Abstract
Small interfering RNA molecules (siRNA) hold great promise to specifically target cytoprotective factors to enhance cancer therapy. Like antisense RNA strategies, however, the use of siRNA is limited because of in vivo instability. As a first step to overcome delivery issues, a series of graft copolymers of polyethylene glycol and polyethylenimine (PEI-g-PEG) were synthesized and investigated as nontoxic carriers for delivery of siRNA targeting the signaling peptide of secretory clusterin (sCLU), a prosurvival factor that protects cells from ionizing radiation (IR) injury, as well as chemotherapeutic agents. Three copolymers with different PEG grafting densities were tested for their abilities to bind and form nanocomplexes with siRNA. A copolymer composed of 10 PEG grafts (2 kDa each) per PEI polymer (2k10 copolymer) gave the highest binding affinity to siRNA by ethidium bromide exclusion assays, and had the smallest nanocomplex size (115 ± 13 nm diameter). In human breast cancer MCF-7 cells, 2k10–siRNA-sCLU nanocomplexes suppressed both basal as well as IR-induced sCLU protein expression, which led to an over 3-fold increase in IR-induced lethality over 2k10–siRNA scrambled controls. In summary, this study demonstrates the proof-of-principle in using nanoparticle-mediated delivery of specific siRNAs to enhance the lethality of IR exposure in vitro, opening the door for siRNA-mediated knockdown of specific cytoprotective factors, such as DNA repair, antiapoptotic, free radical scavenging, and many other proteins.
Keywords: siRNA delivery, polyethylenimine, nanomedicine, secretory clusterin, cancer radiotherapy
Introduction
Small interfering RNA (siRNA) therapy has shown significant therapeutic promise against a variety of diseases such as cancer, HIV, and influenza (Brummelkamp et al 2002b; Novina et al 2002; Ge et al 2004). The ability of siRNA to quickly suppress protein expression in an efficient and highly specific manner makes it a prime candidate for medical applications. This promise is hampered, however, by in vivo siRNA instability, and the fact that agents used for in vitro siRNA delivery tend to be too toxic for in vivo applications. Currently, the most common delivery method of siRNA for in vitro applications is the use of cationic lipids, such as lipofectamine (Elbashir et al 2002). While this system does provide reasonable transfection efficiencies in cell culture in vitro, lipofectamine and its related cationic lipids are, in general, too toxic for use in vivo. One solution to this problem is to use DNA plasmids that direct the synthesis of siRNA. These plasmids have the advantage of inducing more persistent gene suppression, as well as being able to be incorporated into viral vectors (Brummelkamp et al 2002a; Devroe and Silver 2004). Unfortunately, these viral vectors exhibit problems of eliciting immune responses in vivo that hamper their safety for clinical applications.
Cationic polymers have gained prominence in nonviral DNA delivery (De Smedt et al 2000), although their use in siRNA delivery is much more recent. Of these polymers, polyethylenimine (PEI) contains primary, secondary, and tertiary amines, giving this polymer the unique abilities to complex DNA and serve as a low pH (4–5) buffer (Suh et al 1994). Owing to this buffering capacity or “proton sponge” effect, PEI has shown higher transfection efficiency than other cationic polymers (Boussif et al 1995). Recently, Ge et al (2004) reported the use of PEI–siRNA complexes for the treatment of influenza in mice. However, in vivo toxicity has been noted for PEI upon intravenous administration. To reduce PEI toxicity, polyethylene glycol (PEG) was grafted onto PEI, greatly reducing toxicity for the resulting gene delivery complexes (Kunath et al 2002). The PEI component of the copolymer allows for complexation with polynucleotides, and increases endosomal release of the nanocomplexes into the cytoplasm. The PEG component not only reduces toxicity of the PEI component, but also stabilizes the resulting nanocomplex. This modified vector was recently used to deliver siRNA targeting the vascular endothelial growth factor (VEGF) for treatment of angiogenic tumors (Schiffelers et al 2004). To date, polymer-mediated delivery of siRNA has not been used in tandem with radiotherapy.
The clusterin (CLU) gene encodes 2 proteins (1 secreted [sCLU]; 1 intracellular and nuclear [nCLU]) through 2 separate and unique mRNAs. Full-length CLU mRNA encodes sCLU protein, a secreted glycoprotein that was discovered in 1983 from ram testes fluid (Izawa 1998). This protein was subsequently found to be involved in many processes, including Alzheimer’s disease (Laping et al 1994), tissue regeneration (Little and Mirkes 1995), aging, and cancer (Peatfield and Boothman 1991; Welsh 1994; Betuzzi 2003). The CLU gene can be activated in response to many different cell stresses, including ionizing radiation (IR).
Although nCLU has been shown to induce cell death (Yang et al 2000; Klokov et al 2004), sCLU is cytoprotective, possibly because of its chaperone-like function (Criswell et al 2003; Klokov et al 2004). sCLU protein levels are suppressed by p53, resulting in low expression in normal human cells, but are expressed at high levels in cancers with altered p53 function. Since this protein confers IR resistance and is overexpressed in many cancers, including breast, prostate, colon, and esophageal (Welsh 1994; Gleave et al 2001; July et al 2004), sCLU is a prime target for cancer therapy. In fact, phase I clinical trials using antisense oligonucleotide therapy against sCLU are currently ongoing (Gleave et al 2001). Targeting sCLU protein levels using antisense oligonucleotide therapy can increase the lethality of treatments using IR, as well as other chemotherapeutic agents (Zellweger et al 2003).
In this report, a grafted PEG-g-PEI copolymer was used to mediate delivery of siRNA against sCLU (siRNAsCLU). The resulting polymer–siRNA-sCLU nanocomplexes effectively suppressed sCLU protein expression, which led to significantly enhanced IR lethality in MCF-7 cells. The enhanced efficacy of this treatment strategy makes it a promising candidate for target-specific cancer radiotherapy.
Materials and methods
Materials
Branched polyethylenimine (PEI, MW 10 kDa) was purchased from Polysciences (Warrington, PA, USA). Methoxy N-hydroxysuccinimide polyethylene glycol (MeO-PEG-NHS, 2.0 kDa) was purchased from Nektar Therapeutics (San Carlos, CA, USA). Lipofectamine was purchased from Invitrogen (Carlsbad, CA, USA). Deionized water was treated with diethyl pyrocarbonate (DEPC) and autoclaved prior to use. Antibodies to human sCLU (B5) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA) and α-tubulin antibodies were obtained from Calbiochem (La Jolla, CA, USA). The CellTiter 96 Non-Radioactive Cell Proliferation Assay Kit was obtained from Promega (Madison, WI).
Cell culture
Human breast cancer MCF-7 cells were cultured in RPMI 1640 medium containing 5% fetal bovine serum, 2 mM glutamine, and 100 IU/mL penicillin–streptomycin in a 95% air/5% CO2 humidified atmosphere at 37°C. MCF-7 cells were free from mycoplasma contamination. All cell culture reagents were purchased from BioWhittaker (Walkersville, ME, USA).
siRNA oligomers
The double-stranded siRNA 21-mers used for the complexation studies were purchased from Qiagen (Valencia, CA, USA). In addition, double-stranded siRNAs directed against the signal peptide leader sequence within full-length CLU mRNA that encodes sCLU (siRNA-sCLU) or siRNAs generated from nontargeted, scrambled sequences (siRNA-scr) were purchased from Dharmacon (Lafayette, CO, USA). The siRNA-sCLU sequence was:
5’- GCG UGC AAA GAC UCC AGA AdTdT-3’
3’-dTdTCGC ACG UUU CUG AGG UCU U-5’
The siRNA-scr sequence was:
5’-GCG CGC UUU GUA GGA UUC GdTdT-3’
3’-dTdTCGC GCG AAA CAU CCU AAG C-5’
Syntheses of PEG-g-PEI copolymers
PEI polymer was first dissolved in deionized H2O at 25 mg/mL. Different amounts of MeO-PEG-NHS (100, 200, and 300 mg as solid powders) were added to the above solution with magnetic stirring to produce PEG-g-PEI copolymers with different PEG grafting densities. The reactions were performed at room temperature overnight. After reaction, mixtures were purified by membrane dialyses (MW cutoff: 10 kDa) in distilled water for 2 days. The resulting solutions were lyophilized to obtain white solid material (yield 50%–60%). The molecular weights of the graft copolymers were analyzed by gel permeation chromatography on a Zorbax GF-250 column (Agilent technologies, Palo Alto, CA) with HPLC grade water as eluent (0.3 mL/min). PEG grafting densities were characterized by 1H NMR (Varian Gemini NMR spectrometer, Palo Alto, CA, USA) in deuterated chloroform by calculating peak ratios of –CH2CH2-O protons (3.7–3.8 ppm) from PEG over –CH2CH2-NH- protons (2.3–3.0 ppm) from PEI.
Ethidium bromide (EtBr) exclusion assays
First, aliquots of siRNA (7.8 μg) were solubilized in DEPC-treated water at 1.95 μg/mL containing 0.5 μg/mL EtBr. EtBr–siRNA fluorescence intensity (λex = 525 nm, λem = 590 nm) was measured and set to 100% on an LS-45 fluorescence spectrometer (Perkin Elmer, Boston, MA). Different amounts of PEG-g-PEI copolymer were then added in 0.2 N/P increments (each addition had 5 μL volume to minimize dilution effects). N/P ratios were calculated as the number of nitrogen atoms in the PEI or PEI copolymer over that of the phosphate groups in the siRNA. Fluorescence intensities were recorded after each addition until final N/P ratios of 2.4 were reached. For each addition, the fluorescence intensity was measured both immediately after mixing and 10 minutes later to ensure equilibrium complexation. After the experiment, fluorescence intensities were plotted as a function of N/P ratio for different graft copolymers to compare their binding affinities to siRNA.
Preparation of polymer–siRNA nanocomplexes
An aqueous solution of siRNA (300 μg/mL) was added drop-wise to DEPC-treated water containing PEG-g-PEI (150–450 μg/mL) while vortexing. Different copolymer concentrations were used to provide polymer–siRNA nanocomplexes with different N/P ratios. After addition, the mixture was vortexed for an additional 20 seconds and then allowed to incubate at room temperature for 30 minutes before analyses or use. Particle size was determined by dynamic light scattering (DLS) on a 90Plus particle size analyzer (Brookhaven Instruments, Holtzville, NY, USA).
MTT cytotoxicity assays
MCF-7 cells were seeded at a density of 2 × 104 cells per well in 96-well plates 1 day prior to addition of polymers or polymer–siRNA nanocomplexes. After addition, cells were incubated overnight with either nanocomplexes or equivalent concentrations of polymer alone. The CellTiter assay was performed according to the manufacturer’s instructions. At the end of the incubation, medium was aspirated and replaced with 100 μL of diluted MTT dye solution, and cells were incubated in a 5% CO2/95% air atmosphere at 37°C for 4 hours. Solubilization–Stop Solution was added to each well (100 μL) to end reactions, followed by incubation at 37°C for 1 hour. The contents of wells were then mixed by gentle pipetting prior to spectrophotometric analyses at a wavelength of 550 nm with a reference wavelength of 770 nm. All MTT assays were performed in triplicate, and data points were calculated as a percentage of viable cells over untreated control ± standard deviation (SD).
siRNA transfections and western blot analysis
MCF-7 cells were seeded at a density of 3 × 105 cells per well in a 6-well plate 1 day prior to transfection. Cells were washed with RPMI 1640 medium without serum or antibiotics twice prior to the addition of siRNA containing nanocomplexes. siRNA nanocomplexes or aqueous solutions of polymer alone were added to each well, and serum-free RPMI 1640 medium was added to adjust the total volume to 1.0 mL/well. To examine the effects of sCLU knockdown on IR lethality, cells were treated with 2 consecutive doses of nanocomplexes prior to exposure to ionizing radiation. IR was delivered by exposure to a 60Co sealed source at 580 rad/minute as previously described (Yang et al 2000; Leskov et al 2003). After overnight incubation with nanocomplexes, medium was removed and replaced with RPMI 1640 medium (2 mL/well) containing 5% fetal bovine serum for another 48 hours. At harvest, whole-cell extracts were prepared in RIPA buffer (0.1% SDS, 0.5% deoxycholate, 1% NP-40, 150 mM NaCl, 50 mM Tris, pH 8.0). Total protein content was measured using standard Bradford assessments, and 50 μg of each sample was loaded and proteins separated by SDS-PAGE. Western blot analysis was performed as described (Yang et al 2000). Quantification of protein expression levels was performed using ImageJ software (a freeware distributed by the NIH). Western blots shown are representative of experiments performed 3 or more times with equivalent results. Exposure time for autoradiographs was optimized to provide images of adequate quality for analysis.
Clonogenic survival assays
MCF-7 cells (3 × 105 cells/well) were transfected with siRNA-sCLU or siRNA-scr containing (each at 400 pmol) nanocomplexes using the 2k10 PEI-g-PEG copolymer. After incubation for 60–72 hours, cells were trypsinized, and plated onto 60-mm dishes at various cell densities (2000–20000 cells/dish) in triplicate and irradiated for 12 hours at different doses (0–5 Gy). Ten days later, cells were fixed and stained using crystal violet in 20% ethanol as described (Criswell et al 2005). Colonies containing more than 50 normalappearing cells were counted on a Nikon Eclipse TE300 microscope (Nikon, Melville, NY, USA). Percent survival was calculated as the ratio of number of colonies of treated cells over untreated control and performed 3 times. Graphed are means ± SD versus IR dose in Gy.
Results
Syntheses of PEG-g-PEI copolymers
Three different compositions of grafted copolymers were tested for their abilities to complex with siRNA. PEG chains with molecular weights of ~2 kDa were grafted onto hyperbranched PEI (MW 10 kDa) using N-hydroxysuccinimide (NHS) chemistry in grafting densities of 5.7, 9.7, and 15.4 (as estimated by NMR) PEG chains per PEI molecule and named 2k5, 2k10, and 2k15, respectively. Upon each modification, the amino group on PEI was converted to an amide functional group. In all three copolymers, gel permeation chromatography demonstrated a monomodal peak and 1H NMR confirmed the grafting density of PEG in the copolymer (data not shown).
EtBr exclusion assays
In the formation of nanocomplexes, copolymers bind to siRNA molecules primarily through electrostatic interactions of the cationic ammonium groups on PEG-g-PEI with the negatively charged phosphate groups on the siRNA backbone. The polymer–siRNA complexation excludes EtBr from siRNA, which leads to a decreased fluorescence intensity. Figure 1 plots the normalized fluorescence intensity as a function of copolymer addition. The N/P ratio was used to reflect the stoichiometry of copolymer over siRNA. All the PEG-g-PEI copolymers showed an increase in binding affinity over the homopolymer PEI. More specifically, unmodified PEI showed a maximal quenching at an N/P ratio of 1.6. The 2k5 and 2k15 copolymers both showed more effective fluorescence quenching than PEI. The 2k10 copolymer showed the highest binding to siRNA with maximum quenching at an N/P ratio of 1.2.
Figure 1.
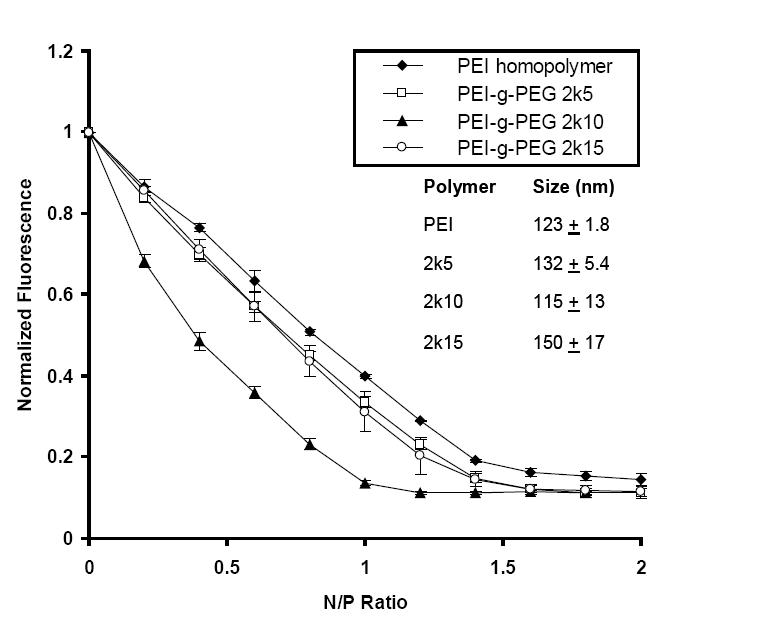
PEI-g-PEG copolymers complex with siRNA and exclude EtBr. The fluorescence intensity was normalized to the siRNA–EtBr complex prior to addition of copolymers. For each copolymer, 2k refers to the molecular weight of the grafting PEG chain and 5, 10, or 15 refer to the number of PEG grafts per PEI molecule. The inset shows the hydrodynamic diameters of the polymer–siRNA nanocomplexes as measured by dynamic light scattering.
Abbreviations: EtBr, ethidium bromide; PEG, polyethylene glycol; PEI, polyethylenimine; siRNA, small interfering RNA.
Nanocomplex size measurement
Dynamic light scattering was used to determine the hydrodynamic diameters of copolymer–siRNA nanocomplexes. An N/P ratio of 7 was used for all the copolymers complexing with siRNA. PEI formed nanocomplexes with siRNA with a diameter of 123 ± 1.8 nm. The 2k5 and 2k15 copolymers formed nanocomplexes of 132 ± 5.4 nm and 150 ± 17 nm in diameter, respectively. Among all tested copolymers, 2k10 formed the smallest nanocomplexes with a diameter of 115 ± 13 nm. Since small particle size has been correlated with nanoparticle uptake and transfection efficiency (Zauner et al 2001; Prabha et al 2002) and binding affinity is an indication of nanoparticle stability, the 2k10 copolymer was chosen for subsequent siRNA studies.
MTT cytotoxicity assessments
The MTT assay (Mosmann 1983) was used to determine the short-term cytotoxicity of the 2k10 copolymer and 2k10/siRNA-sCLU nanocomplexes (Figure 2). Three N/P ratios (5, 7, 14) at 3 siRNA doses (200, 400, 800 pmol, siRNA-scr was used in this study) were examined with siRNA alone as a control. Additionally, the same concentrations of copolymers alone for each nanocomplex condition were examined. Log-phase MCF-7 cells showed no significant cytotoxicity after exposure to 200 pmol siRNA alone, 2k10 alone, or 2k10–siRNA nanocomplexes. At 400 pmol, slightly higher cytotoxicity (ie, 75% cell viability, p < 0.05 from siRNA alone vs control cells) was observed at an N/P ratio of 14 for both 2k10 and 2k10–siRNA nanocomplexes. At 800 pmol, significant cytotoxicity (> 50% cell lethality) was observed at an N/P ratio of 14 for either 2k10 copolymer alone or 2k10–siRNA nanocomplexes. High N/P ratios were correlated with high transfection efficiencies in PEI-based DNA delivery systems (Petersen et al 2002), but high N/P ratios also increased cytotoxicity. Subsequent transfection studies were performed at an N/P ratio of 7 because of cytotoxicity limits.
Figure 2.
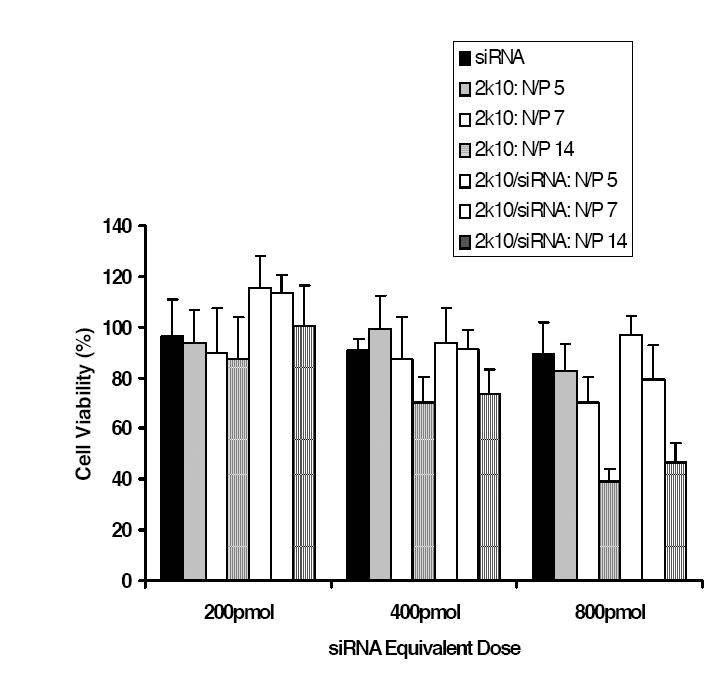
Cell viability after co-incubation with siRNA, 2k10, or 2k10–siRNA nanocomplexes overnight at different N/P ratios and siRNA doses (n = 6; ± SD)
Abbreviations: siRNA, small interfering RNA.
2k10–siRNA-sCLU nanocomplexes inhibit both basal and IR-inducible sCLU expression
We next investigated the ability of 2k10–siRNA-sCLU nanocomplexes to inhibit protein expression of the prosurvival factor, sCLU, in log-phase MCF-7 cells before (basal levels) and after IR treatment. Changes in basal or IR-induced levels of sCLU protein expression were monitored by expression of the intracellular 60 and ~40 kDa forms of sCLU proteins by western blot analyses; we previously showed that changes in the intracellular 60 kDa precursor form of sCLU can be used to monitor overall sCLU levels in the medium of MCF-7 cells (Criswell et al 2003; Klokov et al 2004; Criswell et al 2005). Exposure of MCF-7 cells to 2k10 co-polymer alone that lacked siRNA-sCLU showed no inhibition of sCLU protein expression versus untreated cells (Figure 3a). Similarly, nanocomplexes made with a siRNA-scr also showed no inhibition of sCLU protein expression in exposed MCF-7 cells. In contrast, exposure of MCF-7 cells with nanocomplexes containing siRNA-sCLU showed significant inhibition of sCLU expression. Inhibition of sCLU protein expression in MCF-7 cells showed a semidose-dependence, with little or no inhibition observed after treatment with 200 pmol, but greater inhibition observed with 400 pmol. Image analysis estimated a ~60% knockdown of sCLU protein expression compared to MCF-7 cells exposed to similar nanocomplexes containing siRNA-scr. At 800 pmol, MCF-7 cells showed similar levels of sCLU protein inhibition as observed in MCF-7 cells exposed to 400 pmol siRNA-sCLU nanocomplexes. In all cases, siRNA-sCLU alone at 200, 400, and 800 pmol showed no inhibition of basal sCLU protein expression (data not shown).
Figure 3.
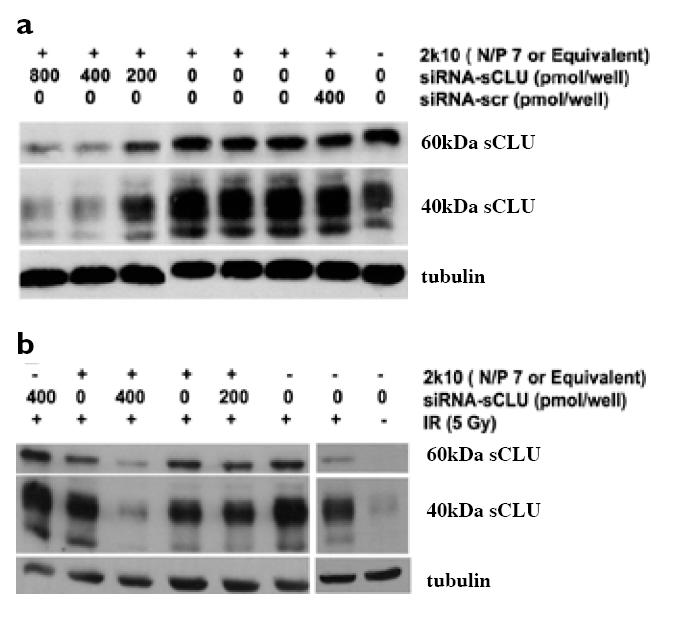
Exposure of MCF-7 cells with 2k10–siRNA-sCLU nanocomplexes is effective in inhibiting basal and IR-inducible sCLU protein expression. Western blot analysis showed basal (a) or IR-inducible (b) sCLU expression was significantly knocked down by 2k10–siRNA-sCLU nanocomplexes. An N/P ratio of 7 was used for nanocomplex formation before addition to cells. In (b), cells were treated with nanocomplexes or vector for 2 consecutive doses prior to exposure to 5 Gy IR.
Abbreviations: IR, ionizing radiation; sCLU, secretory clusterin; scr, scrambled; siRNA, small interfering RNA.
Exposure of MCF-7 cells to IR results in significant upregulation of CLU gene expression, including > 200-fold increases in sCLU protein expression (Boothman et al 1993; Yang et al 2000). IR-inducible sCLU protein expression is also cytoprotective against IR toxicity in various cancer cells (Zellweger et al 2002; Zellweger et al 2003), including in MCF-7 cells (Criswell et al 2005). To investigate the efficacy of nanocomplexes containing siRNA-sCLU to suppress IR-inducible sCLU expression, cells were transfected with siRNA-sCLU nanocomplexes and then exposed to IR (5 Gy). Neither siRNA-scr nanocomplexes nor 2k10 copolymer alone showed protein inhibition versus nonirradiated control cells (Figure 3b). Moreover, free siRNA-sCLU oligomers alone did not inhibit sCLU protein expression. Importantly, the 2k10–siRNA-sCLU nanocomplexes showed marked inhibition (80% knockdown) of sCLU protein expression (Figure 3b).
Knockdown of sCLU expression enhances IR lethality
Finally, we tested the ability of 2k10–siRNA-sCLU nanocomplexes to enhance the radiosensitivity of log-phase MCF-7 cells. The 2k10–siRNA-sCLU nanocomplexes were tested alongside siRNA-scr nanocomplexes as a negative control. As a positive control, siRNA was also delivered using a commercially available transfection agent, Lipofectamine. Lipofectamine-delivered siRNA-sCLU was previously shown to be able to enhance the IR sensitivity of MCF-7 cancer cells via suppression of sCLU protein expression (Criswell et al 2005). Treatment of MCF-7 cells with 2k10–siRNA-sCLU nanocomplexes demonstrated the highest sensitivity to IR treatment, with a 38% ± 3% survival at 0.5 Gy and a 3% ± 1% survival at 3 Gy. All values were normalized to the lethality (95% survival) of unirradiated cells. As previously demonstrated, lipofectamine-delivered siRNA-sCLU increased the lethality of IR in MCF-7 cells (Criswell et al 2005), with 58% ± 4% survival at 0.5 Gy and 6% ± 1% survival at 3 Gy, compared with 93% ± 5% and 54% ± 4% survival of nontransfected cells (Criswell et al 2005). As expected, MCF-7 cells treated with 2k10–siRNA-scr nanocomplexes showed little to no enhancement of IR treatment (Figure 4a). MCF-7 cells treated with siRNA-scr nanocomplexes showed 88% ± 12% survival after 0.5 Gy and decreasing survival at increasing radiation doses down to 12% ± 2% at the 3 Gy dose.
Figure 4.
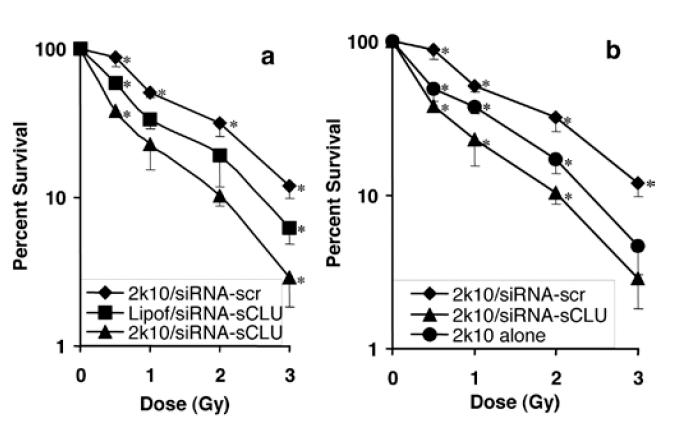
Post-IR clonogenic survival assay of MCF-7 cells. (a) Log scale plot of cell survival after exposure to 2k10–siRNA-sCLU nanocomplexes compared with a positive Lipofectamine control. (b) Cell survival of 2k10 nanocomplexes vs 2k10 copolymer alone. siRNA-scr nanocomplex was used as a negative control in both (a) and (b). Error bars were calculated as the standard deviation from 3 trials. *indicates datapoints that are significantly different (p < 0.05) from others in the series.
Abbreviations: IR, ionizing radiation; lipof, lipofectamine; sCLU, secretory clusterin; scr, scrambled; siRNA, small interfering RNA.
We also compared the effects of 2k10–siRNA nanocomplexes with 2k10 copolymers alone (Figure 4b). The 2k10–siRNA-sCLU nanocomplexes showed significantly greater IR sensitivity over 2k10 alone (p value < 0.05 at all but the highest IR dose), with 2k10 copolymer alone having a 50% ± 9% survival at 0.5 Gy falling to a 5% ± 2% survival at 3 Gy. MCF-7 cells treated with 2k10 alone showed higher IR sensitivity over the MCF-7 cells treated with the 2k10–siRNA-scr nanocomplexes.
Discussion
Various PEG-grafted PEI copolymers were synthesized and examined for binding to siRNA to form nanometersized complexes. The 2k10 copolymer from this series was subsequently shown to be nontoxic and was able to deliver siRNA, effectively knocking down basal and IR-inducible sCLU protein expression in MCF-7 cells. Consequently, these nanocomplexes increased the IR lethality in exposed MCF-7 cells.
Results from this study validate the direct use of EtBr binding assays in evaluating siRNA-copolymer affinity. Previous reports used DNA oligonucleotides to simulate double stranded siRNA to evaluate the interactions of copolymers with oligomers (Itaka et al 2004). Since siRNA is double stranded, EtBr can intercalate between base pairs and sufficient fluorescence was observed to investigate copolymer-siRNA interactions. It is worth noting that the PEG-grafted PEI copolymer showed a greater fluorescence quenching ability than the PEI homopolymer. The enhanced quenching ability of the copolymers suggests a stabilization effect of PEG in the resulting nanocomplexes, shifting the equilibrium towards nanocomplex formation. An optimal grafting density of 10 PEG chains per PEI molecule in 2k10 copolymers was found to give the highest binding affinity and nanocomplex stability. This stability also agrees with DLS studies, with the 2k10 copolymer forming the smallest nanocomplexes. The reduced exclusion of EtBr and larger nanocomplex size of the 2k15 copolymer suggests that the PEG content in this copolymer is nearing a point where PEG chains interfere with siRNA complexation. A similar lessening of particle size and greater stabilization of nanocomplexes as a result of smaller amounts of PEG grafting was previously observed in PEI copolymer nanocomplexes with DNA (Petersen et al 2002). Due to the favorable siRNA complexation behavior, the 2k10 copolymer was subsequently chosen for siRNA studies, as well as its ability to knock down sCLU protein levels to enhance IR lethality.
Data from MTT assays showed the 2k10 vector had little or no toxicity at doses corresponding to 400 pmol or less siRNA with N/P ratio as high as 14 (Figure 2). These data suggest that PEG lessens the toxicity of the heavily cationic PEI portion of the copolymer structure.
Western blot analysis confirmed that 2k10 was an effective carrier for siRNA. The 2k10/siRNA-sCLU nanocomplexes inhibited both basal as well as IR-induced sCLU expression in MCF-7 cells (Figure 3). sCLU upregulation is a very sensitive indicator of cell stress, with induction of sCLU expression at levels where most cytotoxic agents do not generate significant lethality (Criswell et al 2003). Importantly, the slight cytotoxicity of 2k10 nanocomplexes did not trigger sCLU protein up-regulation in MCF-7 cells, as confirmed in all western blot analyses (as in representative western blots shown in Figure 3). Furthermore, the slight cytotoxicity of 2k10 nanocomplexes did not eliminate the copolymer’s ability to effectively inhibit sCLU protein expression. Control nanocomplexes containing siRNA-scr demonstrate the specificity of siRNA-sCLU, in which sCLU protein level was not altered, nor was the IR sensitivity of MCF-7 cells exposed to these nanocomplexes.
siRNA-sCLU delivered by 2k10 nanocomplexes resulted in significant enhancement in IR lethality of MCF-7 cells, consistent with the specific knockdown of sCLU protein expression. The 2k10/siRNA-sCLU nanocomplexes showed enhanced IR lethality greater than 3-fold over complexes containing siRNA-scr (Figure 4). Importantly, siRNA-sCLU nanocomplexes did not affect expression of nCLU protein levels (data not shown) as previously shown for this siRNA that is directed to the Exon II-encoded, signal leader peptide sequence. Thus, the increased IR toxicity is primarily due to the specific RNA interference-mediated knockdown of sCLU protein levels rather than nonspecific effect of siRNA, since this was not observed with the siRNA-scr control. The increased toxicity observed in the 2k10 alone over the 2k10– siRNA-scr nanocomplexes is possibly due to cellular effects as a result of uncompleted polymer. Polycations can disrupt cell membranes resulting in toxicity (De Smedt et al 2000) and the neutralization of that charge by complexation with siRNA can lessen or eliminate this toxicity (Brownlie et al 2004).
It is important to note that the double stranded siRNA used in this study is not the only form of RNA silencing compound. Short hairpin RNAs, siRNA encoding plasmids, and peptide nucleic acids (PNAs) are alternative materials that may require different architectures in polymer carriers. Despite this, the underlying concepts behind polymermediated siRNA delivery would apply in these cases. Meanwhile, polymer carriers and siRNA delivery can show marked cell line dependence and subsequent studies will explore this possible dependence.
In summary, PEG-grafted PEI was shown to bind to siRNA and form nanometer-sized complexes. These nanocomplexes inhibited sCLU expression as well as its IRinduced up-regulation in MCF-7 cells, resulting in increased IR lethality. This study validates the concept of using polymer delivered siRNA to specifically down-regulate cytoprotective factors as an aid to cancer radiotherapy. Still, questions such as nonspecific uptake of these nanocomplexes and their targeting efficiency to tumors remain in regard to the in vivo translation in animals. Although the small nanoparticle size allows for facile i.v. injection, potential barriers may still exist for sufficient interstitial penetration in tumors and efficient cell uptake. Our laboratories are currently working on experiments to address the above issues and aim to establish cancer-specific siRNA delivery in vivo to improve radiotherapy of human cancers.
Acknowledgments
This research was supported by grants R01-CA-90696 (J.G.) and R01-AI34039 (B.R.G.W.) from the National Institutes of Health and grant DE-FG-022179 (D.A.B.) from the Department of Energy.
References
- Bettuzzi S. The new anti-oncogene clusterin and the molecular profiling of prostate cancer progression and prognosis. Acta Biomed Ateneo Parmense. 2003;74:101–4. [PubMed] [Google Scholar]
- Boothman DA, Meyers M, Fukunaga N, et al. Isolation of X-Ray inducible transcripts from radioresistant human-melanoma cells. Proc Natl Acad Sci U S A. 1993;90:7200–4. doi: 10.1073/pnas.90.15.7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussif O, Lezoualc’h F, Zanta MA, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo. Proc Natl Acad Sci U S A. 1995;92:7297–301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlie A, Uchegbu IF, Schätzlein AG. PEI based vesicle-polymer hybrid gene delivery system with improved biocompatibility. Int J Pharm. 2004;274:41–52. doi: 10.1016/j.ijpharm.2003.12.029. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002a;296:550–3. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002b;2:243–7. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- Criswell T, Beman M, Araki S, et al. Delayed activation of insulin-like growth factor-1 receptor/Src/MAPK/Egr-1 signaling regulates clusterin expression, a pro-survival factor. J Biol Chem. 2005;280:14212–21. doi: 10.1074/jbc.M412569200. [DOI] [PubMed] [Google Scholar]
- Criswell T, Klokov D, Beman M, et al. Repression of IR-inducible clusterin expression by the p53 tumor suppressor protein. Cancer Biol Ther. 2003;2:372–80. doi: 10.4161/cbt.2.4.430. [DOI] [PubMed] [Google Scholar]
- De Smedt SC, Demeester J, Hennink WE. Cationic polymer based gene delivery systems. Pharm Res. 2000;17:113–26. doi: 10.1023/a:1007548826495. [DOI] [PubMed] [Google Scholar]
- Devroe E, Silver PA. Therapeutic potential of retroviral RNAi vectors. Expert Opin Biol Ther. 2004;4:319–27. doi: 10.1517/14712598.4.3.319. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, et al. Duplexes of 21 Nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–8. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Weber K, et al. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods. 2002;26:199–213. doi: 10.1016/S1046-2023(02)00023-3. [DOI] [PubMed] [Google Scholar]
- Ge Q, Filip L, Bai A, et al. Inhibition of influenza virus production in virus-infected mice by RNA interference. Proc Natl Acad Sci U S A. 2004;101:8676–81. doi: 10.1073/pnas.0402486101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave ME, Miyake H, Zellweger T, et al. Use of antisense oligonucleotides targeting the antiapoptotic gene, clusterin/testosterone-repressed prostate message 2, to enhance androgen sensitivity and chemosensitivity in prostate cancer. Urology. 2001;58:39–49. doi: 10.1016/s0090-4295(01)01241-9. [DOI] [PubMed] [Google Scholar]
- Itaka K, Kanayama N, Nishiyama N, et al. Supramolecular nanocarrier of siRNA from PEG-based block catiomer carrying diamine side chain with distinctive pKa directed to enhance intracellular gene silencing. J Am Chem Soc. 2004;126:13612–3. doi: 10.1021/ja047174r. [DOI] [PubMed] [Google Scholar]
- Izawa M. Identification of a transcript predicting an alternative form of sulfated glycoprotein-2 (clusterin) in rat tissues. Biochem Mol Biol Int. 1998;44:9–18. doi: 10.1080/15216549800201012. [DOI] [PubMed] [Google Scholar]
- July LV, Beraldi E, So A, et al. Nucleotide-based therapies targeting clusterin chemosensitize human lung adenocarcinoma cells both in vitro and in vivo. Mol Cancer Ther. 2004;3:223–32. [PubMed] [Google Scholar]
- Klokov D, Criswell T, Leskov KS, et al. IR-inducible clusterin gene expression:a protein with potential roles in ionizing radiation-induced adaptive responses, genomic instability, and bystander effects. Mutat Res Fund Mol M. 2004;568:97–110. doi: 10.1016/j.mrfmmm.2004.06.049. [DOI] [PubMed] [Google Scholar]
- Kunath K, von Harpe A, Petersen H, et al. The structure of PEG-modified poly(ethylene imines) influences biodistribution and pharmacokinetics of their complexes with NF-B decoy in mice. Pharm Res. 2002;19:810–17. doi: 10.1023/a:1016152831963. [DOI] [PubMed] [Google Scholar]
- Laping NJ, Morgan TE, et al. Transforming growth factor-beta 1 induces neuronal and astrocyte genes:tubulin alpha 1, glial fibrillary clusterin. Neuroscience. 1994;58:563–72. doi: 10.1016/0306-4522(94)90081-7. [DOI] [PubMed] [Google Scholar]
- Leskov KS, Klokov DY, et al. Synthesis and functional analyses of nuclear clusterin, a cell death protein. J Biol Chem. 2003;278:11590–600. doi: 10.1074/jbc.M209233200. [DOI] [PubMed] [Google Scholar]
- Little SA, Mirkes PE. Clusterin expression during programmed and teratogen-induced cell death in the postimplantation rat embryo. Teratology. 1995;52:41–54. doi: 10.1002/tera.1420520106. [DOI] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Novina CD, Murray MF, Dykxhoorn DM, et al. siRNA-directed inhibition of HIV infection. Nat Med. 2002;8:681–6. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- Peatfield RC, Boothman BR. Transient myoclonus after exposure to oven cleaner. Mov Discord. 1991;6:9–91. doi: 10.1002/mds.870060121. [DOI] [PubMed] [Google Scholar]
- Petersen H, Fechner PM, Martin AL, et al. Polyethylenimine-graft-poly(ethylene glycol) copolymers:influence of copolymer block structure on DNA complexation and biological activities as gene delivery system. Bioconj Chem. 2002;13:845–54. doi: 10.1021/bc025529v. [DOI] [PubMed] [Google Scholar]
- Prabha S, Wne-Zhong Z, Panyam J, et al. Size-dependency of nanoparticle-mediated gene transfection:studies with fractionated nanoparticles. Int J Pharm. 2002;244:105–15. doi: 10.1016/s0378-5173(02)00315-0. [DOI] [PubMed] [Google Scholar]
- Schiffelers RM, Ansari A, Xu J, et al. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticles. Nucleic Acids Res. 2004;32:e149–58. doi: 10.1093/nar/gnh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh J, Paik H, Hwang BK. Ionization of poly(ethylenimine) and poly(allylamine) at various pH’s. Bioorg Chem. 1994;22:318–27. [Google Scholar]
- Welsh J. Induction of apoptosis in breast cancer cells in response to vitamin D and antiestrogens. Biochem Cell Biol. 1994;72:537–45. doi: 10.1139/o94-072. [DOI] [PubMed] [Google Scholar]
- Yang C, Leskov K, Hosley-Eberlein K, et al. Nuclear clusterin/XIP8, an x-ray-induced Ku70-binding protein that signals cell death. Proc Natl Acad Sci U S A. 2000;97:5907–12. doi: 10.1073/pnas.97.11.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zauner W, Farrow NA, Haines AMR. In vitro uptake of polystyrene microspheres:effect of particle size, cell line and cell density. J Control Release. 2001;71:39–51. doi: 10.1016/s0168-3659(00)00358-8. [DOI] [PubMed] [Google Scholar]
- Zellweger T, Chi K, Miyake H, et al. Enhanced radiation sensitivity in prostate cancer by inhibition of the cell survival protein clusterin. Clin Cancer Res. 2002;8:3276–84. [PubMed] [Google Scholar]
- Zellweger T, Kiyama S, Chi K, et al. Overexpression of the cytoprotective protein clusterin decreases radiosensitivity in the human LNCaP prostate tumour model. BJU Int. 2003;92:463–9. doi: 10.1046/j.1464-410x.2003.04349.x. [DOI] [PubMed] [Google Scholar]


