCl/2 mM EDTA/5 mM 2-mercaptoethanol) supplemented with 1.5% triton and protease inhibitors (100 mM benzamidine HCl, 1 μg/ml leupeptin, pepstatin A, and aprotinin; Sigma). Tissues were dounce homogenized, sonicated, and centrifuged (20 min, 14,000 g, 4°C). Protein concentrations of the supernatants were determined by Bradford analysis (Bio-Rad).
3MeA DNA Glycosylase Activity Assay.
3MeA DNA glycosylase activity was measured as described (6). Briefly, extract protein was incubated for 5 hr at 37°C with 27.6 μg of 17.9 Ci/mmol N-[3H]methyl-N-nitrosourea (Amersham)-treated calf thymus DNA substrate (2,492 cpm/μg, prepared as described) (28). Released bases were analyzed by descending paper chromatography as described (29).
Toxicity Assay of Aag +/+ and Aag −/− Primary Mouse Embryo Fibroblasts (MEFs).
Primary MEFs were isolated from 13.5-day-old mouse embryos and maintained as described (30). 104 cells per 96-well were plated 24 hr before treatment. Cells were incubated in serum-free media containing MeOSO2(CH2)2-lexitropsin (Me-Lex) for 1 hr, or exposed to UV in 0.1 ml PBS. After 72-hr incubation in 0.2 ml of media, media was brought to 0.5 mg/ml 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, and cells were incubated an additional 4 hr. Media was replaced with 0.2 ml of dimethyl sulfoxide, and metabolically active cells were quantified by absorbance at 560 nm. Four independent samples were averaged, and their standard deviations are indicated. Each curve was repeated at least three times; representative curves are shown.
Preparation of Oligonucleotide Substrates for ɛA, Hx, and Uracil Glycosylase Activity.
The sequence of the ɛA containing oligonucleotide has been reported (31) and was a gift from O. Scharer and G. Verdine, Harvard University, Boston. An oligo of identical sequence to that of ɛA was purchased from Midland Certified Reagents (Midland, TX) with 8oxoG in place of ɛA at position 13. The sequence of the uracil containing oligonucleotide has been reported (32) and was a gift of B. Demple and R. Bennett, Harvard School of Public Health, Boston. The Hx-containing oligonucleotide has the sequence 5′-GGATCATCGTTTTT(Hx)GCTACATCGC (Amitof, Boston). Oligonucleotides were labeled with [32P]γ-ATP (6,000 Ci/mmol; NEN/DuPont) and 10 units of T4 polynucleotide kinase, and annealed to 2-fold excess complementary strand. Complete annealing was confirmed by 20% nondenaturing PAGE (data not shown).
DNA Glycosylase Assays By Using Oligonucleotide Substrates.
Protein or protein extracts were incubated with 100 fmols of double-stranded oligonucleotides at 37°C in 20 mM Tris⋅HCl, pH 7.2/100 mM KCl/5 mM EDTA/1 mM EGTA/5 mM 2-mercaptoethanol in a total volume of 30 μL. DNA was chemically cleaved at abasic sites (0.1 N NaOH, 90°C, 4 min). Reactions were analyzed by 20% denaturing PAGE, and radioactivity was detected by a Bio-Rad PhosphorImaging system (GS525 Molecular Imager).
RESULTS
Generation of Aag Null Mice.
We cloned a murine 3MeA DNA glycosylase gene (Aag) to generate Aag-deficient cells (12) and mice by targeted homologous recombination (Fig. 1A). The targeting strategy used to generate 3MeA DNA glycosylase-deficient mice has been described (12). Briefly, Aag exon II was disrupted with a neo expression cassette to inactivate Aag and to allow selection for integration, and the herpes simplex virus-tk expression cassette (not shown) was placed adjacent to the Aag gene fragment to allow selection against random integration. E14 mouse ES cell transfectants, enriched for targeted homologous recombinants, were screened by Southern blot analysis to identify Aag +/− heterozygotes (an example of this kind of analysis is shown in Fig. 1B).
Aag +/− ES cells were injected into mouse blastocysts, and germ-line transmission was obtained from two independent ES cell clones. Aag +/− offspring of chimeric mice were intercrossed to generate Aag −/− animals, which were identified by PCR and Southern analysis (an example is shown in Fig. 1B, lane 7). Approximately 19% of the progeny of Aag +/− mice were homozygous Aag mutants (n = 139), which is not significantly different from Mendelian segregation. Thus Aag does not appear to be required for normal embryonic development. To date, Aag homozygous mice are phenotypically normal with no gross locomotory deficiencies that would hint of neurological or muscular defects (up to 8 months). In addition, both male and female homozygous Aag mice are fertile.
Aag Represents the Major 3MeA DNA Glycosylase in Several Mouse Tissues.
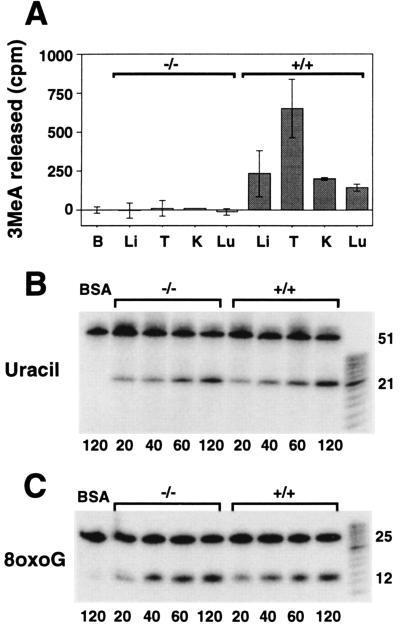
Escherichia coli and Saccharomyces cerevisiae both have at least two different 3MeA DNA glycosylases (19, 33), and although only one class of 3MeA DNA glycosylases has been cloned to date, mammals also may express more than one 3MeA DNA glycosylase activity. Although there is no detectable 3MeA DNA glycosylase activity in pluripotent Aag −/− mouse ES cells (12), other activities could potentially be revealed after differentiation. Indeed, exposure to endogenous and exogenous DNA-damaging agents may vary among tissue types, making it useful to selectively express a different selection of DNA glycosylases in different cell types. We therefore measured 3MeA DNA glycosylase activity in liver, testes, kidney, and lung extracts prepared from Aag +/+ and Aag −/− mice (Fig. 2A). Because mouse to mouse variation may affect our results, we assayed at least two independent mice of each genotype. All four wild-type tissues express robust 3MeA DNA glycosylase activity, whereas all four Aag −/− tissues are devoid of such activity. It is possible that we were unable to detect a minor 3MeA DNA glycosylase activity under these conditions. Nevertheless, these results suggest that these tissues do not constitutively express a 3MeA DNA glycosylase other than Aag.
Figure 2.
3MeA, uracil, and 8-oxoG DNA glycosylase activity in mouse tissue extracts. (A) 1,000 μg (BSA, B; liver, Li; and kidney, K), 500 μg (lung, Lu) or 250 μg (testes, T) of protein extracts from Aag +/+ or Aag −/− tissues were incubated in triplicate reactions for 5 hr at 37°C with [3H]methyl-N-nitrosourea-treated calf thymus DNA. 3MeA was separated from other bases by descending paper chromatography. The cpm associated with 3MeA per mg protein extracts are indicated for the average of two or three independent mice. Error bars represent standard deviations. (B) Time-dependent release of site-specific uracil from 5′ 32P-labeled double-stranded oligonucleotides by Aag −/− and Aag +/+ liver extracts. BSA (100 μg) was incubated with the oligo substrate (first lane). Remaining lanes each contain 100 μg of either Aag −/− or Aag +/+ liver extract. After incubation at 37°C for the times indicated in min beneath each lane, oligonucleotides were chemically cleaved at abasic sites and analyzed by denaturing PAGE. DNA glycosylase activity is indicated by the appearance of a 21-mer. (C) Time-dependent release of site-specific 8oxoG from 5′ 32P-labeled double-stranded oligonucleotides by Aag −/− and Aag +/+ testes extracts. BSA (75 μg) were incubated with the 8oxoG containing oligonucleotides (first lane). Remaining lanes each contain 75 μg of either Aag −/− or Aag +/+ testes extracts. Samples were analyzed as described in B. DNA glycosylase activity is indicated by the appearance of a 12-mer.
We confirmed that the absence of 3MeA DNA glycosylase activity in Aag −/− tissue extracts was not because of a general enzyme inactivation of these extracts, by showing that Aag +/+ and Aag −/− liver extracts have equivalent uracil DNA glycosylase activity (Fig. 2B). Further, because 3MeA DNA glycosylase activity levels have been shown to vary between mouse strains (34), and because Aag −/− mice have a mixed 129 and C57BL/6 genetic background (see Materials and Methods), we measured 3MeA DNA glycosylase activity in 129 and C57BL/6 mice. These mice displayed robust 3MeA DNA glycosylase activity in lung, liver, and testes (data not shown; kidney not tested). Thus, the varied contribution from the 129 and C57BL/6 genetic backgrounds can not account for the complete lack of activity observed in the Aag −/− mice. Taken together these data indicate: (i) Aag activity was successfully eliminated in the Aag homozygous null mice; and (ii) Aag is the major 3MeA DNA glycosylase activity in at least four tissues.
Aag +/+ and Aag −/− testes extracts show essentially identical 8oxoG DNA glycosylase activity (Fig. 2C). Although Aag removes 8oxoG in vitro (8), Aag is not a major contributor toward 8oxoG glycosylase activity, at least in testes. Clearly, (an)other enzyme(s) contribute the majority of the 8oxoG DNA glycosylase activity in testes, which is consistent with the cloning of a novel 8oxoG DNA glycosylase from mammalian cells (35–39, 59).
Cells Derived from Aag Null Mice Are Sensitive to Alkylating Agent.
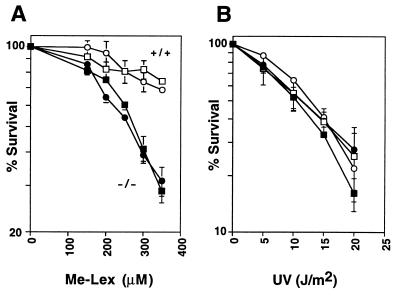
Primary MEFs were derived from five 13.5-day embryos from a cross between two Aag +/− mice. PCR and Southern blot genotyping of these cells are shown in Fig. 1B (lanes 1–5). MEFs from two Aag −/− and two Aag +/+ embryos were treated with the alkylating agent Me-Lex that specifically induces 3MeA DNA lesions (40, 41), or with UV. Aag −/− cells are more sensitive to the toxic effects of 3MeA lesions than Aag +/+ cells (Fig. 3A), as measured by using an 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay (see Materials and Methods). However, as expected, Aag +/+ and Aag −/− cells are equally sensitivity to UV (Fig. 3B), an agent that induces potentially lethal base damages that are not thought to be substrates of Aag.
Figure 3.
Toxicity assay of Aag +/+ or Aag −/− MEFs treated with increasing doses of Me-Lex (A) or UV (B). Percent metabolically active treated cells is expressed relative to untreated control cells. Cells from two Aag −/− embryos (closed symbols) and two Aag +/+ embryos (open symbols) were analyzed.
Mouse Aag Releases ɛA and Hx and Is the Major ɛA and Hx DNA Glycosylase in Several Tissues.
It previously was established that, in addition to 3MeA, the human and rat 3MeA DNA glycosylases can release ɛA and Hx from DNA (7, 10, 42), two lesions that promote base mispairing (13, 16). Here we establish that mouse Aag also repairs ɛA and Hx. Double-stranded 25-mer DNA oligonucleotides, containing a single ɛA, or a single Hx, located at a defined position, were used as substrates for the Aag enzyme that was partially purified from E. coli expressing the Aag cDNA (see Materials and Methods). Similarly prepared fractions from bacteria lacking the Aag cDNA were used as a negative control. Abasic sites were converted to strand breaks to produce either a labeled 14-mer (for the Hx substrate), or a labeled 12-mer (for the ɛA substrate). Fig. 4A clearly shows that Aag can remove Hx and ɛA lesions from DNA.
Figure 4.
Aag is the major Hx and ɛA DNA glycosylase in mouse liver, testes, kidney, and lung. (A) Release of ɛA and Hx by partially purified poly-histidine-Aag fusion protein (see Materials and Methods). 5′ 32P-labeled double-stranded oligonucleotides containing site-specific ɛA or Hx were incubated with either polyhistidine-tagged Aag (lanes 2 and 4) or with negative control extracts from E. coli not expressing the Aag cDNA protein (see Materials and Methods) (lanes 1 and 3). After 1 hr incubation at 37°C to allow release of aberrant bases by Aag glycosylase, oligonucleotides were chemically cleaved at the abasic sites. Fragments were separated by 20% denaturing PAGE. Release of Hx or ɛA is indicated by the appearance of either a 14-mer or a 12-mer, respectively. (B) Liver: time-dependent release of Hx (Upper) and ɛA (Lower) for liver extract proteins from Aag +/+ mice (136 μg) and Aag −/− mice (142 μg). The BSA control or extract proteins were incubated at 37°C for the indicated times, chemically cleaved, and analyzed as described above. (C) Testes: 100 μg of extract proteins, prepared from the testes of two Aag −/− and two Aag +/+ mice were incubated for 5 hr with Hx and ɛA oligonucleotide substrates and analyzed as described above. (D) Kidney: 131 μg of extract proteins, prepared from the kidneys of two Aag −/− and two Aag +/+ mice, were incubated for 6 hr with Hx and ɛA containing oligonucleotide substrates and analyzed as described above. (E) Lung: 95 μg of extract proteins, prepared from the lungs of two Aag −/− and Aag +/+ mice, were incubated for 6 hr with Hx and ɛA oligonucleotide substrates and analyzed as described above.
By using Aag +/+ liver extracts, assay conditions were optimized for ɛA and Hx DNA glycosylase activity while suppressing nonspecific DNA degradation (Fig. 4B). Whereas Aag +/+ liver extracts display robust ɛA and Hx DNA glycosylase activity that increases with increasing incubation time, Aag −/− liver extracts are completely devoid of these activities. The same liver extracts show apparently equal uracil DNA glycosylase activity (Fig. 2B). Similar results were observed for liver extracts from two other mice (Aag −/− and Aag +/+; data not shown). We infer that Aag is the major ɛA and Hx DNA glycosylase in mouse liver. It should be noted that the ɛA lesion has been shown to degrade at a slow rate during storage to form 4-amino-5-(imidazol-2-yl)imidazole (β), which is even more mutagenic and toxic than ɛA itself (at least in E. coli) (43). Such degradation also is likely to occur in vivo. It is possible that in the results shown here, a small fraction of the observed cleavage may be because of Aag mediated excision of the highly mutagenic β adduct; this possibility is currently under investigation.
Although Aag appears to be the major ɛA and Hx DNA glycosylase in mouse liver, it was possible that other activities could be revealed in other differentiated tissues. Testes, kidney, and lung tissues from two Aag +/+ and two Aag −/− mice were examined for activity on each substrate. Fig. 4 C–E shows that all three Aag +/+ tissues display both Hx and ɛA DNA glycosylase activity. However, Aag −/− testes, kidney, and lung do not express measurable levels of Hx DNA glycosylase activity, once again implicating Aag as the major DNA glycosylase for repair of the mutagenic Hx lesion. In contrast, the ɛA DNA glycosylase assays displayed a more complex picture. Whereas Aag −/− testes and kidney do not express measurable ɛA DNA glycosylase, there was some detectable ɛA DNA glycosylase activity in Aag −/− lung (Fig. 4 C–E). However, because Aag represents all of the measurable ɛA DNA glycosylase activity in liver, kidney, and testes, and more than 50% of that in lung (as judged by densitometry, data not shown), we conclude that Aag represents the major ɛA DNA glycosylase activity in all four tissues.
DISCUSSION
The Aag null mice described here are viable animals specifically mutant in a BER enzyme, albeit in just one of at least six mammalian DNA glycosylases that can initiate BER. Attempts to generate mice that are wholly deficient in BER (by inactivating enzymes acting downstream of DNA glycosylases) revealed that the major mammalian AP-endonuclease gene (APE/APEX/REF-1/HAP1) and a major DNA repair polymerase gene (β-pol) are essential for normal mouse development (44, 45). Because all six or more DNA glycosylases presumably continue to remove aberrant bases in these animals, one interpretation of these findings is that the accumulation of abasic sites (in the case of APE homozygous mutants) or nucleotide gaps (in the case of β-pol homozygous mutants) is toxic (of course there are other possible explanations for this embryonic lethality). Our objective is to investigate the biological consequences of DNA base lesions in the genome, and this goal may be accomplished by disrupting the enzymes that initiate BER. We therefore generated Aag homozygous mutant mice.
Here we show that, like its human and rat homologues, the mouse Aag 3MeA DNA glycosylase is proficient in releasing ɛA and Hx in addition to 3MeA. Furthermore, biochemical characterization of the Aag null mice identify Aag as a major DNA glycosylase for repair of 3MeA, Hx, and ɛA DNA lesions in several major tissues. Aag is not, however, the major DNA glycosylase for removal of all of the substrates identified in biochemical assays in vitro, because the Aag −/− testes extracts maintain wild-type levels of 8oxoG DNA glycosylase activity. Because Aag homozygous mutant mice are viable, we infer that: (i) the spontaneous DNA lesions repaired by Aag are not lethal when left unrepaired; or (ii) other DNA repair enzyme(s) can take over the repair of potentially lethal lesions normally repaired by Aag. As a first step toward revealing the biological consequences of an Aag deficiency in mammals, we have shown that primary fibroblasts derived from Aag −/− embryos are more sensitive to the toxic effects of 3MeA lesions than their wild-type counterparts. We therefore anticipate that the phenotype of Aag −/− animals likewise will include sensitivity to DNA alkylating agents.
The importance of removing damaged DNA bases from the genome of mammals has been amply demonstrated by the observation that mice and people specifically deficient in nucleotide excision repair have increased susceptibility to UV-induced tumorigenesis (46–50). The precise biological effects of the lesions repaired by Aag in animals are not yet known. However, we recently demonstrated that 3MeA DNA lesions specifically induce sister chromatid exchanges and chromosome aberrations (unpublished data), and strong evidence implicates ɛA and Hx as miscoding lesions (13, 16). It therefore seems likely that Aag may not only protect animals against toxicity, but also may limit mutation and, thus, the appearance of cancers.
3MeA, ɛA, and Hx are endogenously produced DNA lesions. The cellular metabolite S-adenosyl-l-methionine, used for methyl transfer to numerous cellular constituents, inadvertently may alkylate DNA (17, 18). Lipid peroxidation products, like 2,3-epoxy-4-hydroxynonanal, react with DNA to form ɛA (51, 52), and the formation of Hx occurs by spontaneous deamination of adenine and by exposure to NO⋅ (produced by activated macrophages) (19, 53). 3MeA, ɛA, and Hx also are induced by exposure to certain carcinogens in our environment (23, 24, 54, 55). The Aag −/− animals are not yet old enough to determine if Aag has an influence on spontaneous cancer rates, and moreover, the Aag null allele must be backcrossed at least 10 generations to produce the appropriately inbred genetic background for an accurate comparison of the relative sensitivities of Aag −/−, Aag +/− and Aag +/+ mice to genotoxic and carcinogenic agents. However, the striking biochemical phenotype of the Aag −/− genotype reported here suggests that Aag is likely to be an important contributor to genomic stability.
The mammalian 3MeA DNA glycosylases release a wide variety of damaged DNA bases in vitro, but the in vivo repair of these lesions may or may not be Aag-dependent. Because the in vitro assays used in this study are specific for the activity of DNA glycosylases, studies of the in vivo rate of removal of damaged bases will establish if the Aag substrates can be acted on by other repair systems. Although nucleotide excision repair (NER) removes a broad spectrum of DNA lesions and mismatch repair (MMR) can act on at least one methylated base (O6 MeG), various studies indicate that neither NER nor MMR play a predominant role in 3MeA repair (56–58). However, whether or not the mammalian NER and MMR pathways can repair Hx and ɛA lesions has not yet been addressed, and the possibility that these pathways play a major role in their repair cannot be eliminated. Monitoring the in vivo rate of removal of all three lesions in Aag null mice, and in these mice carrying additional defects in NER or MMR, will help to reveal the relative importance of the three repair systems. The appropriate crosses of Aag −/−, XPA −/−, and Msh2 −/− mice currently are underway.
In summary, the viable and fertile Aag null mice described here will allow us to explore the role of this repair enzyme in protecting animals against endogenously produced damaged DNA bases and base adducts created by cancer chemotherapeutics or by toxic agents in our food and in our environment. Given the broad range of substrates repaired by mammalian 3MeA DNA glycosylases, the biological role of Aag may be multifaceted and may vary among cell types. Further, by crossing Aag null mice with other genetically engineered mouse strains, we will be able to study the interplay between Aag initiated BER and other DNA damage response pathways, such as those involved in triggering cell cycle arrest and apoptosis.
Acknowledgments
We thank Drs. Orlando D. Scharer and Gregory L. Verdine of the Department of Chemistry at Harvard University for the kind gift of oligonucleotides containing ɛA, and Drs. John Cairns and John Essigmann for invaluable comments. This work was supported by National Institutes of Health Grants R01 CA55042 and P01-ES03926, and by grants from the Dutch Cancer Society (projects EUR 90–20, 94-793) and a Human Frontiers Science Programme. The animals used in this study were cared for according to Institutional guidelines. The research of G.W. has been made possible by a fellowship of the Royal Netherlands Academy of Arts and Sciences. M.W. was supported by a National Institutes of Health Radiation Biology Training Grant 5T32CA09078 and National Institutes of Health Individual Fellowship Award CA73135-01. B.E. was supported by a Pharmaceutical Manufacturers Association Foundation Advanced Predoctoral Fellowship in Pharmacology/Toxicology, a Graduate Student Research Fellowship Award from the Society of Toxicology sponsored by Hoffmann-LaRoche, Inc., and a National Institutes of Health Training Program in Environmental Health Sciences 5T32ES07155. J.A. is a Leukemia Society of America Fellow. L.S. was supported by a Burroughs Wellcome Toxicology Scholar Award.
Note added in proof
Recently, viable Aag −/− mice that appear to be similar to those described here have been generated independently by another group (R. H. Elder and G. P. Margison, personal communication).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: Aag, alkyladenine DNA glycosylase; BER, base excision repair; 3MeA, 3-methyladenine; Hx, hypoxanthine; ɛA, 1,N6-ethenoadenine; 8oxoG, 8-oxoguanine; ES, embryonic stem; MEFs, mouse embryo fibroblasts; Me-Lex, MeOSO2(CH2)2-lexitropsin.
A commentary on this article begins on page 12754.
References
- 1.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 2.Lindahl T. Nature (London) 1976;259:64–66. doi: 10.1038/259064a0. [DOI] [PubMed] [Google Scholar]
- 3.Chakravarti D, Ibeanu G C, Tano K, Mitra S. J Biol Chem. 1991;266:15710–15715. [PubMed] [Google Scholar]
- 4.O’Connor T R, Laval F. EMBO J. 1990;9:3337–3342. doi: 10.1002/j.1460-2075.1990.tb07534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Connor T R, Laval J. Biochem Biophys Res Commun. 1991;176:1170–1177. doi: 10.1016/0006-291x(91)90408-y. [DOI] [PubMed] [Google Scholar]
- 6.Samson L, Derfler B, Boosalis M, Call K. Proc Natl Acad Sci USA. 1991;88:9127–9131. doi: 10.1073/pnas.88.20.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saparbaev M, Laval J. Proc Natl Acad Sci USA. 1994;91:5873–5877. doi: 10.1073/pnas.91.13.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bessho T, Roy R, Yamamoto K, Kasai H, Nishimura S, Tano K, Mitra S. Proc Natl Acad Sci USA. 1993;90:8901–8904. doi: 10.1073/pnas.90.19.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dosanjh M K, Chenna A, Kin E, Fraenkel-Conrat H, Samson L, Singer B. Proc Natl Acad Sci USA. 1994;91:1024–1028. doi: 10.1073/pnas.91.3.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saparbaev M, Kleibl K, Laval J. Nucleic Acids Res. 1995;23:3750–3755. doi: 10.1093/nar/23.18.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engelward B P, Boosalis M S, Chen B J, Deng Z, Siciliano M J, Samson L D. Carcinogenesis. 1993;14:175–181. doi: 10.1093/carcin/14.2.175. [DOI] [PubMed] [Google Scholar]
- 12.Engelward B, Dreslin A, Christensen J, Huszar D, Kurahara C, Samson L. EMBO J. 1996;15:945–952. [PMC free article] [PubMed] [Google Scholar]
- 13.Kamiya H, Miura H, Kato H, Nishimura S, Ohtsuka E. Cancer Res. 1992;52:1836–1839. [PubMed] [Google Scholar]
- 14.Moriya M. Proc Natl Acad Sci USA. 1993;90:1122–1126. doi: 10.1073/pnas.90.3.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuda T, Yagi T, Kawanishi M, Matsui S, Takebe H. Carcinogenesis. 1995;16:2389–2394. doi: 10.1093/carcin/16.10.2389. [DOI] [PubMed] [Google Scholar]
- 16.Pandya G A, Moriya M. Biochemistry. 1996;35:11487–11492. doi: 10.1021/bi960170h. [DOI] [PubMed] [Google Scholar]
- 17.Barrows L R, Magee P N. Carcinogenesis. 1982;3:349–351. doi: 10.1093/carcin/3.3.349. [DOI] [PubMed] [Google Scholar]
- 18.Rydberg B, Lindahl T. EMBO J. 1982;1:211–216. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karran P, Lindahl T. Biochemistry. 1980;19:6005–6011. doi: 10.1021/bi00567a010. [DOI] [PubMed] [Google Scholar]
- 20.Nair J, Barbin A, Guichard Y, Bartsch H. Carcinogenesis. 1995;16:613–617. doi: 10.1093/carcin/16.3.613. [DOI] [PubMed] [Google Scholar]
- 21.Loft S, Poulsen H E. J Mol Med. 1996;74:297–312. doi: 10.1007/BF00207507. [DOI] [PubMed] [Google Scholar]
- 22.Leanderson P, Tagesson C. Chem Biol Interact. 1992;81:197–208. doi: 10.1016/0009-2797(92)90034-i. [DOI] [PubMed] [Google Scholar]
- 23.Spencer J P, Jenner A, Chimel K, Aruoma O I, Cross C E, Wu R, Halliwell B. FEBS Lett. 1995;375:179–182. doi: 10.1016/0014-5793(95)01199-o. [DOI] [PubMed] [Google Scholar]
- 24.Swenberg J A, Fedtke N, Ciroussel F, Barbin A, Bartsch H. Carcinogenesis. 1992;13:727–729. doi: 10.1093/carcin/13.4.727. [DOI] [PubMed] [Google Scholar]
- 25.Smith A G, Hooper M L. Dev Biol. 1987;121:1–9. doi: 10.1016/0012-1606(87)90132-1. [DOI] [PubMed] [Google Scholar]
- 26.Laird P W, Zijderveld A, Linders K, Rudnicki M A, Jaenisch R, Berns A. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen S, Evans G A. In: Methods in Molecular Biology. White B A, editor. Vol. 15. Totowa, NJ: Humana; 1993. pp. 75–80. [DOI] [PubMed] [Google Scholar]
- 28.Samson L, Linn S. Carcinogenesis. 1987;8:227–230. doi: 10.1093/carcin/8.2.227. [DOI] [PubMed] [Google Scholar]
- 29.Chen J, Derfler B, Maskati A, Samson L. Proc Natl Acad Sci USA. 1989;86:7961–7965. doi: 10.1073/pnas.86.20.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freshney R. Culture of Animal Cells: A Manual of Basic Techniques. New York: Wiley–Liss; 1994. [Google Scholar]
- 31.Scharer O D, Verdine G L. J Am Chem Soc. 1995;117:10781–10782. [Google Scholar]
- 32.Singhal R K, Prasad R, Wilson S H. J Biol Chem. 1995;270:949–957. doi: 10.1074/jbc.270.2.949. [DOI] [PubMed] [Google Scholar]
- 33.Chen J, Derfler B, Samson L. EMBO J. 1990;9:4569–4575. doi: 10.1002/j.1460-2075.1990.tb07910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Washington W J, Dunn W C, Jr, Generoso W M, Mitra S. Mutat Res. 1988;207:165–169. doi: 10.1016/0165-7992(88)90082-6. [DOI] [PubMed] [Google Scholar]
- 35.Roldan-Arjona T, Wei Y F, Carter K C, Klungland A, Anselmino C, Wang R P, Augustus M, Lindahl T. Proc Natl Acad Sci USA. 1997;94:8016–8020. doi: 10.1073/pnas.94.15.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arai K, Morishita K, Shinmura K, Kohno T, Kim S R, Nohmi T, Taniwaki M, Ohwada S, Yokota J. Oncogene. 1997;14:2857–2861. doi: 10.1038/sj.onc.1201139. [DOI] [PubMed] [Google Scholar]
- 37.Aburatani H, Hippo Y, Ishida T, Takashima R, Matsuba C, Kodama T, Takao M, Yasui A, Yamamoto K, Asano M. Cancer Res. 1997;57:2151–2156. [PubMed] [Google Scholar]
- 38.Radicella J P, Dherin C, Desmaze C, Fox M S, Boiteux S. Proc Natl Acad Sci USA. 1997;94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenquist T A, Zharkov D O, Grollman A P. Proc Natl Acad Sci USA. 1997;94:7429–7434. doi: 10.1073/pnas.94.14.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Chen F-X, Mehta P, Gold B. Biochemistry. 1993;32:7954–7965. doi: 10.1021/bi00082a017. [DOI] [PubMed] [Google Scholar]
- 41.Encell L, Shuker D E G, Foiles P G, Gold B. Chem Res Toxicol. 1996;9:563–567. doi: 10.1021/tx9501849. [DOI] [PubMed] [Google Scholar]
- 42.Singer B, Antoccia A, Basu A K, Dosanjh M K, Fraenkel-Conrat H, Gallagher P E, Kusmierek J T, Qiu Z H, Rydberg B. Proc Natl Acad Sci USA. 1992;89:9386–9390. doi: 10.1073/pnas.89.20.9386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Basu A K, Wood M L, Niedernhofer L J, Ramos L A, Essigmann J M. Biochemistry. 1993;32:12793–12801. doi: 10.1021/bi00210a031. [DOI] [PubMed] [Google Scholar]
- 44.Gu H, Marth J D, Orban P C, Mossmann H, Rajewsky K. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 45.Xanthoudakis S, Smeyne R J, Wallace J D, Curran T. Proc Natl Acad Sci USA. 1996;93:8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Vries A, van Oostrom C T, Hofhuis F M, Dortant P M, Berg R J, de Gruijl F R, Wester P W, van Kreijl C F, Capel P J, van Steeg H, Verbeek S J. Nature (London) 1995;377:169–173. doi: 10.1038/377169a0. [DOI] [PubMed] [Google Scholar]
- 47.Nakane H, Takeuchi S, Yuba S, Saijo M, Nakatsu Y, Murai H, Nakatsuru Y, Ishikawa T, Hirota S, Kitamura Y, Kato Y, Tsunoda Y, Miyauchi H, Horio T, Tokunaga T, Matsunaga T, Nikaido O, Nishimune Y, Okada Y, Tanaka K. Nature (London) 1995;377:165–168. doi: 10.1038/377165a0. [DOI] [PubMed] [Google Scholar]
- 48.Sands A T, Abuin A, Sanchez A, Conti C J, Bradley A. Nature (London) 1995;377:162–165. doi: 10.1038/377162a0. [DOI] [PubMed] [Google Scholar]
- 49.Van der Horst G T J, van Steeg H, Berg R J W, van Gool A J, de Wit J, Weeda G, Morreau H, Beems R B, van Kreijl C F, de Gruijl F R, Bootsma D, Hoeijmakers J H J. Cell. 1997;89:425–435. doi: 10.1016/s0092-8674(00)80223-8. [DOI] [PubMed] [Google Scholar]
- 50.Bootsma D, Cleaver J E, Kraemer K H, Hoeijmakers J H J. In: The Metabolic Basis of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Vall D, editors. New York: McGraw-Hill; 1997. , in press. [Google Scholar]
- 51.El Ghissassi F, Barbin A, Nair J, Bartsch H. Chem Res Toxicol. 1995;8:278–283. doi: 10.1021/tx00044a013. [DOI] [PubMed] [Google Scholar]
- 52.Chung F L, Chen H J, Nath R G. Carcinogenesis. 1996;17:2105–2111. doi: 10.1093/carcin/17.10.2105. [DOI] [PubMed] [Google Scholar]
- 53.Nguyen T, Brunson D, Crespi C L, Penman B W, Wishnok J S, Tannenbaum S R. Proc Natl Acad Sci USA. 1992;89:3030–3034. doi: 10.1073/pnas.89.7.3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stillwell W G, Glogowski J, Xu H-X, Wishnok J S, Zavala D, Montes G, Correa P, Tannenbaum S R. Cancer Res. 1991;51:190–194. [PubMed] [Google Scholar]
- 55.Fernando R C, Nair J, Barbin A, Miller J A, Bartsch H. Carcinogenesis. 1996;17:1711–1718. doi: 10.1093/carcin/17.8.1711. [DOI] [PubMed] [Google Scholar]
- 56.Cleaver J E. Mutat Res. 1971;12:453–462. doi: 10.1016/0027-5107(71)90095-9. [DOI] [PubMed] [Google Scholar]
- 57.Altamirano-Dimas M, Sklar R, Strauss B. Mutat Res. 1979;60:197–206. doi: 10.1016/0027-5107(79)90184-2. [DOI] [PubMed] [Google Scholar]
- 58.Ceccotti S, Aquilina G, Macpherson P, Yamada M, Karran P, Bignami M. Curr Biol. 1996;6:1528–1531. doi: 10.1016/s0960-9822(96)00758-0. [DOI] [PubMed] [Google Scholar]
- 59.Lu R, Nash H M, Verdine G L. Curr Biol. 1997;7:397–407. doi: 10.1016/s0960-9822(06)00187-4. [DOI] [PubMed] [Google Scholar]