Abstract
Ribosome-associated chaperone Trigger Factor (TF) initiates folding of newly synthesized proteins in bacteria. Here, we pinpoint by site-specific crosslinking the sequence of molecular interactions of Escherichia coli TF and nascent chains during translation. Furthermore, we provide the first full-length structure of TF associated with ribosome–nascent chain complexes by using cryo-electron microscopy. In its active state, TF arches over the ribosomal exit tunnel accepting nascent chains in a protective void. The growing nascent chain initially follows a predefined path through the entire interior of TF in an unfolded conformation, and even after folding into a domain it remains accommodated inside the protective cavity of ribosome-bound TF. The adaptability to accept nascent chains of different length and folding states may explain how TF is able to assist co-translational folding of all kinds of nascent polypeptides during ongoing synthesis. Moreover, we suggest a model of how TF's chaperoning function can be coordinated with the co-translational processing and membrane targeting of nascent polypeptides by other ribosome-associated factors.
Keywords: chaperone, nascent chains, protein folding, ribosome, Trigger Factor
Introduction
The folding of newly synthesized proteins critically depends on a network of molecular chaperones that acts already during protein synthesis. In bacteria, the initial folding steps are assisted by Trigger Factor (TF), which associates with ribosomes and interacts with virtually all emerging nascent chains. Downstream of ribosome-associated TF, the ATP-dependent DnaK and GroEL chaperone systems continue the de novo folding of proteins (Bukau et al, 2000; Frydman, 2001; Hartl and Hayer-Hartl, 2002). TF is not essential, but the loss of TF in cells lacking the cooperating DnaK chaperone results in synthetic lethality at temperatures ⩾30°C and causes misfolding and aggregation of several hundred different newly synthesized proteins (Deuerling et al, 1999; Teter et al, 1999).
TF consists of three domains and folds into an extended three-dimensional structure (Ferbitz et al, 2004) (Figure 2A). The N domain of TF harbours a ‘signature motif' (43-GFRxGxxP-50) in an exposed loop, which mediates binding to the ribosomal tunnel exit protein L23 (Kramer et al, 2002). The N domain is connected by means of a long linker to the second domain (peptidyl-prolyl cis/trans isomerase (PPIase) domain), which is located at the opposite end of the molecule and displays PPIase activity (Hesterkamp and Bukau, 1996; Stoller et al, 1996), whose relevance in vivo is still unclear (Kramer et al, 2004a). The third domain is built by the C-terminal region, which forms the body of TF with two protruding arms and was shown to constitute the main module for its chaperone activity (Merz et al, 2006).
Figure 2.
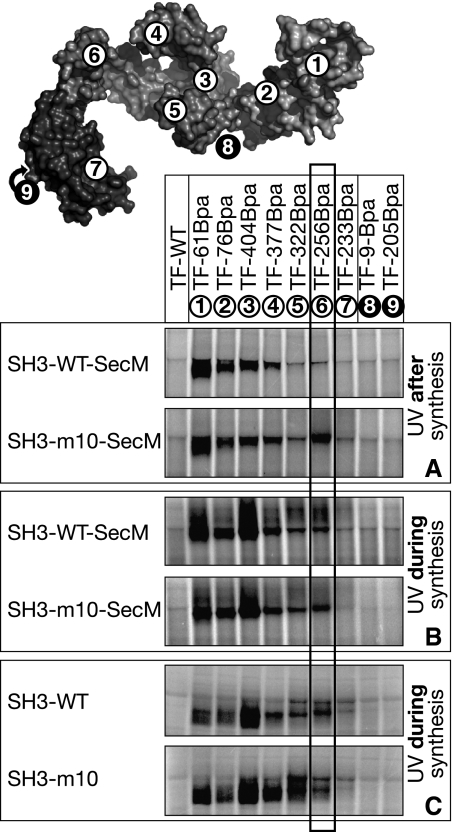
Site-specific crosslinking of TF-Bpa variants to arrested nascent ICDH. (A) Structural model of ribosome-associated TF (Ferbitz et al, 2004). The ribosome (grey) is cut in half to visualize the ribosomal tunnel with a modelled nascent chain (magenta). TF (blue) is shown in ribbon representation docked to the ribosomal protein L23 (green) and in surface representation rotated by 90° to allow one to view the interior. Positions of UV-activatable Bpa crosslinker are indicated in green; red numbers label Bpa positions on the interior of TF; exterior positions are labelled with white numbers. (B) SecM-arrested nascent ICDH polypeptides of varying length were synthesized in vitro in the presence of TF variants. After translation, UV light was applied to crosslink TF-Bpa variants to nascent chains. Crosslinks were detected by SDS–PAGE and autoradiography. Only crosslinking products are depicted. ICDH-16-SecM (with a total length of 47 aa including the SecM linker, marked by a star) represents the minimal length of a nascent chain required for site-specific TF crosslinking.
To date, structures of three different complexes between an N-terminal fragment of TF and a large ribosomal subunit have been determined. The ribosome-binding domain of Escherichia coli TF was co-crystallized with the 50S ribosomal subunit from Haloarcula marismortui, and two different structures of homologous complexes of Deinococcus radiodurans were obtained (Ferbitz et al, 2004; Baram et al, 2005; Schlunzen et al, 2005). Superposition of full-length TF onto the co-crystallized N-terminal fragments demonstrated that TF binds to L23 and hunches over the ribosomal exit site orienting its hydrophobic inner surface towards the emerging nascent polypeptides. The models, however, differ in size and shape of the predicted space between TF and the ribosome. Previously, we suggested that TF forms a cradle-like structure that provides a protective environment supportive for co-translational folding of small domains (Ferbitz et al, 2004). On the other hand, it was proposed that the N domain of TF is located much closer to the ribosomal surface and that a loop of the ribosomal protein L24 restricts the space between TF and the ribosome. Therefore, the N domain of TF would rather form a narrow crevice, which would only allow binding of unfolded extended stretches of nascent chains (Schlunzen et al, 2005).
Insights into the molecular mechanism of action of TF have been provided by several recent studies. Based on activity measurements of newly synthesized model enzymes, it was suggested that TF in cooperation with DnaK delays folding of newly synthesized multidomain proteins to increase the folding yield (Agashe et al, 2004). On the other hand, TF was shown to shield nascent chains of the size of protein domains and larger from proteolysis, suggesting that ribosome-bound TF provides a protective environment by accommodation of large portions of nascent polypeptides (Hoffmann et al, 2006; Tomic et al, 2006). A recent study showed that arrested luciferase chains of 60 and 164 amino acids (aa) contact all three domains of TF, whereas an arrested luciferase chain of intermediate length (77 aa) interacts only with the N and C domains of TF (Lakshmipathy et al, 2007). Generally, the chaperone activity of TF was described as a dynamic cycle of ribosome association and dissociation as well as nascent polypeptide binding and release (Bukau et al, 2000; Kaiser et al, 2006). The affinity of TF to ribosome–nascent chain complexes (RNCs) was shown to increase with length and hydrophobicity of the nascent polypeptide (Kaiser et al, 2006; Raine et al, 2006; Rutkowska et al, 2008).
Despite extensive research on TF, two critical aspects of the mechanism of TF action, namely the sequence of interactions between TF and the emerging nascent chain, and the conformation of the chaperone on the translating ribosome, are not yet clear. Here, we address these questions: first, by a site-specific crosslinking approach, we analyse the interactions between TF and nascent polypeptides by monitoring the very initial contacts upon emergence of the polypeptides at the ribosomal tunnel exit and following the interactions during polypeptide elongation and folding; second, we use the information from the crosslinking analysis to generate stable homogenous TF–RNCs and determine the positioning and topology of full-length TF bound to a translating 70S ribosome by cryo-electron microscopy (Cryo-EM).
Results
Ribosome-bound TF interacts with various lengths of nascent polypeptides
To characterize the interaction of TF with growing nascent polypeptides, we generated ribosome-arrested nascent chain constructs, which gradually increased in length and thus provided snapshots of the ongoing protein synthesis (Figure 1). The nascent polypeptides were derived from the natural TF substrate isocitrate dehydrogenase (ICDH) and were shown to exist in rather unfolded conformations (Hoffmann et al, 2006). In frame to the ICDH fragments, we fused the SecM peptide (31 aa) (Nakatogawa and Ito, 2002), which stalls translation and spans the ribosomal tunnel to expose the nascent ICDH portion outside the ribosome. By using an in vitro transcription/translation system derived from E. coli cells lacking TF, we generated 35S-methionine-labelled SecM-arrested nascent ICDH fragments. Purified wild-type (wt) TF was added before translation and interaction of TF with nascent chains was monitored after synthesis by adding the chemical crosslinker disuccinimidyl suberate (DSS) (Supplementary Figure S1A). Crosslinking adducts were analysed by SDS–PAGE and autoradiography. On exposure of nine N-terminal residues of ICDH (ICDH-9-SecM, total length 40 aa) at the ribosomal surface, a crosslink to TF was observed (Figure 1), which persisted for all longer nascent ICDH constructs. The longest nascent chain (ICDH-146-SecM, total length 177 aa) gave rise to multiple crosslinking products with TF, which may reflect crosslinking at different positions within the nascent polypeptide. By using L23-VSE/AAA-mutant ribosomes and mutant TF-FRK/AAA (Kramer et al, 2002), which are mutated in residues responsible for TF–ribosome binding, no DSS crosslink could be observed (Supplementary Figure S2A). Thus, the interaction of TF with nascent ICDH, irrespective of the length, depends on the association of TF with the ribosome.
Figure 1.
Chemical crosslinking of TF to arrested nascent ICDH polypeptides of varying length. SecM-arrested 35S-labelled nascent polypeptides were synthesized in vitro exposing different lengths of ICDH polypeptides outside the ribosomal tunnel (4–146 aa, indicated by a filled circle). The interaction of TF with these nascent chains was probed by chemical crosslinking with DSS. Crosslinking products (indicated by an asterisk) were visualized by SDS–PAGE and autoradiography.
Emerging nascent polypeptides traverse the interior of TF in a sequential and length-dependent manner
The visualization of interactions by chemical DSS crosslinking does not provide information about the interaction sites of a nascent chain within TF nor about whether different binding sites are used depending on the length of the nascent chain. To precisely follow a growing nascent polypeptide upon its exit from the ribosomal tunnel, we generated TF variants (TF-Bpa) that carry an incorporated modified amino acid (p-benzoyl-phenylalanine (Bpa)) (Ryu and Schultz, 2006) at one particular position. Following UV exposure, Bpa acts as a zero-space crosslinker and can covalently link TF to nascent polypeptides in the immediate vicinity. We positioned Bpa crosslinker on the entire interior as well as on the exterior of the TF cavity according to the three-dimensional structure (Figure 2A) (Ferbitz et al, 2004). All TF-Bpa variants showed wt-like activity in refolding of denatured glyceraldehyde-3-phosphate dehydrogenase in vitro (Supplementary Figure S2B), which demonstrates their functionality (Merz et al, 2006). The TF-Bpa mutants were added to the in vitro translation reaction before translation of ribosome-arrested ICDH constructs. After synthesis, the TF-Bpa variants were activated by UV light for crosslinking (Supplementary Figure S1A).
The first site-specific crosslink could be detected between ICDH-16-SecM exposing 16 aa of ICDH and TF-61Bpa having the crosslinker positioned in the N domain of TF very close to the tunnel exit (Figure 2). In contrast, the addition of DSS resulted in crosslinking adducts of TF-Bpa variants and ICDH-13-SecM, indicating that all TF-Bpa mutants efficiently associated with RNCs containing ICDH-13-SecM (data not shown). Notably, DSS includes a 10 Å spacer whereas Bpa is a zero-space crosslinker. Thus, nascent chains of such length are too short to contact any of the site-specific Bpa-crosslinker positions within TF.
On exposure of 32 aa of ICDH (ICDH-32-SecM), additional crosslinking appeared with TF-76Bpa harbouring the crosslinker in the N domain more distant from the tunnel exit than TF-61Bpa. Thus, emerging nascent polypeptides initially contact the N domain of TF before any other TF region. A further extension of nascent ICDH by more than 5 aa (ICDH-37-SecM, ICDH-40-SecM) resulted in crosslinks of TF-404Bpa, which has the crosslinker positioned at the bottom of the internal cavity between the two C-terminal arms. Furthermore, on exposing nascent ICDH of 40 aa and longer, we detected crosslinking to the tips of the TF arms (first TF-377Bpa, then TF-322Bpa), as well as to the region of the C-terminal domain that is located close to the PPIase domain (TF-256Bpa), indicating that the nascent polypeptide traverses further through the TF arms by using the entire C domain. Finally, the exposed nascent ICDH chains of a length of 59 residues or longer (ICDH-59-SecM, ICDH-79-SecM, ICDH-146-SecM) crosslinked strongly to all positions within TF, including the inside of the PPIase domain (TF-233Bpa). Importantly, neither wt TF nor the two TF variants with a crosslinker positioned at the exterior (N domain: TF-9Bpa; PPIase domain: TF-205Bpa) gave rise to significant crosslinking products with nascent ICDH (Figure 2B).
We conclude that the growing nascent ICDH chain contacts the entire interior of TF in a sequential manner, first by contacting the N terminus and then passing through the arms engaging the entire C domain to finally progress towards and into the PPIase domain.
Nascent ICDH engages the complete interior of TF during ongoing translation
To exclude possible artificial effects of ribosome stalling and to monitor TF interactions under physiological conditions without compromising the dynamics of protein synthesis, we synthesized full-length ICDH in vitro without translation arrest in the presence of the TF-Bpa constructs and applied UV light continuously during synthesis (Supplementary Figure S1C). Thereby, the Bpa crosslinker is activated for immediate co-translational crosslinking upon first contact with a nascent peptide. Upon termination of translation, the crosslinked complex of TF and full-length ICDH is released from the ribosome. After the synthesis reaction, the translation mixture was subjected to ultracentrifugation to separate released crosslinking products of TF and full-length ICDH (supernatant) from ribosome-bound complexes of TF crosslinked to incompletely translated nascent ICDH fragments (pellet).
Analysis of the supernatant fractions revealed that all TF-Bpa variants with Bpa in the interior of TF crosslinked to nascent ICDH, whereas Bpa positioned at the exterior of TF did not crosslink (Figure 3A). In a control experiment, UV light was applied to the supernatant fraction to verify that none of the TF-Bpa variants crosslinked post-translationally to soluble ICDH (Figure 3C). Analysis of the pellet fractions of co-translationally irradiated translation reactions revealed crosslinks between incompletely synthesized nascent ICDH fragments and TF-Bpa variants. Again, crosslinking to ICDH occurred exclusively within the interior of TF (Figure 3B). The crosslinking products appeared as a smear, reflecting the crosslinking of TF to all lengths of ICDH at different stages of elongation. Interestingly, the size spectrum of this crosslinking smear differed for different TF-Bpa variants. The closer a crosslinker within TF was positioned towards the ribosomal tunnel, the more smaller-sized crosslinking adducts appeared, suggesting that more-distant positions within TF need longer nascent ICDH chains for co-translational interaction. This is in agreement with the results obtained using arrested nascent ICDH chains (Figure 2B). Importantly, similar crosslinking patterns were obtained translating full-length E. coli methionine adenosyltransferase (MetK) or E. coli outer membrane proteins A (OmpA) (Supplementary Figure S2C). We conclude that during ongoing translation, an emerging nascent chain traverses the entire interior of TF in a length-dependent manner.
Figure 3.
Interactions of non-arrested ICDH with TF during ongoing translation. TF-Bpa variants were analysed for crosslinking to non-arrested ICDH during ongoing translation by applying UV light during synthesis. The released full-length ICDH (A, supernatant) was separated from ribosome-associated incompletely synthesized ICDH nascent polypeptides (B, pellet) by ultracentrifugation. TF–ICDH crosslinking products are indicated by asterisks. As control, UV irradiation was applied after synthesis to the supernatant fraction (C).
TF accommodates folded and unfolded domains
Next, we addressed the question of whether the ability of a nascent polypeptide to fold into a native-like structure influences the interactions with TF. To this end, we used the folded SH3 domain of α-spectrin (62 aa) and the mutated variant SH3-m10 (62 aa), which harbours two point mutations that result in random coil conformation of the protein (Blanco et al, 1999; Hoffmann et al, 2006). Both variants were fused by means of a short linker (11 aa) to the SecM arrest sequence. In these SecM-stalled constructs, the nascent SH3 domain (SH3-wt-SecM) adopts a compactly folded, protease-resistant conformation on the ribosomal surface, whereas the random coil SH3-m10 (SH3-m10-SecM) remains unfolded and protease-sensitive as demonstrated earlier (Hoffmann et al, 2006).
By using the UV-activatable TF-Bpa variants (Supplementary Figure S1A), we found that the crosslinking pattern of the unfolded SH3-m10-SecM (Figure 4A) was comparable to that of nascent ICDH of similar length (ICDH-59-SecM; Figure 2B). The reduced crosslinking efficiency of positions 322 (TF-322Bpa) and 233 (TF-233Bpa) might be due to conformational differences between SH3-m10 and ICDH nascent polypeptides. Similar to nascent SH3-m10-SecM, the folded SH3-wt-SecM (Figure 4A) revealed strong crosslinking adducts to the interior of TF at positions 61 and 76 in the N domain and positions 404 and 377 between and within the C-terminal arms, but in contrast no crosslinking to position 256 in the area beyond the C-terminal arms was observed. The absence of the crosslink between the folded SH3-wt domain and TF-256Bpa had been reproducibly observed in independent experiments. These results suggest that the SH3-wt domain (7 kDa), which is capable of folding into a compact structure, resides predominantly within the region of the N domain and the C-terminal arms of TF. The results furthermore suggest that the conformation of the nascent polypeptide may alter the interaction with TF and that TF can accommodate substrates of different folding states in its interior including small folded domains such as SH3.
Figure 4.
Localization of nascent chains with different folding states within TF. The folding-competent SH3-wt domain and the unfolded mutant SH3-m10 (see text) were synthesized in vitro and analysed for their interactions with TF-Bpa variants. (A, B) The SH3 variants were stalled by SecM arrest (SH3-wt-SecM, SH3-m10-SecM) and UV irradiation was applied (A) after synthesis or (B) during ongoing synthesis. (C) Non-arrested SH3 and SH3-m10 were generated, UV light was applied during ongoing synthesis and the ribosome-dissociated crosslinking products of TF-Bpa variants and released full-length SH3-wt or SH3-m10 were analysed.
Nascent chains traverse through the arms of TF before folding
The difference in the interaction of TF with the folded SH3 domain and its unfolded SH3-m10 variant can be explained by two different scenarios: (i) either the SH3 nascent chain folds rapidly upon leaving the ribosomal tunnel and, thus, is not able to contact TF in the region beyond the arms, or (ii) the nascent SH3 domain initially traverses through TF in a length-dependent manner but repositions upon folding. To discriminate between these possibilities, we applied UV light co-translationally during ongoing synthesis (Supplementary Figure S1B) of arrested SH3-wt-SecM and SH3-m10-SecM to allow crosslinking of TF to the nascent chain immediately upon first contact. In this experimental set-up, all TF-Bpa variants that crosslinked to SH3-m10-SecM also revealed crosslinking to SH3-wt-SecM with comparable efficiencies (Figure 4B). Most importantly, this includes strong crosslinking to TF-256Bpa and TF-322Bpa (compare Figure 4A and B). Additionally, we translated non-arrested SH3-wt and SH3-m10 without the SecM stalling motif and applied UV irradiation co-translationally (Supplementary Figure S1C). Analysis of the released full-length products in the supernatant (Figure 4C) confirmed that nascent SH3-wt and SH3-m10 revealed similar crosslinking patterns to the interior of TF during ongoing translation. Thus, the unfolded SH3-wt nascent chain initially traverses through the C-terminal arms towards the PPIase of TF and, as a consequence of its folding, the contact to position 256 in TF is lost. The loss of crosslinking to this position may be either due a repositioning of the entire SH3-wt domain after folding or it could be due to a local conformational change of SH3 position upon folding such that no crosslink to TF-256Bpa occurs while all other positions are unaffected.
Generation of stable TF–RNCs
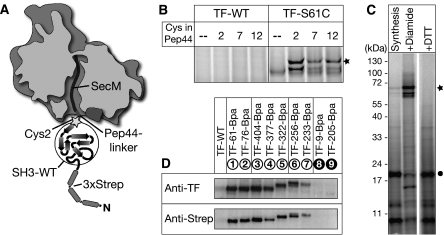
To gain insights into the active conformation of operating TF, which interacts with both the ribosome and the emerging nascent polypeptide, and to validate the conclusions drawn from our crosslinking analysis, we analysed full-length TF bound to RNCs by Cryo-EM. As TF binds to RNCs with relatively low affinity and the interaction is transient with a lifetime in the range of seconds (Kaiser et al, 2006), it was essential to stabilize this interaction. Our strategy was to covalently attach TF to the nascent chain by means of a single disulphide bond. To introduce a cysteine in TF, we chose residue Ser61 (TF-S61C), which locates close to the tunnel exit and interacts early with the emerging nascent chains as demonstrated above. Consequently, the interacting cysteine in the nascent peptide was placed within the first few residues being exposed outside the ribosomal tunnel. We designed the following model nascent chain (Figure 5A): to stall the nascent chain and to span the ribosomal exit tunnel, we used a SecM peptide (36 aa) at the C-terminal end. We fused, in frame, the peptide Pep44 (QRKLFFNLRKTKQ) that binds to TF with high affinity (Patzelt et al, 2001). In this Pep44-linker region, we introduced a cysteine residue for efficient disulphide bridge formation with TF-S61C. Successively, an SH3-wt domain was attached, which would restrict the conformational flexibility of the nascent chain upon folding into a compact structure. N-terminally, the model polypeptide contained a triple Strep tag for affinity purification of the RNCs. To test for the optimal position of the cysteine residue, we designed three nascent chain constructs harbouring the cysteine at aa 2, 7 or 12 within the Pep44-linker (Strep-SH3-2Cys-SecM, Strep-SH3-7Cys-SecM, Strep-SH3-12Cys-SecM). These constructs were synthesized in vitro in the presence of the TF-S61C variant and, after synthesis, diamide was added to induce oxidizing conditions. The formation of the disulphide bridge was highly efficient for TF-S61C and Strep-SH3-2Cys-SecM (Figure 5B, C) and could be completely resolved upon treatment with the reducing agent dithiothreitol (Figure 5C). No linkage products could be observed in control reactions with wt TF or cysteine-free nascent chains.
Figure 5.
Generation of stabilized TF–RNCs. TF–RNCs were generated and stabilized by disulphide bond formation. (A) Schematic description of the model nascent chain, Strep-SH3-2Cys-SecM, composed of a triple Strep tag (3xStrep), a folded SH3-wt domain (SH3-wt), a peptide-linker of 13 aa (Pep44-linker) harbouring a single cysteine (Cys2, asterisk) and the SecM arrest sequence (SecM). (B) Analysis of disulphide bond formation of TF-wt (cysteine-free) and TF-S61C variant with model nascent chains harbouring the single cysteine at different positions in the Pep44-linker (Cys2, Cys7, Cys12). Formation of disulphide bridges (asterisk) was induced by the addition of diamide after in vitro synthesis. (C) TF-S61C and nascent Strep-SH3-2Cys-SecM (filled circle) efficiently build disulphide bonds (asterisk) upon diamide addition, which can be resolved by the reducing agent dithiothreitol. (D) Site-specific crosslinking of purified RNCs of Strep-SH3-2Cys-SecM and TF-Bpa variants. Crosslinking products were analysed by western blot analysis using antibodies specific for TF or the Strep tag of the model nascent chain.
To ensure that Strep-SH3-2Cys-SecM nascent chains localize underneath TF, we analysed crosslinking of the TF-Bpa variants to purified RNCs of Strep-SH3-2Cys-SecM by UV irradiation. Crosslinking adducts were investigated by western blot analysis using antibodies specific for TF or the Strep tag of the nascent peptide. Although all TF-Bpa variants with a crosslinker in the interior crosslinked to stalled Strep-SH3-2Cys-SecM nascent chains, positions on the outside of TF did not crosslink (Figure 5D). In comparison with the nascent SH3-wt-SecM construct (Figure 4A), Strep-SH3-2Cys-SecM additionally crosslinked to TF positions located beyond the C-terminal arms (TF-256Bpa, TF-322Bpa, TF-233Bpa). This is most likely due to the additional Strep tag (40 aa) at the N terminus, which is long enough to extend to the PPIase domain. In summary, the Strep-SH3-2Cys-SecM nascent chain is accommodated in the interior of ribosome-bound TF and efficiently forms a covalent linkage to TF-S61C under oxidizing conditions.
Structure of TF in its active state bound to an RNC
The Cryo-EM structure of the E. coli TF in complex with the E. coli RNC containing Strep-SH3-2Cys-SecM was determined at 19 Å resolution. Our reconstruction was determined with a low-pass-filtered E. coli RNC as an initial model (Schaffitzel et al, 2006) without using any structural data from crystallographic experiments.
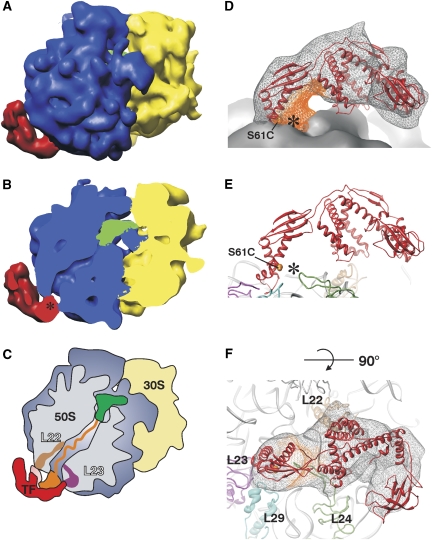
It is directly evident from the electron density that compared to vacant ribosomes, the tunnel exit is covered by an additional density of 45 Å by 100 Å representing the E. coli TF interacting with the nascent chain (Figure 6A–C). The EM density of the ribosome was interpreted by fitting the crystal structures of the E. coli ribosomal subunits (Schuwirth et al, 2005) (Supplementary Figure S4). The structure of the ribosome is very similar to the published structures of SecM-stalled RNCs (Schaffitzel et al, 2006) including the position of the P-site and E-site tRNA. The additional density at the tunnel exit accounts for all domains of TF including the PPIase domain. It was interpreted by fitting the E. coli TF crystal structure (Ferbitz et al, 2004) (Figure 6D–F). The density fitting was performed by an exhaustive search for all possible TF orientations using the signature motif bound to protein L23 as the fixed anchor point. Subsequently, the position of the PPIase domain was adjusted by a 24° rotation towards the C-terminal arms of TF. The density on the tip of the first arm and the PPIase domain is weaker than the density for other portions of TF, suggesting some flexibility within these regions of the TF molecule. This is in agreement with the crystal structure of TF (Ferbitz et al, 2004), in which a similar level of flexibility can be observed when comparing the conformations of the two molecules in the crystal. Superimposing the N-terminal domains of the two TF molecules in the crystal results in an 11 Å displacement of the PPIase domain.
Figure 6.
Cryo-EM analysis of TF bound to an RNC. (A) Structure of the E. coli RNC (50S subunit, blue; 30S subunit, yellow; P-site tRNA, green) in complex with TF interacting with the nascent chain (red). (B) Same perspective as in (A) but with the ribosome sliced along the tunnel. The tunnel exit is marked with a star. (C) Schematic drawing of the TF interacting with the nascent chain. The 50S subunit is shown in blue and the 30S subunit is in yellow. The nascent chain (orange) connected to the peptidyl P-tRNA (green) is shielded by TF (red). (D) Crystal structure of the E. coli TF (red ribbon, crosslinked residue 61 is shown as orange spheres) fitted into the EM density (grey mesh). The density next to the tunnel exit (black star) attributed to the nascent chain is shown in orange. The ribosome is visualized as a grey surface. (E) Orientation of the TF (red ribbon) on the ribosome (rRNA, grey ribbon). The proteins in the vicinity of the tunnel exit (black star) are highlighted in different colours (L23, magenta; L22, light brown; L29, cyan; L24, green). (F) Same as (E) but tilted by 90° towards the viewer and including the TF density shown as a grey mesh.
The EM reconstruction shows that TF binds to the ribosomal protein L23 and forms an arch over the tunnel exit extending above the ribosomal surface (Figure 6D–F). In the ribosome-bound state, the hydrophobic inner surfaces of the N domain and the entire C domain of TF face the ribosome and create a protective environment around the tunnel. In contrast to earlier reports (Schlunzen et al, 2005), this conformation of TF would not allow the loop of protein L24 to reach the inside of the cradle (Supplementary Figure S4). The space between the ribosome and TF is large enough to accommodate a folded protein domain, which is in agreement with our crosslinking data showing that the folded SH3 is accommodated underneath TF (Figures 4 and 5D). Indeed, the tunnel exit is partially filled with density, which could be attributed to parts of the SH3 domain of the nascent chain, as this domain is separated by only two amino acids from the cysteine forming a disulphide bond with TF-S61C (Figure 6D–F). We conclude that, as suggested earlier by the model of Ferbitz et al (2004), TF arches over the exit site of the ribosome, thereby forming a cradle-like structure that can accommodate unfolded nascent chains as well as small folded domains.
Discussion
In this study, we delineate the sequence of events between TF and emerging nascent chains by following the molecular interactions that significantly change depending on the length and folding state of nascent polypeptides. We corroborate these findings with the first structure of operating TF bound to a translating ribosome.
The crosslinking analysis showed that all three domains of TF are able to interact with nascent polypeptides, which is in line with a recent report (Lakshmipathy et al, 2007). In addition to this finding, we dissected the chronology of interactions between TF and the nascent chain during peptide elongation and co-translational folding to provide an insight into the molecular mechanism of the action of TF on translating ribosomes. We found that emerging nascent polypeptides first contact the N domain of TF, then use the entire C domain by passing through the narrow region formed by the C-terminal arms and finally reach the area of the distally located PPIase domain. This is in agreement with a recent in vitro study showing that all three domains of TF contribute to the chaperone activity, whereby the C-terminal domain of TF is essential and constitutes with its two protruding arms the central structural module for the chaperone activity (Merz et al, 2006). The consecutive order, in which the crosslinks between TF and nascent chains of increasing lengths were observed, implies that the elongating nascent chain uses multiple binding sites along the TF interior in a sequential and directed manner. The minimal length of an extended nascent chain (3 Å per amino acid) to reach a distinct crosslinker position can be estimated on the basis of the TF crystal structure docked on the ribosome: theoretically, 43 residues are necessary to contact the position of the first crosslinker within TF's N domain (TF-61Bpa), approximately 63 residues are necessary to proceed to the C domain between the arms (TF-404Bpa) and around 85 residues are required to reach the PPIase domain (TF-233Bpa). Our experimental data fit well with these theoretical values: nascent ICDH-SecM chains of 47 aa (ICDH-16-SecM) crosslinked to TF-61Bpa, chains of 71 aa (ICDH-40-SecM) revealed crosslinking to TF-404Bpa and, after translation of 90 aa (ICDH-59-SecM), nascent chains crosslinked to all tested positions within TF's interior including position 233 in the PPIase pocket. This implies that nascent chains initially follow in an unfolded conformation a rather defined path alongside the TF interior, most likely headed by their N terminus (Figure 7A and B). Such a length-dependent progression of nascent chains through TF was shown not only for arrested nascent ICDH constructs, but also for non-arrested nascent polypeptides tested during ongoing translation (ICDH, MetK and OmpA; pellet fractions of Figure 3B and Supplementary Figure S2C). It is not clear how nascent chains are relocated underneath the TF cradle. Both two-dimensional diffusion along the hydrophobic surface of TF and a series of fast dissociation and re-association steps, in the millisecond range, can be considered as possibilities. When the polypeptide is further elongated (⩾90 aa), two different scenarios could be envisioned (Figure 7C). In the first scenario, the polypeptide may continue to traverse through the TF interior upon elongation and perhaps exit in the area of the PPIase domain, whereas in the second scenario, the polypeptide could start to accumulate inside TF. The observation that nascent polypeptides longer than 90 aa tested herein exclusively crosslinked to the inside of TF might argue for an accumulation of the polypeptide underneath TF, and the appearance of multiple crosslinking adducts of longer nascent chains could reflect crosslinking of the TF-Bpa variants to different positions of such an accumulated polypeptide. Additionally, previous studies have shown that ribosome-bound TF protects nascent polypeptides up to a size of about 41 kDa against proteolysis in vitro (Hoffmann et al, 2006; Tomic et al, 2006). However, both scenarios are not mutually exclusive and may occur depending on the nature of a nascent chain including its hydrophobicity and folding kinetics. The binding of an unfolded polypeptide stretch could delay folding and allow binding of downstream factors such as DnaK or GroEL. On the other hand, the accumulation of a nascent polypeptide with multiple transient interactions to TF would significantly reduce the available conformational freedom of the unfolded polypeptide and could thereby promote the formation of intramolecular contacts within the nascent chain, which are known to drive the folding of extended polypeptides towards their native structure (Jahn and Radford, 2007).
Figure 7.
Model of the passage of nascent polypeptides through TF. TF directs the nascent chains through its interior in a sequential and length-dependent manner. (A) Initially (with a length of 40–60 aa), the N terminus of the nascent chain slides along the N domain, where it might be accessible by means of the lateral openings from both sides for processing factors such as PDF, MAP or SRP. (B) Up to a length of 90 aa, the nascent chain traverses through the C-terminal arms towards the PPIase domain and engages the entire interior. (C) Upon further elongation, the nascent chain might leave TF or, alternatively, may accumulate and perhaps fold in the interior of the TF chaperone. On demand, the folding of a subset of newly synthesized proteins is further assisted by cytosolic chaperones.
Our suggested model of TF interaction with nascent polypeptides (Figure 7) can be reconciled with TF's dynamic cycling on and off the ribosome. Two recent studies have determined the kinetics of TF association with ribosomes and RNCs, showing that the half-life of the TF–RNC ranges between approximately 15 and 50 s depending on the length and hydrophobicity of the nascent polypeptide exposed at the ribosomal exit (Kaiser et al, 2006; Rutkowska et al, 2008). Such a long residence time of TF on the ribosome allows for synthesis of at least 150–200 residues of a nascent chain. It is therefore indeed possible that a nascent chain moves through or accumulates beneath TF while this chaperone remains associated.
To analyse how the interactions between a nascent chain and TF change upon folding, we performed crosslinking experiments using nascent chains of identical length but of different folding capability (SH3-wt, SH3-m10). Initially, both nascent SH3-wt and permanently unfolded SH3-m10 traversed through the TF arms towards the area of the PPIase domain. However, after attaining the native conformation, the SH3-wt domain probably contacted TF only in the region of the N domain and the C-terminal arms (Figure 4), which has been proposed to form a protective cavity for folding of the nascent chain. Given the dynamic cycling of TF on and off the ribosome (t1/2=15 s; Maier et al, 2003; Kaiser et al, 2006; Rutkowska et al, 2008), folding may occur underneath ribosome-bound TF or after the departure of TF followed by subsequent rebinding. In either case, our data demonstrate that ribosome-bound TF accommodates and protects small folded domains of the size of SH3 (25 × 28 Å) within its internal substrate-binding cavity and thus may allow folding to occur. The ability of TF to protect nascent polypeptides of the size of 41 kDa against proteolytic attack (Hoffmann et al, 2006) suggests that TF possesses a rather flexible binding mode perhaps allowing the enlargement of the cavity size for accommodation of larger domains as well.
To provide a structural insight into the physiological state of TF and to test our model suggested earlier (Ferbitz et al, 2004), we used Cryo-EM analysis to determine the orientation and overall conformation of TF operating on a translating E. coli ribosome displaying a folded SH3 domain. To generate homogenous TF–RNCs, we stabilized the transient interaction between TF and the translating ribosome by covalently fixing the chaperone to the nascent peptide by means of a disulphide bridge. This covalent attachment is not expected to change the orientation and the conformation of bound TF as (i) it occurs at a natural contact point between the nascent chain and the chaperone as determined in this study, (ii) the flexible nature of the nascent chain cannot in itself impose a particular orientation of the TF and (iii) the crosslink was formed with high efficiency suggesting that it did not involve an energetically unfavourable interaction nor an unusual orientation of the chaperone on the ribosome. Consistent with our crosslinking data, the electron density (Figure 6) showed that TF forms an arch over the ribosomal tunnel exit, thereby providing a space that is large enough to accommodate a small folded domain. Indeed, additional density near the tunnel exit might be assigned to parts of the SH3 domain covalently linked to TF (TF-S61C) (Figure 6D). As the reconstruction was obtained without using any structural data from crystallographic experiments, the EM structure independently supports the model of a protective cavity beneath TF (Ferbitz et al, 2004).
Comparing the EM-derived structure with the crystal structure of full-length TF (Ferbitz et al, 2004) shows significant differences in the orientation of the PPIase domain, which rotates towards the C-terminal domain, although the overall conformation is very similar. Nevertheless, we cannot exclude that ribosome binding causes minor conformational rearrangements as suggested by FRET experiments with TF labelled at the N-terminal domain and C-terminal arms (Kaiser et al, 2006). The FRET efficiency decreased upon ribosome binding, indicating a conformational rearrangement in the range of 5–10 Å.
To accommodate nascent chains of different size and shape, it would be advantageous for TF to exhibit some degree of flexibility, which is indicated by the EM density. Consistent with swinging motion stemming from the ribosome-binding site, the TF density is less well defined with increasing distance to the N-terminal domain. Thus, our obtained model for TF bound to the nascent chain represents the median most populated conformation under physiological conditions and does not exclude the existence of minor, less frequent states of ribosome-bound TF. The crystal structures of N-terminal TF fragments bound to 50S ribosomal subunits of different organisms showed the fragment extending in different directions from the attachment point on the ribosome. The crystal structure of the E. coli N-terminal TF fragment in complex with the H. marismortui 50S ribosomal subunit (Ferbitz et al, 2004) showed this domain in a similar orientation as in our EM structure of TF. However, in the D. radiodurans TF-50S ribosomal subunit complexes (Baram et al, 2005; Schlunzen et al, 2005), TF's N domain is rotated towards L24 such that its β-sheet would be located partially outside the EM density obtained here and derived models of full-length TF bound to the 50S ribosomal subunit are less consistent with the EM density (see Supplementary Figure S3). In these two structures, the N-terminal domain of TF might be constrained in its conformational freedom by crystal lattice contacts.
The structural evidence that TF in its active state forms an arch over the ribosomal exit tunnel in combination with the information about the precise localization of nascent polypeptides during the initial passage through TF provides insights into how the action of TF on the ribosome can be spatially and temporally coordinated with other co-translational processes. Nascent chains need to be processed by enzymes, including the removal of the N-terminal formyl group by protein deformylase (PDF) in bacteria and the cleavage of the N-terminal methionine by aminopeptidases (MAPs). This processing has been shown to occur co-translationally as soon as the nascent chains reach a length of 40–60 aa (Housman et al, 1972; Ball and Kaesberg, 1973). We localized nascent chains of such a length exclusively in the area of TF's N domain, which is accessible through large lateral gaps on both sides and therefore could allow PDF and MAP to approach nascent polypeptides during their progression through the TF interior (Figure 7A). The same entry might be used by the signal recognition particle (SRP) as SRP co-translationally binds nascent chains as soon as their signal sequence has a distance of 40–60 aa from the ribosomal peptidyl-transferase centre (Ullers et al, 2006). The conformation of TF seen in our Cryo-EM analysis would not allow the simultaneous binding of SRP in a manner as described for SRP bound to a nascent signal sequence at the ribosome (Schaffitzel et al, 2006). Nevertheless, as previously shown by crosslinking (Buskiewicz et al, 2004), concomitant binding of TF and SRP might be possible. In the presence of TF, SRP might be flexibly attached only to L23 and L29, as observed for the 70S ribosome complex without a nascent chain (Schaffitzel et al, 2006). Such a mode might allow SRP to sample the nascent chain even in the presence of TF. Upon recognition of the signal sequence, SRP would rearrange and TF would be replaced. Alternatively, SRP and the other factors might bind to their targets while TF is dissociated from the ribosome and thus temporarily disengaged from the nascent polypeptide. However, we consider this scenario less likely, given the rather long half-life of TF–ribosome complexes and its high association rates for RNCs.
The combination of biochemical and structural results described here reveals the mechanism of TF action during co-translational protein folding through a series of snapshots at a molecular level. Moreover, this work provides the basis for further experiments aimed at revealing the complex network of co-translational processes.
Materials and methods
In vitro transcription/translation and crosslinking analysis
The cloning of plasmids encoding nascent chains and TF variants is summarized in Supplementary Table S1. TF variants were generated and purified according to published protocols (Kramer et al, 2004b; Ryu and Schultz, 2006). The E. coli-based in vitro transcription/translation system used in this study was derived from MC4100Δtig cells (Schaffitzel et al, 2001) and was complemented with 0.5 μM ribosomes and 1.5 μM of wt TF, TF-Bpa variants or TF-S61C. The synthesis was stopped after 15 min at 37°C by adding 2 mM chloramphenicol. Chemical crosslinking was achieved by the addition of 2.5 mM DSS and a 30 min incubation at 25°C. The crosslinker reaction was quenched with 50 mM Tris–HCl (pH 7.5) for 15 min at 25°C. In the case of site-specific crosslinking, UV light (UVP BLAK-RAY B100AP) was applied either post-translationally for 3 min on ice, or co-translationally during synthesis for 15 min at 37°C. For SecM-arrested nascent chains, ribosomes were centrifuged through a sucrose cushion (30% in reaction buffer) at 250 000 g for 70 min at 4°C. In the case of the non-arrested full-length nascent chain constructs, centrifugation was performed without a sucrose cushion at 350 000 g for 12 min at 4°C. 35S-methionine labelled polypeptides and crosslinking products were separated by SDS–PAGE and visualized by autoradiography. Every crosslinking experiment was independently repeated at least three times. For generation and detection of disulphide bonds between TF and nascent chains, 1 mM diamide was added after translation and the sample was incubated for 5 min at 20°C (when indicated, 5 mM dithiothreitol was added subsequent to diamide treatment). Complex formation was analysed by non-reducing SDS–PAGE and autoradiography.
Cryo-EM analysis
RNCs were purified using the construct Strep-SH3-2Cys-SecM as described previously (Schaffitzel and Ban, 2007). TF–RNCs were formed by adding a 20-fold molar excess of TF-S61C and subsequently analysed by Cryo-EM. For a detailed description of the procedure, see Supplementary data.
Supplementary Material
Supplementary Data
Acknowledgments
We thank members of the Ban, Bukau and Deuerling laboratories for discussions and comments, the Electron Microscopy Center Zurich (EMEZ) for support, Peter Schultz for donation of plasmids, Lars Ferbitz for his valuable expertise and Beate Zachmann-Brand for technical assistance. This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) to BB and ED (SFB638), a Heisenberg Fellowship and a grant from the DFG to ED, by the Swiss National Science Foundation (SNSF), the National Center of Excellence in Research (NCCR) Structural Biology programme of the SNSF and the ETH Research Grant TH -3/04-1 to NB, a Human Frontiers in Science Program Grant to ED and NB, the Boehringer Ingelheim Fonds to AH and a Federation of European Biochemical Societies long-term fellowship to DB.
Accession numbers The TF–RNC Cryo-EM map and coordinates for the docked TF have been deposited in the EM Data Bank, accession number EMD-1499, and in the Protein Data Bank, accession number 2VRH, respectively.
References
- Agashe VR, Guha S, Chang HC, Genevaux P, Hayer-Hartl M, Stemp M, Georgopoulos C, Hartl FU, Barral JM (2004) Function of trigger factor and DnaK in multidomain protein folding: increase in yield at the expense of folding speed. Cell 117: 199–209 [DOI] [PubMed] [Google Scholar]
- Ball LA, Kaesberg P (1973) Cleavage of the N-terminal formylmethionine residue from a bacteriophage coat protein in vitro. J Mol Biol 79: 531–537 [DOI] [PubMed] [Google Scholar]
- Baram D, Pyetan E, Sittner A, Auerbach-Nevo T, Bashan A, Yonath A (2005) Structure of trigger factor binding domain in biologically homologous complex with eubacterial ribosome reveals its chaperone action. Proc Natl Acad Sci USA 102: 12017–12022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco FJ, Angrand I, Serrano L (1999) Exploring the conformational properties of the sequence space between two proteins with different folds: an experimental study. J Mol Biol 285: 741–753 [DOI] [PubMed] [Google Scholar]
- Bukau B, Deuerling E, Pfund C, Craig EA (2000) Getting newly synthesized proteins into shape. Cell 101: 119–122 [DOI] [PubMed] [Google Scholar]
- Buskiewicz I, Deuerling E, Gu SQ, Jockel J, Rodnina MV, Bukau B, Wintermeyer W (2004) Trigger factor binds to ribosome-signal-recognition particle (SRP) complexes and is excluded by binding of the SRP receptor. Proc Natl Acad Sci USA 101: 7902–7906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B (1999) Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 400: 693–696 [DOI] [PubMed] [Google Scholar]
- Ferbitz L, Maier T, Patzelt H, Bukau B, Deuerling E, Ban N (2004) Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature 431: 590–596 [DOI] [PubMed] [Google Scholar]
- Frydman J (2001) Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem 70: 603–647 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295: 1852–1858 [DOI] [PubMed] [Google Scholar]
- Hesterkamp T, Bukau B (1996) Identification of the prolyl isomerase domain of Escherichia coli trigger factor. FEBS Lett 385: 67–71 [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Merz F, Rutkowska A, Zachmann-Brand B, Deuerling E, Bukau B (2006) Trigger factor forms a protective shield for nascent polypeptides at the ribosome. J Biol Chem 281: 6539–6545 [DOI] [PubMed] [Google Scholar]
- Housman D, Gillespie D, Lodish HF (1972) Removal of formyl-methionine residue from nascent bacteriophage f2 protein. J Mol Biol 65: 163–166 [DOI] [PubMed] [Google Scholar]
- Jahn TR, Radford SE (2008) Folding versus aggregation: polypeptide conformations on competing pathways. Arch Biochem Biophys 469: 100–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser CM, Chang HC, Agashe VR, Lakshmipathy SK, Etchells SA, Hayer-Hartl M, Hartl FU, Barral JM (2006) Real-time observation of trigger factor function on translating ribosomes. Nature 444: 455–460 [DOI] [PubMed] [Google Scholar]
- Kramer G, Patzelt H, Rauch T, Kurz TA, Vorderwulbecke S, Bukau B, Deuerling E (2004a) Trigger factor peptidyl-prolyl cis/trans isomerase activity is not essential for the folding of cytosolic proteins in Escherichia coli. J Biol Chem 279: 14165–14170 [DOI] [PubMed] [Google Scholar]
- Kramer G, Rauch T, Rist W, Vorderwülbecke S, Patzelt H, Schulze-Specking A, Ban N, Deuerling E, Bukau B (2002) L23 protein functions as a chaperone docking site on the ribosome. Nature 419: 171–174 [DOI] [PubMed] [Google Scholar]
- Kramer G, Rutkowska A, Wegrzyn RD, Patzelt H, Kurz TA, Merz F, Rauch T, Vorderwülbecke S, Deuerling E, Bukau B (2004b) Functional dissection of Escherichia coli trigger factor: unraveling the function of individual domains. J Bacteriol 186: 3777–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmipathy SK, Tomic S, Kaiser CM, Chang HC, Genevaux P, Georgopoulos C, Barral JM, Johnson AE, Hartl FU, Etchells SA (2007) Identification of nascent chain interaction sites on trigger factor. J Biol Chem 282: 12186–12193 [DOI] [PubMed] [Google Scholar]
- Maier R, Eckert B, Scholz C, Lilie H, Schmid FX (2003) Interaction of trigger factor with the ribosome. J Mol Biol 326: 585–592 [DOI] [PubMed] [Google Scholar]
- Merz F, Hoffmann A, Rutkowska A, Zachmann-Brand B, Bukau B, Deuerling E (2006) The C-terminal domain of Escherichia coli trigger factor represents the central module of its chaperone activity. J Biol Chem 281: 31963–31971 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Ito K (2002) The ribosomal exit tunnel functions as a discriminating gate. Cell 108: 629–636 [DOI] [PubMed] [Google Scholar]
- Patzelt H, Rudiger S, Brehmer D, Kramer G, Vorderwulbecke S, Schaffitzel E, Waitz A, Hesterkamp T, Dong L, Schneider-Mergener J, Bukau B, Deuerling E (2001) Binding specificity of Escherichia coli trigger factor. Proc Natl Acad Sci USA 98: 14244–14249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raine A, Lovmar M, Wikberg J, Ehrenberg M (2006) Trigger factor binding to ribosomes with nascent peptide chains of varying lengths and sequences. J Biol Chem 281: 28033–28038 [DOI] [PubMed] [Google Scholar]
- Rutkowska A, Mayer MP, Hoffmann A, Merz F, Zachmann-Brand B, Schaffitzel C, Ban N, Deuerling E, Bukau B (2008) Dynamics of trigger factor interaction with translating ribosomes. J Biol Chem 283: 4124–4132 [DOI] [PubMed] [Google Scholar]
- Ryu Y, Schultz PG (2006) Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nat Methods 3: 263–265 [DOI] [PubMed] [Google Scholar]
- Schaffitzel C, Ban N (2007) Generation of ribosome nascent chain complexes for structural and functional studies. J Struct Biol 158: 463–471 [DOI] [PubMed] [Google Scholar]
- Schaffitzel C, Oswald M, Berger I, Ishikawa T, Abrahams JP, Koerten HK, Koning RI, Ban N (2006) Structure of the E. coli signal recognition particle bound to a translating ribosome. Nature 444: 503–506 [DOI] [PubMed] [Google Scholar]
- Schaffitzel E, Rüdiger S, Bukau B, Deuerling E (2001) Functional dissection of Trigger Factor and DnaK: interactions with nascent polypeptides and thermally denatured proteins. Biol Chem 382: 1235–1243 [DOI] [PubMed] [Google Scholar]
- Schlunzen F, Wilson DN, Tian P, Harms JM, McInnes SJ, Hansen HA, Albrecht R, Buerger J, Wilbanks SM, Fucini P (2005) The binding mode of the Trigger Factor on the ribosome: implications for protein folding and SRP interaction. Structure (Camb) 13: 1685–1694 [DOI] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH (2005) Structures of the bacterial ribosome at 3.5 Å resolution. Science 310: 827–834 [DOI] [PubMed] [Google Scholar]
- Stoller G, Tradler T, Rucknagel KP, Rahfeld J-U, Fischer G (1996) An 11.8 kDa proteolytic fragment of the E. coli trigger factor represents the domain carrying the peptidyl-prolyl cis/trans isomerase activity. FEBS Lett 384: 117–122 [DOI] [PubMed] [Google Scholar]
- Teter SA, Houry WA, Ang D, Tradler T, Rockabrand D, Fischer G, Blum P, Georgopoulos C, Hartl FU (1999) Polypeptide flux through bacterial Hsp70: DnaK cooperates with Trigger Factor in chaperoning nascent chains. Cell 97: 755–765 [DOI] [PubMed] [Google Scholar]
- Tomic S, Johnson AE, Hartl FU, Etchells SA (2006) Exploring the capacity of trigger factor to function as a shield for ribosome bound polypeptide chains. FEBS Lett 580: 72–76 [DOI] [PubMed] [Google Scholar]
- Ullers RS, Houben EN, Brunner J, Oudega B, Harms N, Luirink J (2006) Sequence-specific interactions of nascent Escherichia coli polypeptides with trigger factor and signal recognition particle. J Biol Chem 281: 13999–14005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data