Abstract
Rap1 (repressor-activator protein 1) is a multifunctional protein that controls telomere function, silencing and the activation of glycolytic and ribosomal protein genes. We have identified a novel function for Rap1, regulating the ribonucleotide reductase (RNR) genes that are required for DNA repair and telomere expansion. Both the C terminus and DNA-binding domain of Rap1 are required for the activation of the RNR genes, and the phenotypes of different Rap1 mutants suggest that it utilizes both regions to carry out distinct steps in the activation process. Recruitment of Rap1 to the RNR3 gene is dependent on activation of the DNA damage checkpoint and chromatin remodelling by SWI/SNF. The dependence on SWI/SNF for binding suggests that Rap1 acts after remodelling to prevent the repositioning of nucleosomes back to the repressed state. Furthermore, the recruitment of Rap1 requires TAFIIs, suggesting a role for TFIID in stabilizing activator binding in vivo. We propose that Rap1 acts as a rheostat controlling nucleotide pools in response to shortened telomeres and DNA damage, providing a mechanism for fine-tuning the RNR genes during checkpoint activation.
Keywords: chromatin, Rap1, RNR3, SWI/SNF, TFIID
Introduction
Rap1 (repressor-activator protein 1) is an essential DNA binding protein that regulates transcriptional activation, silencing and the maintenance of telomeres (Morse, 2000; Pina et al, 2003). Its ability to maintain telomeres and repress the silent mating type loci is dependent on the recruitment of corepressors to these regions, which includes the histone deacetylase Sir2 and structural proteins to form heterochromatic-like states (Grunstein, 1997; Shore, 2001). Thus, Rap1 is implicated in regulating chromatin structure at repressed loci. It binds to approximately 5% of yeast genes under optimal growth conditions and is enriched at glycolytic enzyme and ribosomal protein (RP) genes (Lieb et al, 2001). Rap1 binds to a wide variety of DNA binding sites, and it is difficult to identify sites based on sequence information alone. This property is believed to be attributed to its functioning with other DNA binding proteins and cofactors to stimulate transcription in a context-dependent manner (Lieb et al, 2001; Pina et al, 2003; Del Vescovo et al, 2004).
Yeast TFIID is composed of the TATA-binding protein (TBP) and at least 14 associated factors, collectively referred to as TAFIIs (Reese et al, 1994; Sanders et al, 2002). Surprisingly, mutating or depleting multiple yeast TAFIIs individually affected only a fraction of the genome (Green, 2000). Computational analysis of gene expression profiles in TAFII mutants found that a fraction of the genome utilizes TFIID (TAFIIs) to deliver TBP to promoters, whereas the remainder utilizes the TAFII-containing SAGA histone acetyltransferase complex (Lee et al, 2000; Huisinga and Pugh, 2004). Based on these works, genes are classified as TAFII sensitive and TAFII insensitive. Studies evaluating what specifies the TAFII sensitivity found that core promoters have a role and they generally lacked a canonical TATA box (Mencia et al, 2002; Basehoar et al, 2004). However, it was later found that upstream regulatory sequences also caused TAFII dependency, and regulation by TFIID has been correlated to the presence of Rap1 binding sites within promoters. Most RP genes are TAFII dependent and UASs from RP genes can recruit TAFIIs to heterologous promoters in vivo and templates in vitro (Li et al, 2002; Mencia et al, 2002; Garbett et al, 2007). The mechanism involves a direct interaction between TFIID and the C terminus and DNA-binding domain (DBD) of Rap1. Rap1 also regulates glycolytic genes, which are considered TAFII-independent, indicating that the regulation by Rap1 is insufficient to designate a gene as TFIID-dependent.
RNR3 (ribonucleotide reductase 3) encodes one of the subunits of RNR, and it is strongly induced upon DNA damage (Huang et al, 1998; Li and Reese, 2000). In the absence of DNA damage, the sequence-specific DNA binding protein Crt1 (constitutive RNR transcription 1) binds to three binding sites (X-boxes) located in the upstream repression sequences (URS) of the gene (Huang et al, 1998). Crt1 represses transcription by recruiting the Ssn6–Tup1 corepressor complex to the promoter, and activation of RNR3 is caused by the checkpoint-dependent phosphorylation and accompanying release of Crt1 from the promoter (Huang et al, 1998; Zhang and Reese, 2004a, 2004b, 2005). RNR3 is a TAFII-dependent gene, and its requirement for TAFIIs maps to the Crt1 binding sites within the promoter and Crt1 itself (Li and Reese, 2000). Recently, we showed that Crt1 has a role in activation, by recruiting TFIID and SWI/SNF to the promoter (Zhang and Reese, 2005). Thus, Crt1 acts as a transient activator that may allow initial access of transcription factors to the promoter, but disassociates after the promoter is ‘opened'. A distinguishing feature of RNR3 is that the remodelling of its nucleosomes and SWI/SNF recruitment to the promoter are dependent on TAFIIs and general transcription factors (Sharma et al, 2003). Mutating TAFIIs, RPB1 or mediator blocks remodelling, which indicates that the preinitiation complex (PIC) has a role in retaining these activities at the promoter, possibly in conjunction with an unidentified activator. Thus, activation of RNR3 requires two steps, an initial opening of the promoter that requires Crt1 and a maintenance phase that requires the PIC and another factor.
The correlation between Rap1 and TFIID dependence led us to explore the role of Rap1 in the activation of the TAFII-dependent RNR3 gene. Here we show that Rap1 is required for the activation of the RNR genes. Rap1 recruitment requires the DNA damage checkpoint pathway, TFIID and chromatin remodelling. Interestingly, characterization of different types of Rap1 mutants suggests that it uses multiple domains to carryout distinct steps in the activation process. Furthermore, our results indicate that Rap1 has a novel role in maintaining genomic integrity and in the regulation of DNA repair genes.
Results
Rap1 is required for the activation of RNR genes
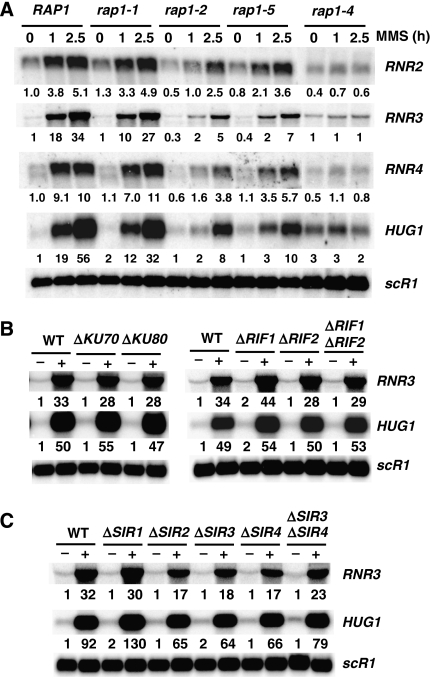
RNR2 and RNR4 have been identified as potential targets of Rap1 by ChIP-on-CHIP analysis conducted on uninduced cells (Lieb et al, 2001; Buck and Lieb, 2006) and Rap1 binds to a site within the regulatory region of RNR2 (Elledge and Davis, 1989; Hurd and Roberts, 1989). However, it has not been demonstrated that Rap1 is required for the activation of the RNR genes, and the most strongly induced RNR gene, RNR3, is not known to bind Rap1 (Lieb et al, 2001). Given that the RNR genes are TAFII-dependent (Li and Reese, 2000) and that Rap1 recruits TAFIIs to promoters (Mencia et al, 2002; Garbett et al, 2007), we examined the requirement for Rap1 in the activation of RNR genes and a coregulated gene HUG1. The expression of these genes was analysed in four different conditional mutants of Rap1, each displaying different growth and transcriptional activation defects (Kurtz and Shore, 1991). The mutants are temperature sensitive, but inductions were carried out at the semi-permissive temperature (30°C) to prevent secondary effects caused by heat shock. Even at the semi-permissive temperature, clear defects in RNR and HUG1 activation were observed in the different alleles. The rap1-4 allele showed the strongest phenotypes, followed by rap1-2, rap1-5 and then rap1-1 (Figure 1A). The rap1-4 mutant grows very slowly at 30°C (Kurtz and Shore, 1991; data not shown), but activation defects do not correlate with the slow-growth phenotypes. The rap1-1 allele has the second most severe slow-growth phenotype of all the alleles, yet it has a comparatively weaker RNR activation defect. Thus, the slow growth cannot fully account for the reduced activation of the RNR genes. Furthermore, rap1-1 has the strongest silencing defects (Kurtz and Shore, 1991) but weaker effects on the activation of the RNR genes. Thus, the activation defects are not intimately linked to the slow-growth or silencing-defect phenotypes of the mutants. Finally, Rap1 mutants with strong reductions in RNR gene expression are hypersensitive to hydroxyurea and methyl methanesulphonate (MMS) (Supplementary Figure S1).
Figure 1.
Rap1 regulates RNR gene expression independent of its telomeric and silencing functions. (A) Northern blot of mRNA isolated from conditional mutants of RAP1 (Kurtz and Shore, 1991). Cells were grown at 30°C and treated, or not (0 h), with MMS for 1 and 2.5 h. The levels of RNA were normalized to the signal of scR1, a loading control. The numbers below each panel are RNA levels relative to the amount in untreated wild-type cells, which was set at 1.0. (B, C) As in panel A except that telomere function mutants were examined. Cells were treated with MMS for 2.5 h in these panels.
Rap1 is an essential component of yeast telomeres and has a role in the repression of the silent mating type loci (Grunstein, 1997; Shore, 2001; Pina et al, 2003). Ample evidence suggests that Rap1 is also involved in masking telomeres and regulating non-homologous end joining (NHEJ) at sites of double-stranded breaks (Tsukamoto et al, 1997; Martin et al, 1999; McAinsh et al, 1999; Mills et al, 1999; Negrini et al, 2007). We set out to determine if altered telomere functions and/or mating type loci silencing causes RNR gene activation defects. We approached this question by examining the activation of RNR genes in strains with deletions in factors that participate in the maintenance of telomeres, NHEJ and silent mating type loci repression. We deleted RIF1 and RIF2 individually and in combination, and found that activation of RNR3 and HUG1 was unaffected (Figure 1B). Likewise, deleting KU70 or KU80 failed to impair DNA damage-induced transcription. Thus, alterations in telomere function and NHEJ pathways do not account for the reduced activation of the RNR genes. Next, we examined the effects of deleting components of the silent information regulator (SIR) pathway. Sir2/3/4 are required for telomeric silencing and Sir1 is additionally required for suppressing the silent mating type loci (Grunstein, 1997; Shore, 2001). Interestingly, deleting SIR2, SIR3 or SIR4 led to an ∼40–50% reduction in the activation of RNR3 and HUG1 (Figure 1C), indicating that they may have a lesser role in the activation of these genes. However, deleting SIR1 had no appreciable effects on the activation of DNA damage-inducible genes. Even though deleting SIR2, SIR3 or SIR4 reduced the expression of the RNR genes, the effect of the RAP1 mutations was significantly stronger (compare Figure 1A and C). Furthermore, as mutations in Ku70/80 or RIF1/2 proteins failed to result in the same phenotype, this argues that the role of the SIRs in regulating the RNR is not directly linked to their functions at the telomeres or NHEJ.
The C terminus of Rap1 is required for activation of RNR3 and binds TFIID and SWI/SNF
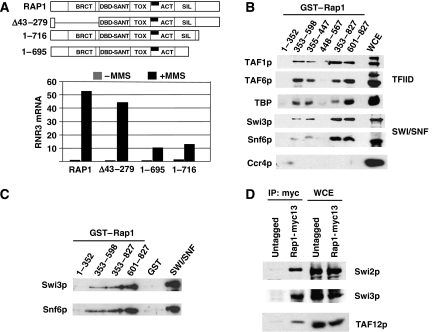
Rap1 contains a number of functional domains, including a myb-like DBD, a silencing and activation domain within the C terminus and a BRCT domain in the N terminus (Morse, 2000; Pina et al, 2003). All have been implicated in activating transcription and chromatin remodelling. We analysed the domain(s) of Rap1 required for the expression of RNR3 and found that deleting the N terminus containing the BRCT domain (Δ43–279) had little to no effect on the activation of RNR3 (Figure 2A). Thus, the BRCT domain, which is also found in a number of DNA damage repair proteins, is not essential for the expression of DNA damage-inducible genes. However, deleting the C terminus clearly impaired activation of RNR3 (rap1 1-716 and rap 1-695 mutants). The level of activation in the C-terminal mutants was about 20% of that of wild-type cells. This was not due to reduced protein levels because western blotting of extracts from these mutants indicated that the Rap1 mutants were expressed to similar levels as wild-type Rap1 (not shown). Both the rap1 1-716 and rap1 1-695 mutants are sensitive to DNA-damaging agents (Supplementary Figure S1).
Figure 2.
The C-terminal domain of Rap1 is required for activation of DNA damage-inducible genes and contributes to TFIID and SWI/SNF binding. (A) Analysis of RNR3 expression in Rap1 deletion mutants. A schematic of the known protein domains in Rap1 is indicated above and was adapted from Morse (2000). DBD-SANT is the myb-like DBD (353–598); TOX is the region identified to relieve the toxicity caused by Rap1 overexpression; SIL is the silencing domain, which is known to bind Sirs and Rif1/2; ACT is the domain that can activate transcription when fused to the DBD of LexA. The graph shows the relative amounts of RNR3 mRNA normalized to scR1, which are expressed relative to the amount of RNA in untreated wild-type cells. (B) Affinity chromatography of WCE on GST–Rap1 columns. The amino acids contained in the fusion proteins are indicated above. Proteins eluting with high-NaCl buffer were analysed by western blotting using polyclonal antibodies. (C) GST pulldown assays using tandem affinity-purified SWI/SNF complex (Snf6-TAP). (D) Co-immunoprecipitation of Rap1 and TFIID and SWI/SNF from WCE. WCE were prepared from strains expressing untagged and myc-tagged versions of Rap1. Anti-myc antibody was used for IP. After extensive washing, the bound proteins were analysed by western blotting.
Expression of RNR3 requires TFIID and the SWI/SNF chromatin remodelling complex (Sharma et al, 2003). Thus, we verified that Rap1 binds to TFIID and examined if it can bind SWI/SNF. Yeast whole-cell extracts (WCE) were passed over glutathione-S-transferase (GST)–Rap1 affinity columns and western blotting was used to detect the bound proteins. Consistent with previous results (Garbett et al, 2007), fragments of Rap1 containing the DBD (353–598) and the C terminus (601–827) retained TFIID from extracts (Figure 2B). Furthermore, the binding of TFIID was further mapped to the N-terminal portion of the DBD containing the myb-like DBD (355–447). Blotting for subunits of the SWI/SNF complex revealed that it also bound to the same fragments of Rap1 (Figure 2B). However, Ccr4, a component of the Ccr4–Not complex that also regulates the expression of RNR3 (Mulder et al, 2005), was not retained, suggesting selective binding of Rap1 to TFIID and SWI/SNF. It has been confirmed that purified TFIID can directly bind to the C terminus and the DBD of Rap1 in vitro (Garbett et al, 2007). To demonstrate that the interaction between Rap1 and SWI/SNF is direct, we repeated the binding assays using highly purified SWI/SNF (Snf6-TAP; Supplementary Figure S2). Figure 2C shows that purified SWI/SNF binds to both the DBD and ‘C terminus of Rap1, but not to the N terminus or GST alone. Finally, we preformed co-immunoprecipitation studies in WCE from untagged and myc-tagged Rap1 strains and found that SWI/SNF and TFIID subunits copurify with Rap1 using the myc antiserum (Figure 2D). Thus, three different binding assays confirm that Rap1 is a novel target of SWI/SNF.
Checkpoint-dependent recruitment of Rap1 to RNR3
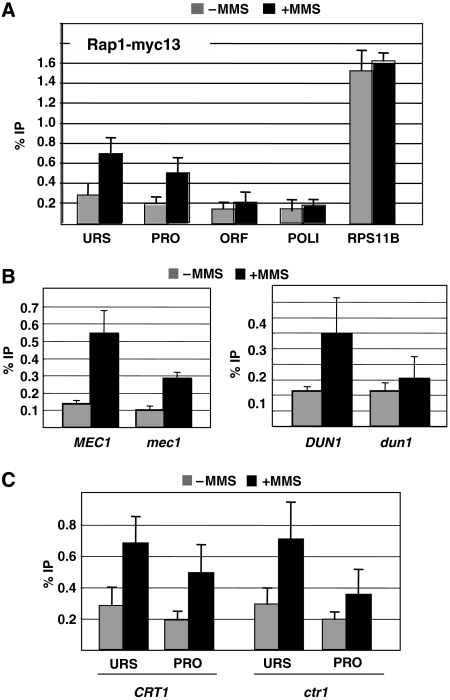
To verify that Rap1 regulates RNR genes directly, we examined its recruitment to RNR3 under the repressed and activated conditions. A strain containing a Rap1-myc13 gene was constructed and subjected to chromatin immunoprecipitation (ChIP) assay. Rap1 crosslinking was examined over the promoter, URS and open reading frame (ORF) of RNR3 after a 2 h treatment with MMS (0.03%). Figure 3A shows that treating cells with MMS caused a 2.2- to 3-fold increase in Rap1 crosslinking over the promoter and URS region of RNR3, consistent with it having a direct role in the activation of this gene. Crosslinking was consistently better over the URS, suggesting that these sequences have a role in Rap1 recruitment. Importantly, low crosslinking was detected over the ORF and the POLI gene, and DNA damage did not significantly increase Rap1 crosslinking to the UAS of the RP gene RPS11B (Figure 3A). This suggests that Rap1 does not associate with chromatin nonspecifically after MMS treatment, which theoretically could occur because Rap1 is known to relocalize from telomeres to sites of DNA damage (Martin et al, 1999).
Figure 3.
Checkpoint-dependent recruitment of Rap1 to RNR3. (A) ChIP assay in untreated (grey bars) and MMS-treated (black bars) strains containing Rap1-myc13. The immunoprecipitated DNA was amplified using primers directed to the URS, promoter (PRO) and the ORF of RNR3. The crosslinking of Rap1 to RPS11B, a target gene, and POLI, a negative control, was analysed in parallel. The data are presented as percentage IP and are the means and standard deviations of at least three independent chromatin preparations and IPs. (B) Analysis of Rap1 recruitment in checkpoint mutants. As in panel A except that crosslinking was examined over the promoter. The Δmec1 strain has a deletion of SML1 to sustain viability. (C) Rap1 crosslinking in a Δcrt1 mutant. Crosslinking was detected over the URS and promoter.
Activation of the RNR genes requires the checkpoint-dependent release of Crt1 from the promoter (Huang et al, 1998; Zhang and Reese, 2005). We therefore examined if the recruitment of Rap1 is checkpoint dependent using strains containing a deletion of MEC1 and DUN1, which are required for the activation and chromatin remodelling of RNR3 (Huang et al, 1998; Li and Reese, 2001). Deleting DUN1 or MEC1 (the viability of the Δmec1 strain was maintained by a Δsml1 deletion) strongly reduced the crosslinking of Rap1, indicating that its recruitment is checkpoint-dependent (Figure 3B). The cell cycle checkpoint pathway regulates RNR expression by causing the phosphorylation and release of the repressor Crt1 from the promoter (Huang et al, 1998). To test if the release of Crt1–Ssn6–Tup1 from the promoter is sufficient for Rap1 recruitment, we examined the crosslinking of Rap1 to RNR3 in a Δcrt1 mutant. Interestingly, we found that deletion of CRT1 was not sufficient to cause Rap1 recruitment and MMS treatment was required (Figure 3C). Thus, the checkpoint pathway regulates Rap1 recruitment independent of Crt1, and suggests that Rap1 may be a direct target of the checkpoint. Deleting CRT1 causes constitutive transcription and chromatin remodelling (Huang et al, 1998; Li and Reese, 2001); thus, it is surprising that transcription is high in the absence of Rap1 recruitment. A possible explanation for this is that Rap1 may be required for activating the repressed promoter, but is less important when the promoter is open, similar to the requirement for the activation domain of Crt1 (Zhang and Reese, 2005).
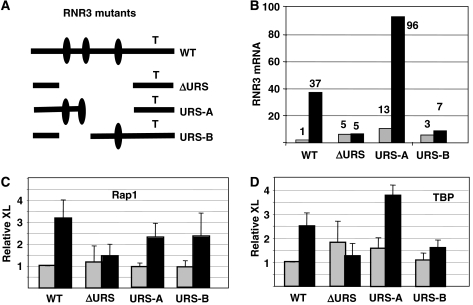
Examination of the promoter region of RNR3 revealed only one potential binding site for Rap1, located a few base pairs upstream of the translation start codon. However, mutation of that site did not affect RNR3 expression or Rap1 recruitment (not shown). Rap1 can bind to many sites within the genome, and most do not match a consensus sequence (Buck and Lieb, 2006), so the inability to identify a binding site for DNA sequence is not surprising. A broader approach was used to identify the regions of the promoter that are required for Rap1 recruitment. As the URS confer DNA damage-dependent transcription to RNR3, we focused on this region. Promoter constructs lacking the entire URS region or the 5′ and 3′ halves were constructed and integrated into the RNR3 locus (Figure 4A). ChIP assays were conducted and the results show that deleting the entire URS region severely compromised both the recruitment of Rap1 and the activation of RNR3 (Figure 4B and C). Deleting either the 5′ or 3′ half of the URS only partially reduced Rap1 crosslinking, suggesting that Rap1 may bind to multiple regions with the URS. Interestingly, deleting the 5′ end of the URS essentially abolished the activation of RNR3, yet some crosslinking of Rap1 was detected. Thus, full Rap1 occupancy is required for activation. Alternatively, deleting this region of the URS may impair the ability of Crt1 to mediate the activation step (Zhang and Reese, 2005).
Figure 4.
The URS mediates Rap1 recruitment. (A) Promoter constructs containing deletions in the URS were constructed in vitro and used to replace the endogenous RNR3 locus. The black ovals indicate the positions of the X-boxes that bind the repressor Crt1. With the start site of translation=+1 and the TATA box (‘T') at −126, the deletions are as follows: ΔURS, Δ−187/−548; URS-A, Δ−187/−259; URS-B, Δ−259/−548. (B) RNR3 mRNA was measured in these cells after a 2.5 h treatment with MMS. (C) Rap1-myc13 and (D) TBP recruitment was examined by ChIP over the promoter. The promoter was not altered by the deletions in the URS region. ChIP data are expressed relative to the signal, corrected for the inputs, of untreated wild-type cells, which was set to a value of 1.0.
Examination of chromatin remodelling at RNR3 reveals multiple functions of Rap1 in gene activation
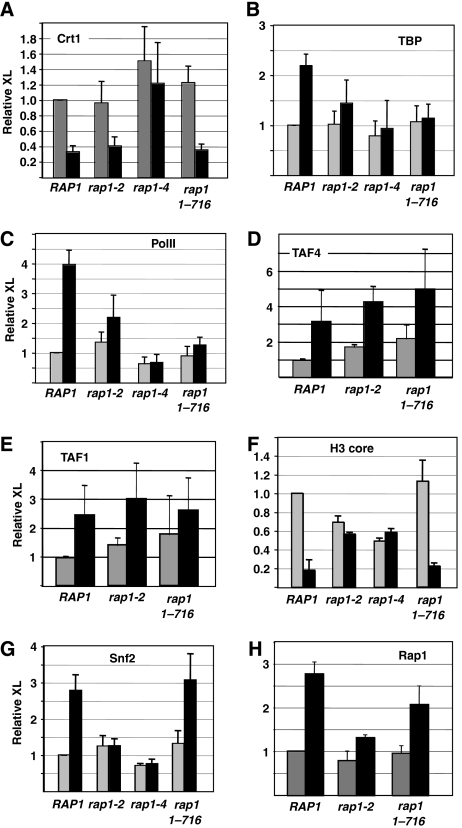
We next examined the integrity of the DNA damage checkpoint pathway in the RAP1 mutants by monitoring the MMS-dependent release of the repressor Crt1 from RNR3. MMS treatment led to reduced crosslinking of Crt1 in wild-type cells, indicating activation of the checkpoint and release of the repressor from the gene (Figure 5A). Likewise, Crt1 was released from the URS to a similar extent in all but one of the Rap1 mutants, rap1-4. The rap1-4 mutant shows stronger growth defects, and may have additional effects on the cells. It is unlikely that indirect effects explain the reduced activation of the RNR genes in the Rap1 mutants because other mutants with strong transcription defects show normal Crt1 release (Figure 5A). As Crt1 was not released in the rap1-4 mutant, we focused on rap1-2 in our subsequent analysis.
Figure 5.
Rap1 orchestrates multiple steps in activation. Cells were treated or not with MMS for 2.5 h and then crosslinked with formaldehyde. Chromatin was immunoprecipitated with antibodies to the proteins indicated in each panel. The Rap1 mutants are indicated below the panel. The data are the amount of crosslinking (corrected for the input DNA; see Materials and methods) relative to that of untreated wild-type cells. The data are reported as the mean and standard deviations from at least three independent crosslinking and IP experiments. (A) Recruitment of Crt1 to the URS region of RNR3. Recruitment of (B) TBP, (C) RNAPII, (D) TAF4, (E) TAF1, (F) histone H3 and (G) Swi2 was measured at the promoter. Crosslinking of Rap1 to the URS was measured in (H).
We verified the requirement for Rap1 in RNR transcription by examining the formation of the PIC. The recruitment of TBP and RNA polymerase II (RNAPII) was examined in the mutants with the strongest transcription defects (Figure 4B and C). Mutating Rap1 strongly reduced the MMS-induced crosslinking of TBP and RNAPII to the promoter, and the level of PIC formation correlated well with the levels of RNR3 mRNA in each mutant (compare Figures 1A and 2A with Figure 4B and C). Thus, Rap1 is required for PIC formation. Rap1 binding sites can direct recruitment of TFIID to promoters and Rap1 binds directly to TAFIIs (Li et al, 2002; Mencia et al, 2002; Garbett et al, 2007). TBP is not crosslinked to RNR3 in the Rap1 mutants, suggesting that TAFIIs are not recruited. However, TAFIIs can be recruited to Rap1 sites in the absence of functional TBP (Li et al, 2002; Mencia et al, 2002), so we examined the recruitment of two TFIID-specific TAFIIs to RNR3 (Figure 4D and E). Remarkably, TAFII recruitment was only weakly, if at all, affected in the Rap1 mutants. This was unexpected given the belief that Rap1 recruits TFIID. As a control, we examined TAFII recruitment to RPS11B in the Rap1 mutants and found that TAFII recruitment was not affected at this bona fide target gene in the mutants (Supplementary Figure S3). The studies initially reporting that Rap1 recruits TFIID did not examine Rap1 mutants, but rather linked recruitment of TAFIIs to Rap1 binding sites (Li et al, 2002; Mencia et al, 2002). A more recent study examining RAP1 mutants concluded that TAF1 recruitment was not affected after correcting for the binding of Rap1 to the promoters (Garbett et al, 2007). Thus, although Rap1 sites are required for TAFII recruitment to promoters, the requirement for Rap1 itself is unclear.
Activation of RNR3 requires the recruitment of SWI/SNF and eviction of the core promoter nucleosome (Sharma et al, 2003; Zhang and Reese, 2007). As Rap1 binds to SWI/SNF, we examined if it is required for SWI/SNF recruitment and nucleosome remodelling. The data in Figure 5F indicate that treating wild-type cells with MMS caused nucleosome eviction, as indicated by the reduced crosslinking of histone H3, and eviction correlated with an increase in SWI/SNF recruitment (Figure 5G). The crosslinking of H3 was somewhat reduced in the Rap1 conditional mutants in the repressed condition (rap1-2 and rap1-4), but, importantly, additional nucleosome loss was not caused by MMS treatment. The remodelling phenotypes of these mutants were also confirmed by MNase mapping (Supplementary Figure S4). Consistent with a lack of remodelling, SWI/SNF was not recruited to RNR3 in the conditional point mutants. Surprisingly, clear differences were observed between the C-terminal truncation mutant and the conditional mutants. Nucleosome eviction was unaffected in rap1 1-716 mutant, and SWI/SNF was recruited in this mutant to a level equal to that in wild-type cells (Figure 3G). The two conditional mutants, rap1-2 and rap1-4, have amino-acid substitutions in the DBD and may affect DNA binding (Kurtz and Shore, 1991), but the rap1 1-716 mutant is expected to retain DNA binding in vivo and may be able to carry out early steps in the activation process. To confirm that the distinct phenotypes of the point mutants and the truncation mutant are related to differences in DNA binding in vivo, we examined the recruitment of two different classes of mutants to RNR3. The results clearly show that the rap1-2 derivative is recruited less well than the wild-type protein and the rap1 1-716 mutant (Figure 5H). These results indicate that the DBD of Rap1 in the absence of amino acids 717–821 is sufficient to recruit the remodelling machinery and promote nucleosome eviction, whereas TBP and RNAPII recruitment requires an intact C terminus. Thus, the data suggest that Rap1 is required for multiple steps in the activation process, and utilizes different domains of the protein to carry out these functions. Interestingly, the phenotypes of the classes of Rap1 mutants suggest that the ability of Rap1 to activate RNR3 correlates better with SWI/SNF recruitment and PIC formation than TAFII recruitment. The significance of this is discussed below.
TFIID- and SWI/SNF-dependent recruitment of Rap1
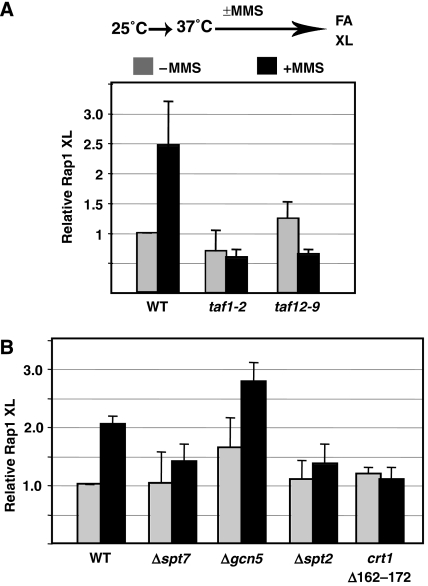
The TAFII subunits of TFIID are required for the recruitment of SWI/SNF and chromatin remodelling of RNR3 (Sharma et al, 2003). We asked if Rap1 recruitment requires TAFIIs and/or chromatin remodelling. A dependence on remodelling would suggest that Rap1 is required for maintaining the remodelled state. We tested this by examining the crosslinking of Rap1 in taf1-2 and taf12-9 temperature-sensitive mutants. These mutants were shown to block PIC formation (TBP, TAFII and RNAPII recruitment) and chromatin remodelling at RNR3 (Sharma et al, 2003). Figure 6A shows that inactivating TAFIIs by shifting the cells to 37°C prevented Rap1 recruitment. Thus, TAFIIs regulate remodelling, at least in part, by aiding the recruitment Rap1 to the promoter or by stabilizing Rap1. This together with our results that TAFII recruitment occurs in the Rap1 mutants indicates that TFIID is required for stabilizing the binding of gene regulatory proteins to promoters, providing an alternative model for how TAFIIs act as coactivators in vivo. Interestingly, inactivation of a temperature-sensitive mutant of TAF1 in hamster cells reduced the binding of Sp1 to the cyclin D1 promoter in vivo, suggesting that mammalian TFIID can stabilize activator binding (Hilton et al, 2005).
Figure 6.
TFIID and chromatin remodelling is required for Rap1 recruitment. (A) Recruitment of Rap1 in TAF1 and TAF12 temperature-sensitive mutants. Cells were grown at the permissive temperature, shifted to 37°C and treated with MMS for 2.5 h. Formaldehyde was added after quickly adjusting the culture to room temperature. Polyclonal antiserum (yN-18) to Rap1 was used in the IP. Crosslinking was measured over the promoter. (B) Crosslinking in SAGA, SWI/SNF and a derepression-defective mutant of Crt1. Cells were grown and crosslinked at 30°C.
We next examined if chromatin remodelling by SWI/SNF and/or histone acetylation by SAGA is required for Rap1 recruitment. ChIP assays were carried out in a Δsnf2 mutant, which is defective for remodelling of RNR3 (Sharma et al, 2003). The results in Figure 6B clearly show that SWI/SNF and chromatin remodelling are required for Rap1 recruitment. Next, we examined the requirement for SAGA by analysing the recruitment of Rap1 in Δgcn5 and Δspt7 mutants. SAGA acetylates nucleosomes through Gcn5 and delivers TBP to promoters through other subunits (Dudley et al, 1999; Sterner et al, 1999). SPT7, and thus SAGA integrity, is required for the recruitment of Rap1 to RNR3 (Figure 6B). However, GCN5 is not. Further, Rap1 recruitment is slightly higher in the Δgcn5 mutant compared with the other strains. Interestingly, we have previously observed increased SWI/SNF recruitment to RNR3 in the Δgcn5 mutant (Sharma et al, 2003), which suggests a striking correlation between SWI/SNF and Rap1 recruitment.
Crt1 and Rap1 are both required for the activation of RNR3. We characterized the interplay between these two proteins by examining the requirement for the activation function of Crt1 in Rap1 recruitment. We have identified derepression-defective Crt1 mutants that are repression competent but fail to activate transcription and recruit TFIID and SWI/SNF (Zhang and Reese, 2005). The recruitment of Rap1 was analysed in one of these mutants, crt1 Δ162–172, and it was found to be blocked in this mutant (Figure 6B). This suggests that the activation function of Crt1 is required for Rap1 recruitment. Together, the data show that chromatin remodelling is required for Rap1 recruitment, suggesting that Rap1 binds after remodelling has occurred. The expression of Rap1 was not significantly altered in any of the regulatory mutants (Supplementary Figure S5).
Discussion
Rap1 is one of the most abundant and ubiquitous transcription factors in yeast. Most of its functions are related to controlling chromatin structure. In addition to regulating heterochromatin-like states, Rap1 is also known to specify the use of TFIID at promoters. The regulation of TAFII-dependent genes by Rap1 has been examined on constitutive, highly expressed genes. How Rap1 functions with TFIID at repressed genes to achieve a remodelled, active state is not known. We used the repressed and highly inducible RNR3 gene to characterize the roles of Rap1, TFIID and the chromatin remodelling machinery in transcription. We provide evidence that Rap1 utilizes multiple domains to carryout distinct steps in opening chromatin at the promoter and maintaining this state to allow for high levels of transcription. Furthermore, its ability to do so requires TFIID, suggesting reciprocal interactions/actions between a sequence-specific DNA binding protein and a general transcription factor. Finally, we have established that Rap1 recruits SWI/SNF to promoters to remodel chromatin.
Rap1 functions with Crt1 to open promoters
The URS region of RNR3 and Crt1 specifies the TAFII-dependent transcription of this gene (Li and Reese, 2000). Although it was unexpected that a repressor, in particular one that leaves the promoter during activation, could specify the dependence on a coactivator, our subsequent analysis revealed that Crt1 acts as a transient activator (Zhang and Reese, 2005). However, what maintains the remodelled state and PIC formation after Crt1 leaves was unknown. We argue that Rap1 plays such a role, and present a more complete picture of how RNR3 is activated and why it is a TAFII-dependent gene. Based on our work to date, we propose the following model: (1) the binding of the Crt1–Ssn6–Tup1 complex establishes a repressive chromatin state, and DNA damage signals convert Crt1 to a form that causes the release of the corepressor, possibly by phosphorylation; (2) the activation domain of Crt1 is exposed, recruiting TFIID and the SWI/SNF complex; (3) remodelling occurs, and Rap1 is then able to engage the promoter; and (4) Rap1 and TFIID undergo reciprocal interactions to maintain SWI/SNF recruitment and PIC assembly. The C terminus of Rap1 is required for PIC formation and transcription. The DBD of Rap1, additionally, maintains the remodelled state by binding to the promoter to resist the reassembly of nucleosomes over its binding site and to recruit SWI/SNF. The first and second steps of this model are supported by our identification of derepression-defective mutants and a physical interaction between Crt1 and the TFIID and SWI/SNF complexes (Zhang and Reese, 2005). The third step is suggested by our data showing that Rap1 recruitment is impaired in the derepression-defective Crt1 mutant that does not remodel the promoter, and in mutants of SWI/SNF and SAGA. The fourth and final part of the model is consistent with our characterization of the roles of the functional domains of Rap1 in RNR3 activation.
Rap1 regulates chromatin and the expression of genes at diverse locations within the genome. Specificity is achieved by the interactions (physical or functional) between Rap1 and other sequence-specific DNA binding proteins and cofactors. For instance, glycolytic genes require Gcr1/2 and ribosomal genes require Fhl1, Ifh1, Hmo1 and others (Pina et al, 2003). Here, we show that Crt1 functions with Rap1 to regulate DNA damage-inducible genes. It is tempting to speculate that Rap1 performs basically the same function at each of these classes of genes. The gene-specific factors, Crt1 in this case, perform unique functions and respond to cellular signals. A common function of Rap1 at all of its target genes may be to prevent the reassembly of nucleosomes or participate in their eviction. The DBD of Rap1 directly binds to SWI/SNF and the different phenotypes observed in DBD mutants versus C-terminal truncation mutants suggest that this domain of Rap1 is required for nucleosome eviction. Rap1 can bind to a site within a nucleosome (Rossetti et al, 2001) or to a partially remodelled state such as loop or ‘bulge' induced by SWI/SNF (Narlikar et al, 2001), destabilizing it and facilitating eviction. Other activators, such as Gal4, can destabilize a partially remodelled state in vitro (Workman and Kingston, 1992). The maintenance of the remodelled state may also require the recruitment of remodelling factors by the DBD. The DBD of glucocorticoid receptor and EKLF can bind SWI/SNF (Yoshinaga et al, 1992; Kadam et al, 2000). Our model is consistent with the work of others showing that the DBD is sufficient for Rap1 to remodel a nucleosome within the HIS4 promoter by destabilizing the remodelled state, but this does not require SWI/SNF (Yu et al, 2001), so the mechanism at RNR3 is different.
Rap1 contributes to the TAFII dependence of RNR3
The URS of RNR3 mediates the recruitment of Rap1 (Figure 5) and fusing this region of the gene to a TAFII-independent core promoter renders it sensitive to TAFII mutations (Li and Reese, 2000). Our observations agree with those of others correlating Rap1 binding sites with TAFII dependency (Mencia et al, 2002; Garbett et al, 2007). However, recruitment of TFIID by Rap1 cannot fully explain the TAFII dependency of RNR3. We found the very surprising result that TAFIIs are recruited to RNR3 and RPS11B in the Rap1 mutants analysed here. It is not difficult to believe that the rap1 1-716 mutant could recruit TAFIIs through the DBD, but this cannot account for the TAFII recruitment in the rap1-2 mutant. The rap1-2 mutant was not recruited well to RNR3. It is a formal possibility that the rap1-2 mutant associates with RNR3 loosely and does not crosslink well, but this seems unlikely. This suggests that Rap1 is not solely responsible for recruiting TAFIIs to this promoter, or an alternative pathway functions when Rap1 is inactive. Artificially recruiting Rap1 to promoters by fusing it to the DBD of LexA could not drive TAFII recruitment to reporter plasmids, and one interpretation of this result is that TAFIIs cannot be recruited by Rap1 alone (Mencia et al, 2002). We have evidence that TFIID recruitment to RNR3 requires an activation domain in Crt1 (Zhang and Reese, 2005), which may have a role. Alternatively, as mentioned above, Rap1 functions with other DNA binding proteins. One such protein is Hmo1. Hmo1 can bind to TAFIIs (Kasahara et al, 2008), and deletion of HMO1 reduced RNR3 activation, but not as strongly as we observed in Rap1 mutants (data not shown). We have not examined whether the effect of the deletion of HMO1 on RNR3 is direct. Although questions about the role of Rap1 in recruiting TAFIIs to RNR and RP genes remain, it is clear that Rap1 has an important function in coordinating the events leading to activation and maintenance of the promoter in the remodelled state.
Furthermore, Rap1 use cannot fully explain the requirement for TFIID because many Rap1-regulated genes are not TFIID-dominant. TFIID-dependent UASs and core promoters work together (Li et al, 2002; Mencia et al, 2002), and features of both the core promoter and the URS are required for TAFII dependence. Considering our model that Rap1 is required for maintaining the promoter in a remodelled state, we propose that the chromatin structure at the core promoter specifies its requirement for Rap1 and TFIID. Specifically, we have shown that the configuration of the core promoter nucleosome specifies the TFIID dependence of RNR3 (Zhang and Reese, 2007). Disrupting this nucleosome by insertion of polynucleotide tracts bypasses the need for TAFIIs. A feature of core promoters responsive to TAFIIs and Rap1 may be their propensity to configure promoter nucleosomes into precise locations, and the role of Rap1 is to evict this nucleosome by exclusion or recruitment of SWI/SNF. The core promoters of many genes, including RNR3, have conserved AT/AA dinucleotide elements that favour nucleosome positioning (Ioshikhes et al, 2006). Thus, combinations of upstream elements and the architecture of the core promoter nucleosome can specify TAFII dependency.
Rap1 and genomic integrity
The role of Rap1 in regulating genomic integrity by maintaining the lengths of telomeres is well known (Shore, 2001). Here we describe a novel function for Rap1, that is, inducing dNTP pools required for DNA repair. This provides an elegant mechanism for coordinating the increase in dNTPs needed for the extension of telomeres, and detection of telomere length. Shortening of telomeres could result in an increase in free Rap1, driving its binding to weaker binding sites within the RNR genes. Conversely, restoration of telomeres could reduce the amount of available Rap1, tuning down RNR expression. The recruitment of Rap1 to RNR3 is dependent on the checkpoint signalling pathway, and inducing Rap1 expression from the GAL1 promoter does not stimulate RNR expression (not shown). Therefore, checkpoint activation is required for removing the repressor complex, and the increase in Rap1 drives its interaction with RNR3. We have not identified any changes in Rap1 levels or modification after DNA damage (not shown), but we cannot rule out that Rap1 is directly impacted by the checkpoint. There are no conserved Rap1 sites in the URS of RNR3, indicating that it interacts with non-consensus lower affinity sites; thus, binding of Rap1 would be sensitive to changes in its concentration once the repressor was removed. Deleting TUP1 and stimulating cells with environmental cues expand the repertoire of sites recognized by Rap1 throughout the genome (Buck and Lieb, 2006), suggesting precedence. RNR genes need to be activated in response to multiple types of damage. Therefore, telomere shortening cannot be the only signal affecting Rap1 dynamics. DNA damage causes the release of Rap1 and SIR proteins from telomeres (Tsukamoto et al, 1997; Martin et al, 1999; Mills et al, 1999), and redistribution of Rap1 from the telomeres could raise the concentration of free Rap1 in the cell. What is the advantage of this mechanism? Regulation of Rap1 would allow the cell to fine-tune the levels of dNTPs during the checkpoint and allow for the initiation of repression before the complete inactivation of the checkpoint.
Materials and methods
Strains and genetic manipulations
Strains used in this study are listed in Supplementary data. Deletion of genes and epitope tagging were carried out by one-step replacement using PCR-generated cassettes (Brachmann et al, 1998; Longtine et al, 1998). Rap1 deletion mutants were constructed by the plasmid shuffle technique using plasmids described previously (Moretti et al, 1994). RNR3 promoter mutations were constructed in an RNR3-LEU2 replacement cassette by restriction digestion of the plasmid to remove portions of the RNR3 URS (Zhang and Reese, 2007). Details about their construction are available on request. In all experiments, cells were grown in 2% peptone, 1% yeast extract, 20 μg/ml adenine sulphate and 2% dextrose (YPAD) at 30°C. The induced cells were treated at an OD600 of 0.6–1.0 with MMS at a concentration of 0.03% for 2–2.5 h.
RNA isolation and northern blot
RNA isolation and northern blotting were carried out as previously reported (Reese and Green, 2003). Fifteen micrograms of total RNA was separated on 1% formaldehyde agarose gels and transferred to a nylon membrane (GE Biosciences) by capillary blotting. After UV crosslinking and 4-h prehybridization at 65°C, radioactively labelled gene-specific probes were added.
Micrococcal nuclease mapping
Nuclei preparation and MNase mapping were carried out essentially as described (Zhang et al, 2006). The digestion products were detected by southern blotting using a 200-bp probe specific for the PstI fragment of RNR3 locus located at +486/725.
ChIP and nucleosome mapping
ChIP was performed as described in previous publications with minor changes (Sharma et al, 2003). A 100 ml portion of cells was grown in YPAD media to an OD600 of 0.5–1.0 and crosslinked for 15 min at 23°C by the addition of 1% formaldehyde. The induced cells were treated at an OD600 of 0.7 with 0.03% MMS and incubated for 2 h before crosslinking. Extracts were prepared by glass bead disruption and the chromatin was sheared into fragments averaging 200–400 bp using a Bioruptor (Diagenode). Chromatin extracts (100 μl) were incubated with 1–3 μl of anti-H3 antibody (AbCam), anti-TBP, anti-SWI2, anti-TAF1, anti-TAF4, anti-Crt1, anti-myc (9E10, Covance) or 8WG16 monoclonal antibody (Covance) overnight. A 10–20 μl portion of purified anti-Rap1 (yN-18 or y-300 from Santa Cruz Biochemical) was used per IP. Polyclonal antibodies are described by Zhang and Reese (2005). The immunoprecipitated DNA and input DNA were analysed by semiquantitative PCR. Multiple dilutions of input and IP DNA were analysed to confirm that the PCR was conducted in the linear range. The PCR products were analysed on agarose gels, stained with ethidium bromide, scanned with a Typhoon system (Molecular Dynamics) and quantified by ImageQuant software (GE Biosciences). The amplified immunoprecipitated DNA was normalized to the input samples. Oligonucleotides are described by Zhang and Reese (2007). The results are averages and standard deviations from at least three independently prepared extracts and IP conducted at separate times.
Affinity chromatography
Portions of Rap1 were fused in-frame with the GST gene and expressed in Escherichia coli from pGEX-6P (GE Biosciences). WCE were prepared and affinity chromatography was performed as described in a previous publication (Reese et al, 1994). Briefly, WCE were adjusted to 3 mg/ml protein in buffer B(0.2) and passed four times over 0.5 ml GST affinity columns containing 3 mg fusion protein per ml of beads. After washing with buffer B0.2, proteins were eluted with buffer B containing 1.5 M NaCl. Bound proteins were detected by western blotting. Tandem affinity-tagged Snf6 was used to purify SWI/SNF as described (Smith et al, 2003). The binding of purified SWI/SNF to GST–Rap1 was carried out in 150 μl using approximately 500 ng of SWI/SNF and 10 μg of GST fusion protein using a batch assay. Beads were washed three times in B0.2 and the bound proteins were eluted with buffer B containing 1.5 M NaCl.
Supplementary Material
Supplementary Figures and Legends
Supplementary data
Acknowledgments
We thank Dr David Shore for providing the Rap1 mutants and discussions. We acknowledge members of the Reese lab and the Center for Gene Regulation at The Pennsylvania State University for advice and comments on this work. V Mitra Sharma and Hesheng Zhang are acknowledged for constructing strains. Marta Mendoza and Jeong-Seon Kim are acknowledged for constructing promoter mutants in Figure 5. This research was supported by funds provided by the National Institutes of Health (GM58672) to JCR.
References
- Basehoar AD, Zanton SJ, Pugh BF (2004) Identification and distinct regulation of yeast TATA box-containing genes. Cell 116: 699–709 [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132 [DOI] [PubMed] [Google Scholar]
- Buck MJ, Lieb JD (2006) A chromatin-mediated mechanism for specification of conditional transcription factor targets. Nat Genet 38: 1446–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Vescovo V, De Sanctis V, Bianchi A, Shore D, Di Mauro E, Negri R (2004) Distinct DNA elements contribute to Rap1p affinity for its binding sites. J Mol Biol 338: 877–893 [DOI] [PubMed] [Google Scholar]
- Dudley AM, Rougeulle C, Winston F (1999) The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev 13: 2940–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge SJ, Davis RW (1989) Identification of the DNA damage-responsive element of RNR2 and evidence that four distinct cellular factors bind it. Mol Cell Biol 9: 5373–5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbett KA, Tripathi MK, Cencki B, Layer JH, Weil PA (2007) Yeast TFIID serves as a coactivator for Rap1p by direct protein–protein interaction. Mol Cell Biol 27: 297–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MR (2000) TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem Sci 25: 59–63 [DOI] [PubMed] [Google Scholar]
- Grunstein M (1997) Molecular model for telomeric heterochromatin in yeast. Curr Opin Cell Biol 9: 383–387 [DOI] [PubMed] [Google Scholar]
- Hilton TL, Li Y, Dunphy EL, Wang EH (2005) TAF1 histone acetyltransferase activity in Sp1 activation of the cyclin D1 promoter. Mol Cell Biol 25: 4321–4332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Zhou Z, Elledge SJ (1998) The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94: 595–605 [DOI] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF (2004) A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell 13: 573–585 [DOI] [PubMed] [Google Scholar]
- Hurd HK, Roberts JW (1989) Upstream regulatory sequences of the yeast RNR2 gene include a repression sequence and an activation site that binds the RAP1 protein. Mol Cell Biol 9: 5359–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioshikhes IP, Albert I, Zanton SJ, Pugh BF (2006) Nucleosome positions predicted through comparative genomics. Nat Genet 38: 1210–1215 [DOI] [PubMed] [Google Scholar]
- Kadam S, McAlpine GS, Phelan ML, Kingston RE, Jones KA, Emerson BM (2000) Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev 14: 2441–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara K, Ki S, Aoyama K, Takahashi H, Kokubo T (2008) Saccharomyces cerevisiae HMO1 interacts with TFIID and participates in start site selection by RNA polymerase II. Nucleic Acids Res 36: 1343–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Shore D (1991) RAP1 protein activates and silences transcription of mating-type genes in yeast. Genes Dev 5: 616–628 [DOI] [PubMed] [Google Scholar]
- Lee TI, Causton HC, Holstege FC, Shen WC, Hannett N, Jennings EG, Winston F, Green MR, Young RA (2000) Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405: 701–704 [DOI] [PubMed] [Google Scholar]
- Li B, Reese JC (2000) Derepression of DNA damage-regulated genes requires yeast TAF(II)s. EMBO J 19: 4091–4100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Reese JC (2001) Ssn6–Tup1 regulates RNR3 by positioning nucleosomes and affecting the chromatin structure at the upstream repression sequence. J Biol Chem 276: 33788–33797 [DOI] [PubMed] [Google Scholar]
- Li XY, Bhaumik SR, Zhu X, Li L, Shen WC, Dixit BL, Green MR (2002) Selective recruitment of TAFs by yeast upstream activating sequences. Implications for eukaryotic promoter structure. Curr Biol 12: 1240–1244 [DOI] [PubMed] [Google Scholar]
- Lieb JD, Liu X, Botstein D, Brown PO (2001) Promoter-specific binding of Rap1 revealed by genome-wide maps of protein–DNA association. Nat Genet 28: 327–334 [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM (1999) Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell 97: 621–633 [DOI] [PubMed] [Google Scholar]
- McAinsh AD, Scott-Drew S, Murray JA, Jackson SP (1999) DNA damage triggers disruption of telomeric silencing and Mec1p-dependent relocation of Sir3p. Curr Biol 9: 963–966 [DOI] [PubMed] [Google Scholar]
- Mencia M, Moqtaderi Z, Geisberg JV, Kuras L, Struh K (2002) Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol Cell 9: 823–833 [DOI] [PubMed] [Google Scholar]
- Mills KD, Sinclair DA, Guarente L (1999) MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell 97: 609–620 [DOI] [PubMed] [Google Scholar]
- Moretti P, Freeman K, Coodly L, Shore D (1994) Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev 8: 2257–2269 [DOI] [PubMed] [Google Scholar]
- Morse RH (2000) RAP, RAP, open up! New wrinkles for RAP1 in yeast. Trends Genet 16: 51–53 [DOI] [PubMed] [Google Scholar]
- Mulder KW, Winkler GS, Timmers HT (2005) DNA damage and replication stress induced transcription of RNR genes is dependent on the Ccr4–Not complex. Nucleic Acids Res 33: 6384–6392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar GJ, Phelan ML, Kingston RE (2001) Generation and interconversion of multiple distinct nucleosomal states as a mechanism for catalyzing chromatin fluidity. Mol Cell 8: 1219–1230 [DOI] [PubMed] [Google Scholar]
- Negrini S, Ribaud V, Bianchi A, Shore D (2007) DNA breaks are masked by multiple Rap1 binding in yeast: implications for telomere capping and telomerase regulation. Genes Dev 21: 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina B, Fernandez-Larrea J, Garcia-Reyero N, Idrissi FZ (2003) The different (sur)faces of Rap1p. Mol Genet Genomics 268: 791–798 [DOI] [PubMed] [Google Scholar]
- Reese JC, Apone L, Walker SS, Griffin LA, Green MR (1994) Yeast TAFIIS in a multisubunit complex required for activated transcription. Nature 371: 523–527 [DOI] [PubMed] [Google Scholar]
- Reese JC, Green MR (2003) Functional analysis of TFIID components using conditional mutants. Methods Enzymol 370: 415–430 [DOI] [PubMed] [Google Scholar]
- Rossetti L, Cacchione S, De Menna A, Chapman L, Rhodes D, Savino M (2001) Specific interactions of the telomeric protein Rap1p with nucleosomal binding sites. J Mol Biol 306: 903–913 [DOI] [PubMed] [Google Scholar]
- Sanders SL, Jennings J, Canutescu A, Link AJ, Weil PA (2002) Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol Cell Biol 22: 4723–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma VM, Li B, Reese JC (2003) SWI/SNF-dependent chromatin remodeling of RNR3 requires TAF(II)s and the general transcription machinery. Genes Dev 17: 502–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D (2001) Telomeric chromatin: replicating and wrapping up chromosome ends. Curr Opin Genet Dev 11: 189–198 [DOI] [PubMed] [Google Scholar]
- Smith CL, Horowitz-Scherer R, Flanagan JF, Woodcock CL, Peterson CL (2003) Structural analysis of the yeast SWI/SNF chromatin remodeling complex. Nat Struct Biol 10: 141–145 [DOI] [PubMed] [Google Scholar]
- Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, Pacella LA, Winston F, Workman JL, Berger SL (1999) Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol 19: 86–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y, Kato J, Ikeda H (1997) Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature 388: 900–903 [DOI] [PubMed] [Google Scholar]
- Workman JL, Kingston RE (1992) Nucleosome core displacement in vitro via a metastable transcription factor–nucleosome complex. Science 258: 1780–1784 [DOI] [PubMed] [Google Scholar]
- Yoshinaga SK, Peterson CL, Herskowitz I, Yamamoto KR (1992) Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science 258: 1598–1604 [DOI] [PubMed] [Google Scholar]
- Yu L, Sabet N, Chambers A, Morse RH (2001) The N-terminal and C-terminal domains of RAP1 are dispensable for chromatin opening and GCN4-mediated HIS4 activation in budding yeast. J Biol Chem 276: 33257–33264 [DOI] [PubMed] [Google Scholar]
- Zhang H, Reese JC (2007) Exposing the core promoter is sufficient to activate transcription and alter coactivator requirement at RNR3. Proc Natl Acad Sci USA 104: 8833–8838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li B, Reese JC (2006) Isolation of yeast nuclei and micrococcal nuclease mapping of nucleosome positioning. Methods Mol Biol 313: 245–256 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Reese JC (2004a) Redundant mechanisms are used by Ssn6–Tup1 in repressing chromosomal gene transcription in Saccharomyces cerevisiae. J Biol Chem 279: 39240–39250 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Reese JC (2004b) Ssn6–Tup1 requires the ISW2 complex to position nucleosomes in Saccharomyces cerevisiae. EMBO J 23: 2246–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Reese JC (2005) Molecular genetic analysis of the yeast repressor Rfx1/Crt1 reveals a novel two-step regulatory mechanism. Mol Cell Biol 25: 7399–7411 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures and Legends
Supplementary data