Abstract
This review describes recent work in cell separation using micro- and nanoscale technologies. These devices offer several advantages over conventional, macroscale separation systems in terms of sample volumes, low cost, portability, and potential for integration with other analytical techniques. More importantly, and in the context of modern medicine, these technologies provide tools for point-of-care diagnostics, drug discovery, and chemical or biological agent detection. This review describes work in five broad categories of cell separation based on (1) size, (2) magnetic attraction, (3) fluorescence, (4) adhesion to surfaces, and (5) new emerging technologies. The examples in each category were selected to illustrate separation principles and technical solutions as well as challenges facing this rapidly emerging field.
Keywords: cell separation, microfluidics, DEP, FACS, magnetic, MACS, bioMEMS
Introduction
The isolation of pure populations of cells from heterogeneous cell suspensions is an essential part of clinical as well as basic research. The diagnostic test for HIV infection, for example, relies on the separation of human lymphocytes from whole blood. Isolation of pure cell populations is an essential part of research efforts to understand fundamental aspects of the body’s response to injury (Feezor et al 2004; Cobb et al 2005). More recently, with major advances in stem cell biology, the isolation of rare cells has become an active area of research. For example, islet 1+ cells that appear to be resident cardiac progenitor cells have been identified in mouse and human hearts (Laugwitz et al 2005). These cells are present in very small numbers, ~100 islet 1+ cells among 109 heart cells. The progenitors are currently isolated by pre-plating of the heart cell suspension for 1 hour, after which fibroblasts and some islet 1+ cells remain attached to the tissue culture plastic. After 4–6 days in culture, the progenitors make up only 0.5% of the total cell population found in the plates. Development of robust and efficient methods for isolating rare cells such as these would clearly represent a major contribution to stem cell biology.
Miniaturized cell separation devices offer many advantages over conventional separation techniques (eg, density gradient centrifugation), such as small sample volumes, portability, low cost, improved sterility, and potential for integration with analytical techniques. Many of these technologies are derived from the semiconductor industry or from advances in synthetic chemistry.
Cell separation techniques can be broadly classified into two categories: techniques based on size and density, and techniques based on affinity (chemical, electrical, or magnetic). In this review, rather than providing a comprehensive review of all cell separation techniques, we provide representative examples from each category with emphasis on micro- and nanotechnology. Since cells are of the order of microns in size and usually handled in suspensions, nearly all of the methods described here are microfluidic. However, some of these techniques, in particular the magnetic separation techniques, also incorporate nanoscale elements. Purity and throughput are important metrics for any separation process, and it is often impossible to attain high levels of both. The description of each technique includes a discussion of these metrics.
Size-based separation
Several novel size-based separation processes are being employed in micro- and nanoscale devices. These devices are compact, simple, and usually do not require additional energy sources or additional equipment. Furthermore, they are extremely effective for low-throughput small-scale applications. Most of the devices force a fluid containing a heterogeneous particle population through a series of channels or obstacles of varied size. Unlike macroscale size-based separation approaches such as the use of cell strainers, the microscale geometry of the flow channels in these devices ensures that fluid flow is laminar. This, in turn, results in predictable and reproducible cell movement. The main advantage of the size-based approach is that it does not require the presence of cell-specific markers or proteins to achieve separation. Hence this approach can be used to isolate stem cells and other rare cells that do not express known markers. Furthermore, since the devices do not contain any biological markers or proteins, they have long shelf-life and are easily transportable. However, one of the major concerns with this approach is maintaining cell viability as the cells pass through the microfluidic device.
Huang et al (2004) describe a technique that allows for separation of micron-sized particles with a resolution of 0.1 μm. In this approach, individual fluid streams can be made to flow along confined lanes in a deterministic manner simply by allowing them to pass through an array of obstacles. The assumption is that a fluid stream will bifurcate when it encounters an obstacle, and thereby sort the particles it brings with it (Figure 1a). As indicated by the arrows in the figure, the fluid streams travel around the obstacles and become confined to one of three lanes. Depending on the size of the particles in the stream and the obstacle parameters, the stream can enter one of two modes of displacement. If the particles are smaller than the width of the lane they are flowing in, the particles zigzag back and forth around the obstacles (zigzag mode), whereas if they are larger than the lane width, they collide with the obstacles and keep displacing in one direction (displacement mode). By manipulating the obstacle parameters, such as their spacing and diameter, as well as the amount by which they are staggered in each subsequent row of the patterned array, micron-sized particles can be sorted rapidly (in a few minutes) with high resolution and with an uncertainty less than 1% of the particle size, or, of the order of tens of nanometers. Such resolution is similar to that of other sorting techniques such as hydrodynamic chromatography and quasi-elastic laser light scattering, and considerably better than that of techniques such as cell straining and density gradient centrifugation. As a proof of concept, the authors used the technique to sort fluorescent polystyrene spheres, as well as circular plasmid DNA. Some limitations of the technique include the requirement of laminar flow and the possibility of random diffusion between adjacent lanes. Nanofabrication may allow for improved resolution and sorting of supramacromolecules such as viral particles and protein complexes.
Figure 1.
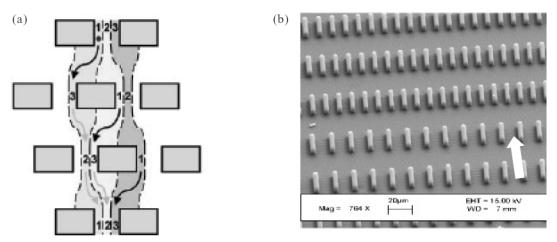
Size-based separation. (a) A stream of fluid flowing at low Re (~10−3) is forced perpendicular to a series of obstacles of defined size and spacing (see arrows). The flow is confined to one of three fluid streamlines or “lanes” (denoted 1, 2, and 3). As shown above, if particles are smaller than the lane width, they continually zigzag between the obstacles, returning to their original lane assignment after traversing several rows of obstacles (zigzag mode). However, when the particles are larger than the lane width, they collide with the obstacles and displace only in one direction (displacement mode – not shown), allowing for separation to occur. Source: Huang LR, Cox EC, Austin RH, et al. 2004. Continuous particle separation through deterministic lateral displacement. Science, 304:987–90.
Reproduced with permission. Copyright © 2004 AAAS. (b) A series of channels of successively smaller width is microfabricated thus creating a cell sieve. The flow is applied parallel to the channels as denoted by the solid arrow. Adapted from Mohamed H, McCurdy LD, Szarowski DH, et al. 2004. Development of a rare cell fractionation device: Application for cancer detection. IEEE Trans Nanobioscience, 3:251–6. Copyright © 2004 IEEE.
Mohamed et al (2004) have designed a device that uses a relatively simple size-based separation approach to isolate rare cells, such as metastatic cancer cells, from peripheral blood. The device contains four successively narrow arrays of parallel channels ranging from 20 μm down to 2.5 μm in thickness (Figure 1b). The device essentially achieves separation by acting like a sieve, trapping larger cells such as mononuclear cells and neuroblastoma cells further upstream in the larger channels, and allowing smaller cells such as erythrocytes to pass through. The authors experimented with several designs of varied channel width and height before identifying the critical dimensions required to trap the cells of interest. The channel width and height were tuned to ensure that the larger, rare cells (in this case, neuroblastoma cells or mononuclear cells) were always trapped while allowing the smaller, bulk cells (in this case, erythrocytes) to always pass through. Thus, the approach can be applied to separation systems of mixed cell populations provided that the two cell types being separated are sufficiently different in size.
To make the process more cost effective, a combination of soft lithography (PDMS, poly(dimethylsiloxane)) and polymers (polyurethanes) were used for fabrication. When cultured neuroblastoma cells were mixed with either plain medium or peripheral blood and passed through the device, the larger neuroblastoma cells were consistently retained in the channels 10 μm wide and 20 μm deep while erythrocytes traversed through the device completely. Similarly, mononuclear cells in a sample of whole blood were trapped in channels 2.5 μm wide and 5 μm deep. While the technique is not without its limitations, including the issue of nonspecific cell adhesion to the channel walls, the technique is more effective in terms of cost and time than conventional cell sorting techniques such as fluorescence-activated cell sorting (FACS) or magnetic-activated cell sorting (MACS).
Besides using a series of obstacles to achieve size-based cell separation, cells can also be separated based on their behavior in laminar flow. Shevkoplyas et al (2005) have developed a biomimetic microfluidic device to facilitate separation of leukocytes from erythrocytes in whole blood samples. The device does not require a power source (except for a small pressure gradient created by a water column to facilitate blood flow), and achieves efficient autoseparation in a single pass using low sample volumes (~μL). It is biomimetic in that it contains a series of bifurcating, rectangular microchannels, emulating the natural structure of blood vessels in vivo. Furthermore, it relies on the natural phenomenon of plasma skimming within a 2D channel to enrich the leukocyte population. When the erythrocytes and leukocytes interact and collide, the erythrocytes, which are smaller and more deformable than leukocytes, preferentially migrate toward the center of the channel, forcing the leukocytes to migrate to the outer wall of the microchannel. By diverting the leukocyte-rich plasma layer through several bifurcations, the authors achieved a 34-fold amplification in the leukocyte-to-erythrocyte ratio compared with the inlet concentration. The device can prove particularly useful in applications where high concentrations of leukocytes or their DNA and RNA are required for analysis.
Differential motility in laminar flow was used by Cho et al (2003) to separate motile sperm cells from non-motile sperm cells. Central to their design is the assumption that viable sperm cells are motile and will therefore migrate against a laminar fluid stream. In contrast, non-motile sperm simply follow the fluid stream lines along with other debris or round cells in the sample. The motile sperm will move across the fluid stream and exit via a different outlet at the bottom of the device, distributing themselves along the width of this outlet, while the non-motile sperm will exit at the outlet situated at the top of the device. The design does not require a power source since it utilizes a height difference between the inlet and outlet as well as surface tension of the fluid stream to create a gravity-driven pump, facilitating a constant fluid flow rate, regardless of the fluid reservoir volume. The authors demonstrated that motile sperm can be purified to levels of nearly 100%. Additionally, the yields of motile sperm at the outlet compared with the inlet were significantly improved and comparable with other conventional separation techniques such as direct swim-up, swim-up from centrifuged pellet, and density gradient separation. Cho et al have proposed several useful applications for the technique, such as home-based screening test for infertility, vasectomy, vasectomy reversal, and sperm toxicology tests. Further improvements to the geometry and configuration of the chip, such as serial connections, may lead to even higher yields.
Fluorescence-based separation
Conventional macroscale FACS systems have emerged as important tools in modern biological laboratories. These systems typically create a hydrodynamically focused stream of cell suspension which is interrogated using a laser beam and an array of photodetectors. Fluorescent labels are attached to one or more cell types in a heterogeneous suspension and the cells are sorted individually based on (a) how they scatter the incident laser light and (b) the wavelength of light that they emit. A recent review by Huh et al (2005) describes a number of microfluidic systems based on this principle. An important advantage of microfluidic FACS systems is their low cost compared with conventional systems. In the context of medicine and point-of-care diagnostics, another attractive characteristic is their small size and portability. Furthermore, microscale FACS systems have been shown to operate comparably to their macroscale analogs in terms of purity and throughput (Wolff et al 2003).
Fu et al (1999) have described a device made of silicone elastomer and glass with embedded platinum electrodes to direct the cells by electro-osmotic flow. An external laser and photomultiplier tube (PMT) interfaced with a computer are used to excite and detect fluorescence. Flow of the cell suspension is controlled by changing the electro-osmotic potentials in response to the PMT signal. The authors operated this device using two algorithms. With the “forward” algorithm, cells that normally flow from the inlet to the waste reservoir are redirected to a collection channel if the fluorescence is above a preset threshold. With the “reverse” algorithm, cells flow at a high rate from the inlet to waste until the detection of fluorescence from a target cell induces the flow to reverse direction to allow a second detection step, followed by flow to the collection channel. The “forward” algorithm is analogous to the mode of operation of most conventional FACS systems, whereas the “reverse” is a novel algorithm that cannot be implemented in conventional systems. A key advantage of the “reverse” algorithm is that it can be used to make more than one measurement on individual cells. This device was used to separate a mixed population of fluorescent and non-fluorescent Escherichia coli cells. In a subsequent publication, the same group (Fu et al 2002) described a more advanced version of this device which consists of integrated microvalves and micropumps. The incorporation of these components “on-chip” allows for the elimination of some macroscopic components and better control of functions such as sample dispensing and recovery, and flushing. With the same sample of mixed cell populations (fluorescent and non-fluorescent mixture of E. coli cells), this device allowed enrichments of up to 90-fold with a throughput of 106 cells/hour.
A further step towards a fully integrated microfluidic FACS system has been taken by Krüger et al (2002). In their device, the laser and detectors are incorporated along with the microfluidic components in a single chip. The only macroscopic parts of this system are a laptop computer, charge-coupled device (CCD) camera, and syringe pumps. Another example of an advanced microfluidic FACS system with integrated waveguides and a cell culture chamber has been described by Wolff et al (2003). With this device, the authors were able to separate fluorescent latex beads from chicken red blood cells at high throughput (12 000 cells/second). This high rate was made possible by a high-speed hydrodynamic valve with a response time of 2.5 milliseconds, which was interfaced with the PMT. The enrichment of fluorescent beads achieved using this device was 100-fold. The authors also developed a second-generation device (Figure 2) designed for improved hydrodynamic focusing and “on-chip” cell culture. This device was used to separate normal yeast cells from yeast cells containing green fluorescent protein. The detected cells were directed to the culture chamber by the high-speed valve and allowed to grow and divide for several days with continuous flow of fresh medium. The incorporation of the culture chamber illustrates how microfluidic FACS systems can reduce the risk of sample loss due to external handling or dead volumes, a feature that would be particularly relevant for the isolation of rare cells.
Figure 2.
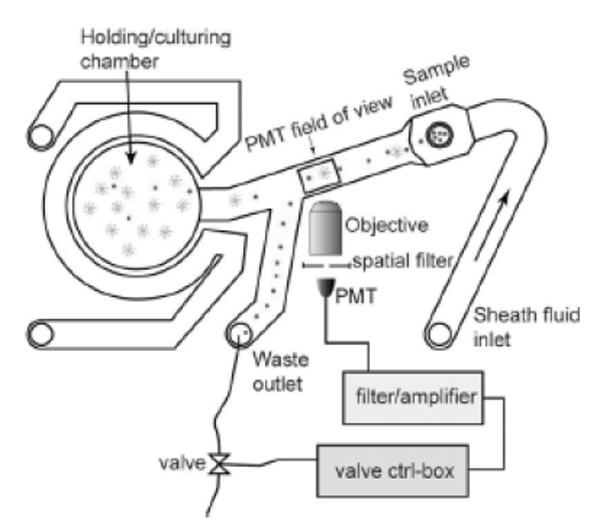
Microfluidic FACS system. Cells flow through a channel into an observation region. Upon detection of a target cell, a high-speed hydrodynamic valve switches fluid flow to send the cell into the holding/culturing chamber. Source: Wolff A, Perch-Nielsen IR, Larsen UD, et al. 2003. Integrating advanced functionality in a microfabricated high-throughput fluorescent-activated cell sorter. Lab Chip, 3:22–7. Reproduced with permission of The Royal Society of Chemistry. Abbreviations: PMT, photomultiplier tube; FACS, fluorescence-activated cell sorting.
Magnetic separation
Cell separation using commercially available magnetic beads is a common laboratory protocol. The IMag Cell Separation System® (BD Biosciences, San Jose, CA, USA) is one example of several available separation systems. Magnetic beads are coated with antibodies specific for surface antigens of the desired cells. When exposed to a mixed cell population, the magnetic beads attach to the surface of desired cells via antibody–antigen interaction. The desired cell subpopulation can then be separated in the presence of a strong magnetic field either in separation columns or on integrated microchips. Magnetic separation is applicable only to cells that can be separated by specific surface antigens. For cells that are commonly distinguished by intercellular proteins (eg, contractile proteins in myocytes) alternative separation methods have to be utilized.
Magnetic sorting is of particular interest since it can easily be miniaturized and utilized in applications where small sample volume is required. This is the main advantage of microscale magnetic separation over the well established and efficient macroscale analogs. The major challenge in the design of microscale magnetic separation systems, which is the focus of most studies, is achieving efficient separation in a continuous (flow through) system. While several examples of microscale magnetic separation systems have been described in recent years, macroscale systems remain more attractive in terms of purity (> 95%) and throughput (~ 1011 cells/hour) (Thiel et al 1998).
Deng et al (2002) have developed a magnetic micro-filtration system for separation of magnetic beads from non-magnetic beads with up to 95% efficiency. In this set-up, nickel posts 10 μm in diameter were positioned in a microfluidic channel. A heterogeneous suspension of beads on the order of 1 μm was flowed into the channel in the presence of an external magnetic field that magnetized the nickel posts. As a result, the magnetic beads adhered to the posts while nonmagnetic beads were removed by the fluid flow. Upon removal of the external magnet and while keeping flow velocity unchanged, the magnetic beads were removed from the microfluidic channel.
A heterogeneous particle population can be separated using a combination of flow and magnetic field in the process called “magnetophoresis” (Pamme and Manz 2004). In this work, a particle suspension was pumped into a laminar flow chamber with several outlets. A nonhomogeneous magnetic field was applied perpendicular to the direction of the flow. Depending on the particle size and magnetic properties, the particles were deflected into the magnetic field and carried into different outlet channels. Although the two systems described were not tested with cells, they appear very promising for cell separation applications.
The combination of magnetic microbeads and external magnetic field was utilized in a Y-shaped microfluidic device by Furdui and Harrison (2004) to separate immunogenic T cells from whole blood with an efficiency of up to 40% (Figure 3a). In the first step, paramagnetic beads coated with Protein-A/anti-human CD3 were flowed into the micro-device and immobilized using an external magnet. Subsequently, whole blood was flowed over the bed of magnetic beads, resulting in the capture of T cells. Upon removal of the external magnet, the T cells were flushed out of the microfluidic device and utilized for PCR analysis. While this purity appears low in comparison with standard methods, such as separation using antibody-coated erythrocytes in conjunction with Ficoll-Hypaque density gradient centrifugation (> 90% efficiency; Wilson et al 1976), the microfluidic approach has the advantage of being easy to integrate with downstream analysis systems.
Figure 3.
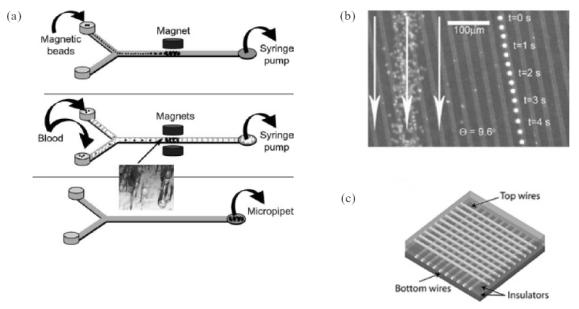
Magnetic cell separation and sorting. (a) Y-shaped device used to separate immunogenic T cells from whole blood in three steps. Paramagnetic Protein A/anti-CD3-coated paramagnetic beads are flowed into the microdevice and immobilized using an external magnet. Subsequently, whole blood is introduced over the bed of magnetic beads, resulting in the capture of T cells. Upon removal of the external magnet, the T cells are flushed out of the microfluidic channel. Source: Furdui VI, Harrison DJ. 2004. Immunomagnetic T cell capture from blood for PCR analysis using microfluidic systems. Lab Chip, 4:614–8. Reproduced with permission of The Royal Society of Chemistry. (b) Combination of fluid flow (arrows) and ferromagnetic strips used to separate leukocytes on planar surfaces. Bright dots: time-lapse image of a tagged leukocyte moving along a ferromagnetic strip (left). Untagged red blood cells are moving along the direction of the fluid flow (right). Source: Inglis DW, Riehn R, Austin RH, et al. 2004. Continuous microfluidic immunomagnetic cell separation. Appl Phys Lett, 85:5093–5. Reproduced with permission. Copyright © 2004 American Institute of Physics. (c) Schematic diagram of a microelectromagnetic matrix for cell manipulation. The matrix consists of two wire meshes superimposed at 180°. Source: Lee H, Purdon AM, Westervelt RM. 2004. Manipulation of biological cells using a microelectromagnet matrix. Appl Phys Lett, 85:1063–5. Reproduced with permission. Copyright © 2004 American Institute of Physics.
Magnetic cell separation can also be achieved on planar surfaces, as demonstrated by Inglis et al (2004). Leukocytes were separated from red blood cells by flowing the blood over the surface coated with ferromagnetic strips. The cells were labeled with anti-CD45 conjugated superparamagnetic nanoparticles (20–100 nm diameter) such that each positive cell carried around 5000 particles. The strips were positioned at an angle of 9.6° relative to the direction of fluid flow, causing the deflection of labeled cells (Figure 3b).
Magnetic labeling can also potentially enable precise manipulation and positioning of cells at the desired location. To prove this principle, Lee et al (2004) constructed a microelectromagnetic matrix by positioning two wire meshes on top of one another in a cross-pattern (Figure 3c). The electric current in each wire was independently controlled via a computer, enabling precise control of the magnetic field at each point in the grid. With this set-up, the authors were able to separate nonviable yeast cells from viable cells by moving the latter individually to a distant location within the grid in a controlled fashion. The solution containing the live/dead-stained, magnetic bead-bound yeast cells was introduced into the microfluidic channel, integrated with the wire mesh. One viable yeast cell and two nonviable cells were initially trapped by a single magnetic field peak. Subsequently, the single peak was split into two smaller peaks; one of the peaks held the nonviable cells and the other one held the viable cell. The entire process was observed by an epifluorescence microscope equipped with a CCD camera. The magnetic peak that held the viable cell was then moved by adjusting the current in the wires, so that the viable cell was moved away from the nonviable cells.
Adhesion-based separation
Adhesion-based cell separation systems are akin to chromatography columns where a mixture is passed through a column packed with beads or other materials capable of binding to the undesired constituents of the feed. In cell separation, antibodies immobilized on surfaces are used for binding. An important advantage of this technique is that it can be used to separate cell populations that have the same size and/or density, such as subpopulations of human lymphocytes. Another advantage of this approach is that there is no need for preprocessing incubation of the starting cell mixture with fluorescent or magnetic antibody tags. This is especially important in separations where it is essential to minimize cell activation.
Conventional macroscale adhesion based systems are described by the term cell affinity chromatography (CAC). These systems can generally provide high purity (> 95%) and high throughput (108–109 cells/hour) (Mandrusov et al 1995; Putnam et al 2003). A major limitation of these systems is their packed bed design, which maximizes the surface area to volume ratio but also results in long residence times (in the order of 1–2 hours) (Mandrusov et al 1995; Putnam et al 2003; Ujam et al 2003). This limitation can be overcome by designing the separation device in the microscale. Microfluidic CAC systems provide high surface areas per unit volume but their small overall volume keeps residence times short (order of minutes or less) (Murthy et al 2004; Sin et al 2005).
Microfluidic CAC systems designed to separate T and B lymphocytes from mixed suspensions have been described by Murthy et al (2004) and Sin et al (2005). These investigations employed microfluidic chambers made of glass and PDMS, whose interior surfaces were functionalized with antibodies capable of binding to surface antigens on the surface of the target cell type. Both investigations used T and B lymphocyte cells lines, Molt-3 and Raji, to create the mixed suspensions, and anti-CD5 and anti-CD19 antibodies to capture the respective cell types. In both reports, the cells captured on the surface were extremely pure (> 97% purity). Nonspecific binding of the undesired cell type was overcome by attaching poly(ethylene glycol) (PEG) on the surface along with the antibody molecules (Murthy et al 2004). In these approaches, the emphasis was on purity rather than throughput. While the number of cells that can be captured was limited by the relatively small surface area of each device, the authors proposed that throughput can be increased by designing a device with larger surface area or by running several devices in parallel.
In the above two reports, the antibodies used for separation were specific to the desired cell type. Anti-CD5 binds to the T cell line but not the B cell line; and anti-CD19 binds to the B cell line but not to the T cell line. This specificity ensures effective separation of the T and B lymphocytes. But what about situations where there is no unique cell surface receptor that distinguishes the target cell type from other cells in the mixed suspension?
Chang et al (2005) have described an adhesion-based microfluidic separation technique that offers a way to address this challenge. This technique emulates the physiological process by which leukocytes from the blood stream are captured by blood vessel walls during an inflammatory response. Blood vessel walls achieve this capture by presenting a variety of adhesion proteins (such as selectins) to the rapidly passing blood cells and selectively capturing only the cells that bear complementary ligands. The authors designed microfluidic devices out of silicon and glass where the capture surfaces were square and rectangular posts arranged in square and offset arrays, respectively. In this respect, their device is similar in design to that of Huang et al (2004) and offers an interesting comparison even though the mode of separation is different (adhesion-based in Chang et al and size-based in Huang et al). Chang et al coated the post surfaces with E-selectin IgG molecules and examined cell capture using HL-60 and U-937 myeloid cell lines, both of which express ligands for E-selectin. With the help of flow experiments as well as finite volume simulations, the authors observed that the movement of cells through their device occurs in three phases: (1) rolling, (2) transient capture on the trailing surface of the posts, followed by (3) detachment and subsequent flow. The authors postulate that each of these three phases is influenced by the strength of bonds between the cells and the selectin molecules on the posts. In the square array device, the cells were observed to first roll and attach on a post. They then detached and always moved to the very next post downstream. In the offset array devices, however, the cells traveled arbitrary distances following the capture and release events, not always moving to the very next array. Although this report does not include experiments with mixed cell suspensions containing both cell types, the authors did measure a significant difference in transit time between the two cell types in their square array device: 1.4 ± 0.8 mm/second for HL-60 and 2.7 ± 1.4 mm/second for U-937. This difference could conceivably be utilized to separate cell populations that are indistinguishable in size and surface receptor expression.
Adhesion-based separation can also be performed in a static microfabricated system, as opposed to a fluidic system. Revzin et al (2005) have described the fabrication of microwells made of PEG for the isolation of lymphocyte subpopulations. These wells are functionalized with antibodies and are capable of capturing target cells bearing complementary antigens with high selectivity. CD5+ T lymphocytes (Molt-3 cell line) adhered preferentially to microwells coated with anti-CD5 (95% of the available wells were occupied) and showed only minimal adhesion to microwells coated with avidin and anti-CD19 (well occupancies were 5% and 13%, respectively). With a microwell size of 15 μm × 15 μm, the authors observed that each well contained a single lymphocyte (Figure 4a). The significance of this capability was demonstrated by the selective removal of individual T lymphocytes from an array of microwells by laser capture microdissection (LCM, Figure 4b). LCM is technique whereby individual cells from different array locations are made to adhere to an adhesive film placed on top of the substrate surface by a pulsed and focused laser beam. The adhesive film is mounted on a cap that is subsequently removed from the substrate surface. The cell contents can then be extracted for downstream molecular analysis. The microwell platform offers a simple and effective method of isolating cell subpopulations based on antigen expression. When combined with LCM extraction, this technology could potentially be employed to extract useful proteomic or genomic data from the selected cells.
Figure 4.
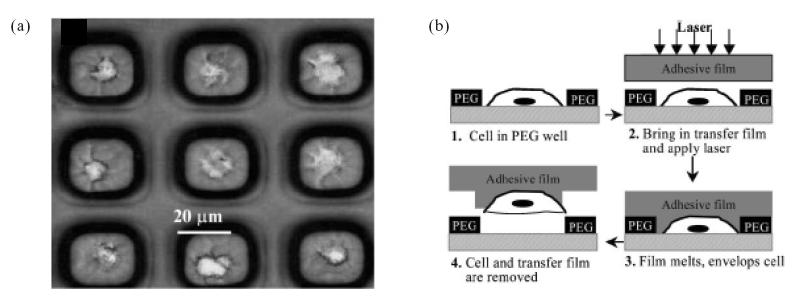
(a) Poly(ethylene glycol) (PEG) microwells containing individual T and B lymphocytes. This type of capture allows subsequent extraction of individual cells by laser capture microdissection (shown schematically in (b)). Source: Revzin A, Sekine K, Sin A, et al. 2005. Development of a microfabricated cytometry platform for characterization and sorting of individual leukocytes. Lab Chip, 5:30–7. Reproduced with permission of The Royal Society of Chemistry.
Electrophoretic separation
Most electrophoretic separation processes employ a process known as dielectrophoresis (DEP), whereby particles with an induced electric polarization, such as dipoles, quadrupoles, and octopoles, can be trapped within a nonuniform electric field. Negative DEP forces cause polarized objects to be repelled away from an electrode, while positive DEP forces attract objects toward an electrode. Since DEP depends on the existence of a gradient in an electric field, an alternating current (AC) voltage is usually employed. The response of a cell to DEP-mediated forces is influenced by cell type, its density, and even its metabolic and physiologic state, making these important parameters for separation. There are essentially three variations of DEP currently being employed for micro- and nanoscale devices: DEP retention, DEP migration, and DEP-field flow fractionation (DEP-FFF). DEP retention (or DEP affinity) is used to hold cells against a fluid flow, so that cells only weakly influenced by DEP forces are carried along with the fluid, while cells held more strongly by DEP forces can be eluted later. DEP migration is similar to DEP retention, but instead it utilizes DEP to cause cells to migrate to different regions of an electrode. Finally, DEP-FFF uses DEP forces to differentially levitate cells against gravity in a fluid flow profile, such as a parabolic flow profile. This causes cells with different dielectric properties to travel at different velocities along the channel. Like magnetic separation, the primary advantage of microscale electrophoretic separation systems is the ability to handle small sample volumes.
Voldman et al (2002) have developed a microfabricated separation system capable of trapping and sorting single cells on the basis of their response to certain stimuli. This device uses DEP retention on individual cells against a fluid flow profile. The authors describe it as a microfabrication-based dynamic array cytometer (μDAC), since it allows cells to be physically arrayed into DEP-based traps, and dynamically probed and sorted thereafter. An array of DEP traps, each comprising four trapezoidally arranged electrodes of opposite polarity, was fabricated to create a non-uniform, quadrupole electric field (Figure 5a). An extruded geometry was used to ensure maximal trapping force. Cells became electrically polarized in the presence of the quadrupole field and were physically trapped within a potential energy well against a fluid flow (Figure 5b). A high frequency (~MHz) AC field was chosen to minimize power dissipation and heat-induced damage to the cells, to prevent transmembrane polarization of cells, and also to minimize corrosion of the electrodes. The cells (HL-60 cell line) were fed into the chip via a reservoir, and then exposed to a stimulus (calcein). They were then individually loaded into the traps and optically interrogated over time for their response to the stimulus (calcein loading) by optical microscopy. Each of the traps was electrically addressable, allowing for manipulation and release of a single cell or multiple cells simultaneously. Thus, based on their responses, the cells could be sorted by turning on or off each individual trap. The authors demonstrated this capability in calcein-labeled HL-60 cells as a proof-of-concept of the technique. A limitation that the authors point out in their μDAC system is that in some cases, more than one cell became confined in a trap.
Figure 5.
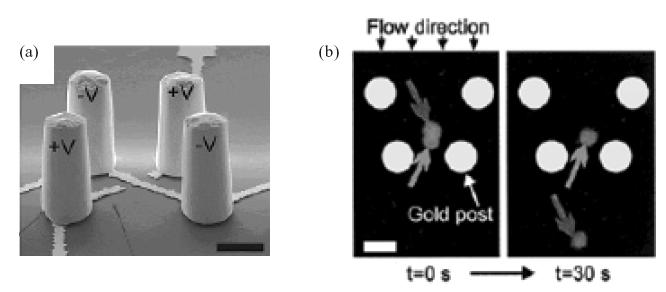
Cell separation using dielectrophoretic traps. (a) A pseudo-colored SEM image showing a dioelectrophoresis (DEP) trap consisting of four trapezoidaly arranged gold electrodes. This configuration induces a dipole moment in the cell in the opposite direction to the electric field. The cell is repelled from the field and stably trapped at the quadrupole’s field minimum. (b) Two cells are loaded into the trap at low flow rate. The application of higher flow rate results in the ejection of one cell (dark grey arrow) from the trap leaving the other cell behind (grey arrow). Source: Voldman J, Gray ML, Toner M, et al. 2002. A microfabrication-based dynamic array cytometer. Anal Chem, 74:3984–90. Reproduced with permission. Copyright © 2002 American Chemical Society.
Huang et al (1999) used DEP-FFF to separate cultured human breast cancer MDA-435 cells from CD34+ hematopoietic stem cells. The device consisted of three layers, including a patterned layer of eight interdigitated electrodes 50 μm in width and spacing, a top glass layer, and a Teflon spacer in between comprising the separation chamber. A syringe pump was used to create a constant flow through the channel, forming a parabolic velocity profile. Since the heights attained in the fluid channel are both a function of the cell dielectric properties and the frequency of the applied voltage, the two cell populations were individually subjected to DEP-FFF over a range of frequencies to obtain information about their elution characteristics as a function of frequency. These initial experiments showed that MDA-435 cells took much longer to elute and even became trapped near the electrodes above 20 kHz, while the CD34+ cells eluted more quickly and completely over a broader range of frequencies. As such, two different frequency protocols were formulated for separation. The first was a trap-and-release protocol, which utilized a fixed, 40 kHz frequency to trap the MDA-435 cells near the electrode, while the second was a frequency sweep between 15 and 35 kHz, which allowed the MDA-435 cells to be levitated slightly, allowing them to flow at a lower speed down the channel. Mixtures of the two cell types were then introduced into the chamber via an injection valve and allowed to equilibrate to different heights in the fluid channel for 5 minutes under an AC voltage of 10 V peak-to-peak. The cells were then subjected to either of the frequency protocols described above for 7 minutes to allow the CD34+ cells to completely elute. The frequency was then switched to 5 kHz to allow the MDA-435 cells to completely elute. The results showed that in both protocols, CD34+ cells were eluted with purity greater than 99% between 3 and 5 minutes, while MDA-435 cells were eluted with 96% purity between 9 and 12 minutes for the trap-and-release protocol and with 99% purity for the frequency-swept protocol. These levels of purity and throughput are comparable to those of macroscale fluorescence-based and magnetic separation systems and better than that of macroscale electrophoretic systems.
In another study by members of the same group (Yang et al 1999), DEP-FFF was employed for the separation of MDA-435 cells from erythrocytes. The authors found that the elution time depended on both the frequency and voltage of the AC field. Larger voltages and frequencies resulted in larger DEP forces and, accordingly, longer elution times. There was no significant difference in elution time between the two cell types as a function of flow rate, indicating that the height obtained within the velocity field was mainly a function of the DEP forces and not hydrodynamic lift forces created by flow within the channel. In contrast to the previous study, the authors found that MDA-435 cells were levitated up to 15 μm higher than erythrocytes and were eluted nearly twice as fast. At a frequency of 10 kHz and a flow rate of 1 mL/minute, the authors obtained MDA-435 cell fractions greater than 98% in purity between 10 and 12 minutes, and erythrocyte fractions greater than 99% purity between 18 and 26 minutes.
More recently, Huang et al (2003) also developed a DEP-based device to separate several strains of bacteria from erythrocytes, simulating the removal of biological warfare agents from blood. The separation was achieved through DEP migration, since the two cell types migrated to different parts of the microelectrode structure. The device was constructed by sequential lamination of five layers: a patterned polyimide layer consisting of interdigitated DEP electrodes, two pressure-sensitive acrylic adhesive layers, one of which contained microfluidic channels, a poly-carbonate substrate layer at the bottom, and a glass cover plate at the top. Cell suspension samples of 5 μL were loaded into the device and an AC voltage of 10 V peak-to-peak at 10 kHz was applied for 5 minutes. The cells were then washed, at first with the AC voltage on and then with the voltage off, to collect the eluted samples. The authors observed that the bacteria preferentially migrated and attached to the electrodes in the presence of a DEP force. In contrast, blood cells accumulated in the recessed areas between the electrodes, where field strengths were at a minimum. When the three strains of bacteria were combined and mixed with blood, the DEP device was able to simultaneously separate the multiple strains from blood cells. PCR amplification of bacterial strain-specific genes did not reveal bands prior to separation, but revealed strong bands after separation, indicating that these PCR products were previously masked due to the presence of blood components. In general, higher collection efficiencies were obtained for lower initial ratios of bacteria to erythrocytes, suggesting that the separation efficiency of the device is limited by the available surface area of its electrodes to about 1 × 106 bacteria per run.
Other emerging separation technologies
While the above sections encompass most of the available microscale separation techniques, a number of others approaches are under development. One of these approaches is microfluidic cell lysis. The strategy in this approach is to lyse the undesired cell populations in a mixed suspension while leaving behind the desired cells. Sethu et al (2004) have developed a microfluidic device for the separation of leukocytes from erythrocytes by lysis of the latter. This separation is usually performed by macroscale lysis using osmotic lysing agents (such as deionized water, sodium chloride buffer, ammonium chloride–sodium bicarbonate buffer), by density centrifugation, or some combination of macroscale lysis and centrifugation. An important characteristic of macroscale lysis that the limiting step is not the actual cell lysis (which occurs in 20–30 seconds), but rather the diffusion of the lysing agent in the cell suspension, which can increase the time required for complete lysis to over 5 minutes. In the device designed by Sethu et al, blood cells are introduced into a channel that is also fed from either side by channels carrying lysis buffer (Figure 6). The result is that the blood cells flow in a narrow stream (18–36 μm wide) at the middle of the channel with lysis buffer on either side. This arrangement significantly reduces the time required for diffusion of the lysis buffer into the cell suspension, resulting in complete lysis within 28–40 seconds, depending on the ratio of blood to lysis buffer. The reduction in the total time required for this process also has the benefits of avoiding perturbation or damage to leukocytes resulting from prolonged contact with lysing agents or from centrifugation.
Figure 6.
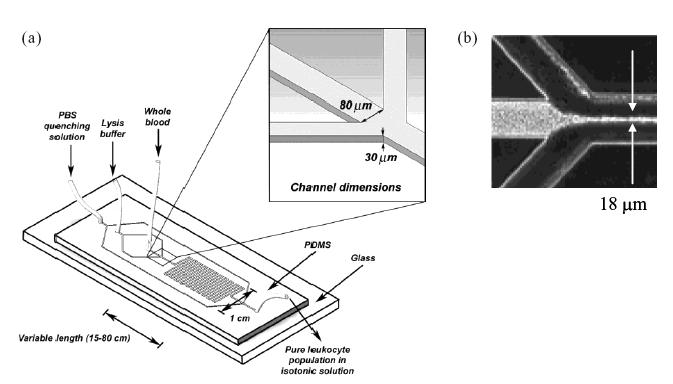
Microfluidic device for red blood cell lysis. (a) A stream of blood cells is induced to flow along the center of the main channel by two adjacent streams of lysis buffer. This narrowing, shown in (b), minimizes the need for lysis buffer diffusion and allows contact with the flowing blood at nearly the single cell level. Source: Sethu P, Anahtar M, Moldawer LL, et al. 2004. Continuous row microfluidic device for rapid erythrocyte lysis. Anal Chem, 76:6247–53. Reproduced with permission. Copyright © 2004 American Chemical Society.
Abbreviations: PBS, phosphate buffered saline; PDMS, poly(dimethylsiloxane).
Sohn et al (2000) found that the capacitance of cells can be related to their DNA content. Using a microfluidic device fabricated from PDMS and glass with embedded electrodes, the capacitance was measured across a portion of the main channel with the cells flowing past one by one. Comparison of capacitance values with measurements of DNA content obtained using conventional techniques yielded a linear graph with different cell types lysing at different points. For example, this technique can distinguish between mammalian erythrocytes (which contain no DNA and therefore have zero capacitance) and leukocytes (whose DNA content, and therefore capacitance, are nonzero).
Gawad et al (2001) have described a microfluidic device that can distinguish between erythrocytes and other cells types by measurements of spectral impedance done on individual cells passing through a channel. Spectral impedance of an individual cell is a function of a number of variables, including cytoplasm conductance, cell size, and membrane capacitance. This device was fabricated with polyimide and glass, and contains integrated channels and electrodes.
Summary
The broad spectrum of cell separation technologies described in this review illustrates the high level of interest and activity in this area. The described size- and density-based approaches offer a great potential for separation of cell subpopulations for which specific markers are not known or cannot be used (eg, to prevent cell activation). Affinity-based approaches (fluorescence-, magnetic-, adhesion-based, and electrophoretic) can be employed for fast (~minutes) and continuous separation with high specificity (~99%). For all of the approaches, the design of the devices is such that they can be operated in a massively parallel fashion to increase scale and throughput without compromising purity and efficacy. Furthermore, micro-fluidic separation systems can be easily incorporated with devices that perform downstream analysis such as single-cell lysis (Irimia et al 2004) and proteomic and genomic analysis (Huang et al 2002; Hashimoto et al 2005; Parano et al 2005; Situma et al 2005). Given the advanced level of design, fabrication, and measurement capabilities, we expect that the focus in the coming years will shift from “proof-of-concept” prototypes to devices that can be economically produced and easily operated in applications such as point-of-care clinical diagnostics, drug discovery, and chemical–biological agent detection.
References
- Chang WC, Lee LP, Liepmann D. Biomimetic technique for adhesion-based collection and separation of cells in a microfluidic channel. Lab Chip. 2005;5:64–73. doi: 10.1039/b400455h. [DOI] [PubMed] [Google Scholar]
- Cho BS, Schuster TG, Zhu XY, et al. Passively driven integrated microfluidic system for separation of motile sperm. Anal Chem. 2003;75:1671–5. doi: 10.1021/ac020579e. [DOI] [PubMed] [Google Scholar]
- Cobb JP, Mindrinos MN, Miller-Graziano C, et al. Application of genome-wide expression analysis to human health and disease. Proc Natl Acad Sci U S A. 2005;102:4801–6. doi: 10.1073/pnas.0409768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng T, Prentiss M, Whitesides GM. Fabrication of magnetic microfiltration systems using soft lithography. Appl Phys Lett. 2002;80:461–3. [Google Scholar]
- Feezor RJ, Baker HV, Mindrinos M, et al. Whole blood and leukocyte RNA isolation for gene expression analyses. Physiol Genomics. 2004;19:247–54. doi: 10.1152/physiolgenomics.00020.2004. [DOI] [PubMed] [Google Scholar]
- Fu AY, Chou HP, Spence C, et al. An integrated microfabricated cell sorter. Anal Chem. 2002;74:2451–7. doi: 10.1021/ac0255330. [DOI] [PubMed] [Google Scholar]
- Fu AY, Spence C, Scherer A, et al. A microfabricated fluorescence-activated cell sorter. Nat Biotechnol. 1999;17:1109–11. doi: 10.1038/15095. [DOI] [PubMed] [Google Scholar]
- Furdui VI, Harrison DJ. Immunomagnetic T cell capture from blood for PCR analysis using microfluidic systems. Lab Chip. 2004;4:614–18. doi: 10.1039/b409366f. [DOI] [PubMed] [Google Scholar]
- Gawad S, Schild L, Renaud P. Micromachined impedance spectroscopy flow cytometer for cell analysis and particle sizing. Lab Chip. 2001;1:76–82. doi: 10.1039/b103933b. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Hupert ML, Murphy MC, et al. Ligase detection reaction/hybridization assays using three-dimensional microfluidic networks for the detection of low-abundant DNA point mutations. Anal Chem. 2005;77:3243–55. doi: 10.1021/ac048184d. [DOI] [PubMed] [Google Scholar]
- Huang LR, Cox EC, Austin RH, et al. Continuous particle separation through deterministic lateral displacement. Science. 2004;304:987–90. doi: 10.1126/science.1094567. [DOI] [PubMed] [Google Scholar]
- Huang Y, Yang J, Wang XB, et al. The removal of human breast cancer cells from hematopoietic CD34(+) stem cells by dielectrophoretic field-flow-fractionation. J Hematother Stem Cell Res. 1999;8:481–90. doi: 10.1089/152581699319939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Mather EL, Bell JL, et al. MEMS-based sample preparation for molecular diagnostics. Anal Bioanal Chem. 2002;372:49–65. doi: 10.1007/s00216-001-1191-9. [DOI] [PubMed] [Google Scholar]
- Huang Y, Yang JM, Hopkins PJ, et al. Separation of simulants of biological warfare agents from blood by a miniaturized dielectrophoresis device. Biomed Microdevices. 2003;5:217–25. [Google Scholar]
- Huh D, Gu W, Kamotani Y, et al. Microfluidics for flow cytometric analysis of cells and particles. Physiol Meas. 2005;26:R73–98. doi: 10.1088/0967-3334/26/3/R02. [DOI] [PubMed] [Google Scholar]
- Inglis DW, Riehn R, Austin RH, et al. Continuous microfluidic immunomagnetic cell separation. Appl Phys Lett. 2004;85:5093–5. [Google Scholar]
- Irimia D, Tompkins RG, Toner M. Single-cell chemical lysis in picoliter-scale closed volumes using a microfabricated device. Anal Chem. 2004;76:6137–43. doi: 10.1021/ac0497508. [DOI] [PubMed] [Google Scholar]
- Krüger J, Singh K, O’Neill A, et al. Development of a microfluidic device for fluorescence activated cell sorting. J Micromech Microeng. 2002;12:486–94. [Google Scholar]
- Laugwitz KL, Moretti A, Lam J, et al. Postnatal isl1+cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–53. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Purdon AM, Westervelt RM. Manipulation of biological cells using a microelectromagnet matrix. Appl Phys Lett. 2004;85:1063–5. [Google Scholar]
- Mandrusov E, Houng A, Klein E, et al. Membrane-based cell affinity-chromatography to retrieve viable cells. Biotechnol Prog. 1995;11:208–13. doi: 10.1021/bp00032a013. [DOI] [PubMed] [Google Scholar]
- Mohamed H, McCurdy LD, Szarowski DH, et al. Development of a rare cell fractionation device: Application for cancer detection. IEEE Trans Nanobioscience. 2004;3:251–6. doi: 10.1109/tnb.2004.837903. [DOI] [PubMed] [Google Scholar]
- Murthy SK, Sin A, Tompkins RG, et al. Effect of flow and surface conditions on human lymphocyte isolation using microfluidic chambers. Langmuir. 2004;20:11649–55. doi: 10.1021/la048047b. [DOI] [PubMed] [Google Scholar]
- Pamme N, Manz A. On-chip free-flow magnetophoresis: Continuous flow separation of magnetic particles and agglomerates. Anal Chem. 2004;76:7250–6. doi: 10.1021/ac049183o. [DOI] [PubMed] [Google Scholar]
- Panaro NJ, Lou XJ, Fortina P, et al. Micropillar array chip for integrated white blood cell isolation and PCR. Biomol Eng. 2005;21:157–62. doi: 10.1016/j.bioeng.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Putnam DD, Namasivayam V, Burns MA. Cell affinity separations using magnetically stabilized fluidized beds - erythrocyte subpopulation fractionation utilizing a lectin-magnetite support. Biotechnol Bioeng. 2003;81:650–65. doi: 10.1002/bit.10511. [DOI] [PubMed] [Google Scholar]
- Revzin A, Sekine K, Sin A, et al. Development of a microfabricated cytometry platform for characterization and sorting of individual leukocytes. Lab Chip. 2005;5:30–7. doi: 10.1039/b405557h. [DOI] [PubMed] [Google Scholar]
- Sethu P, Anahtar M, Moldawer LL, et al. Continuous row microfluidic device for rapid erythrocyte lysis. Anal Chem. 2004;76:6247–53. doi: 10.1021/ac049429p. [DOI] [PubMed] [Google Scholar]
- Shevkoplyas SS, Yoshida T, Munn LL, et al. Biomimetic autoseparation of leukocytes from whole blood in a microfluidic device. Anal Chem. 2005;77:933–7. doi: 10.1021/ac049037i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin A, Murthy SK, Revzin A, et al. Enrichment using antibody-coated microfluidic chambers in shear flow: model mixtures of human lymphocytes. Biotechnol Bioeng. 2005;91:816–26. doi: 10.1002/bit.20556. [DOI] [PubMed] [Google Scholar]
- Situma C, Wang Y, Hupert M, et al. Fabrication of DNA microarrays onto poly(methyl methacrylate) with ultraviolet patterning and microfluidics for the detection of low-abundant point mutations. Anal Biochem. 2005;340:123–35. doi: 10.1016/j.ab.2005.01.044. [DOI] [PubMed] [Google Scholar]
- Sohn LL, Saleh OA, Facer GR, et al. Capacitance cytometry: Measuring biological cells one by one. Proc Natl Acad Sci U S A. 2000;97:10687–90. doi: 10.1073/pnas.200361297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel A, Scheffold A, Radbruch A. Immunomagnetic cell sorting - pushing the limits. Immunotechnology. 1998;4:89–96. doi: 10.1016/s1380-2933(98)00010-4. [DOI] [PubMed] [Google Scholar]
- Ujam LB, Clemmitt RH, Clarke SA, et al. Isolation of monocytes from human peripheral blood using immuno-affinity expanded-bed adsorption. Biotechnol Bioeng. 2003;83:554–66. doi: 10.1002/bit.10703. [DOI] [PubMed] [Google Scholar]
- Voldman J, Gray ML, Toner M, et al. A microfabrication-based dynamic array cytometer. Anal Chem. 2002;74:3984–90. doi: 10.1021/ac0256235. [DOI] [PubMed] [Google Scholar]
- Wilson BS, Teodorescu M, Dray S. Enumeration and isolation of Rabbit T-lymphocyte and B-lymphocyte by using antibody-coated erythrocytes. J Immunol. 1976;116:1306–12. [PubMed] [Google Scholar]
- Wolff A, Perch-Nielsen IR, Larsen UD, et al. Integrating advanced functionality in a microfabricated high-throughput fluorescent-activated cell sorter. Lab Chip. 2003;3:22–7. doi: 10.1039/b209333b. [DOI] [PubMed] [Google Scholar]
- Yang J, Huang Y, Wang XB, et al. Cell separation on microfabricated electrodes using dielectrophoretic/gravitational field flow fractionation. Anal Chem. 1999;71:911–18. doi: 10.1021/ac981250p. [DOI] [PubMed] [Google Scholar]


