Abstract
Neural fate specification in Drosophila is promoted by the products of the proneural genes, such as those of the achaete–scute complex, and antagonized by the products of the Enhancer of split [E(spl)] complex, hairy, and extramacrochaetae. As all these proteins bear a helix-loop-helix (HLH) dimerization domain, we investigated their potential pairwise interactions using the yeast two-hybrid system. The fidelity of the system was established by its ability to closely reproduce the already documented interactions among Da, Ac, Sc, and Extramacrochaetae. We show that the seven E(spl) basic HLH proteins can form homo- and heterodimers inter-se with distinct preferences. We further show that a subset of E(spl) proteins can heterodimerize with Da, another subset can heterodimerize with proneural proteins, and yet another with both, indicating specialization within the E(spl) family. Hairy displays no interactions with any of the HLH proteins tested. It does interact with the non-HLH protein Groucho, which itself interacts with all E(spl) basic HLH proteins, but with none of the proneural proteins or Da. We investigated the structural requirements for some of these interactions by site-specific and deletion mutagenesis.
Cell fate acquisition during development reflects selective activation of transcriptional activators and repressors in different cell types. The fine tuning of this process is achieved by the participation of such regulators in dynamic interaction networks. The helix-loop-helix (HLH) domain, characteristic of a family of transcription factors, mediates both homo- and heterodimeric interactions (1–3), making proteins that contain this domain ideal for participation in a number of complexes, whose formation can be potentially regulated during development.
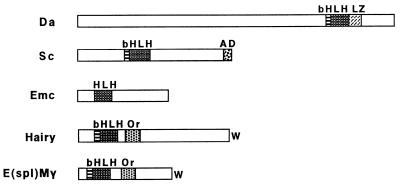
Both activators and repressors are encountered within the HLH family of transcriptional regulators. Based on structural and functional similarities, subclasses of HLH proteins can be discerned (Fig. 1). One class comprises cell-type specific transcriptional activators, whose activity is intimately related with the acquisition of a particular cell type, e.g., the myogenic proteins (4) and the proneural proteins (5). The tissue-specific HLH factors dimerize with ubiquitous HLH proteins, called E-proteins [e.g., E12, Daughterless (2, 6)]. As both of these subclasses contain a basic region next to the HLH domain, they are termed bHLH. The basic domains contact a DNA target site known as the E-box (CANNTG), conferring transcriptional activation. As dimerization is a prerequisite for DNA binding, proteins that prevent the formation of these complexes act as negative regulators of E-box-driven genes. One class of negative regulators contains an HLH domain, but lacks a basic region [e.g., Id, Extramacrochaetae (Emc)], sequestering ubiquitous or activator bHLH proteins into inactive complexes (7–10). A second class of negative HLH regulators comprises Drosophila Hairy (11), Deadpan (12), and the seven Enhancer of split [E(spl)] proteins (13, 14), as well as identified homologues in other species (15, 16). These are distinguished from the Emc/Id class by virtue of having a conserved basic domain and being able to bind specific DNA target sites (17–19). In addition to having structurally distinct bHLH domains, the Hairy/E(spl) proteins are also characterized by two other conserved domains, the Orange domain and the C-terminal conserved tetrapeptide WRPW, neither of which is found in other members of the HLH family. Functional studies have shown that both of these domains, as well as the bHLH domain, are needed for the proper function of these proteins (20, 21).
Figure 1.
Examples of Drosophila HLH proteins. Schematic depiction of Drosophila HLH proteins belonging to different classes. Da is an E-protein and Sc is an activator, encoded by a gene in the ASC. Emc is a negative regulator that lacks a basic domain. Hairy and E(spl)Mγ are two members of the Hairy/E(spl) class of negative regulators. Structural motifs are marked: b, basic domain; HLH, helix-loop-helix domain; LZ, putative leucine zipper; AD, conserved acidic domain among the ASC proteins; Or, Orange domain; and W, terminal WRPW tetrapeptide conserved among Hairy/E(spl) class members.
A developmental process in which all four subclasses of HLH proteins appear to participate is the formation of neural precursors in Drosophila. The proneural proteins of the achaete–scute complex (ASC) (5) and Atonal (22) are activator bHLH proteins that endow groups of cells in the embryonic ectoderm and the imaginal disks with neural potential. Both expression and activity of these proteins are strictly regulated to achieve the correct spatiotemporal specification of neural elements. Their activity is aided by the E-protein Da and antagonized by Emc (10, 23–26). Their expression is under complex regulation by a number of positive and negative factors. Proneural genes are initially expressed in clusters of ectodermal cells (27, 28) under the control of local activators/repressors to yield the “prepattern” (29–31). One of the prepattern repressors of the achaete gene in the adult peripheral nervous system (PNS) is the bHLH protein Hairy (23, 27, 32). After the initial expression in all cells of a cluster, proneural protein expression persists and increases only in the cell that will become the neural precursor, whereas it declines in neighboring cells, which remain ectodermal (27, 28, 33, 34). Different regulators seem to be needed at this refinement step: activation probably involves the proneural proteins themselves, whereas repression necessitates the activity of the Notch signaling pathway (33–35) that acts by inducing the accumulation of E(spl) proteins (36, 37). A likely scenario is that E(spl) proteins repress proneural genes in an analogous fashion to the earlier repression by Hairy. E(spl) bHLH proteins are encoded by seven distinct genes located in a cluster, the E(spl)-complex. Genetic data argue for partially redundant functions of these seven genes, as it has not been possible to isolate point mutations in the complex to date (38, 39). However, the overall structure of the E(spl)-complex has been conserved in the distantly related Drosophila hydei, arguing for the possibility of distinct functions for each of the seven proteins (40).
Although genetic data have revealed the regulatory relationships among the various HLH proteins involved in neural precursor specification, the level at which this regulation takes place has not been fully elucidated. Da, the proneural proteins, and Emc can all form dimers with each other (2, 6, 10). Hairy and E(spl) could also participate in this interacting network or they could form a separate set of DNA-binding oligomers among themselves and repress proneural gene expression, without affecting proneural protein activity. The latter hypothesis is supported by the fact that both Hairy and some of the E(spl) proteins bind specific DNA sites and repress transcription (17–19). Transcriptional silencing by Hairy has been proposed to require the recruitment of a corepressor, the non-HLH protein Groucho (41). Whether this is the only mechanism of repression by Hairy and the E(spl) proteins, which also bind Groucho (41), is an unresolved question. In addition to their role in neurogenesis, some of these same HLH factors appear to participate in other developmental processes, such as the early X/A ratio counting for sex determination (42–44). In this process Dawson et al. (20) have detected a potentially Groucho-independent activity of Hairy.
To begin to address the possible functions of the Hairy/E(spl) class of HLH proteins we have used the yeast two-hybrid system (45, 46) to study interactions among a set of HLH proteins spanning different classes. We detect a large number of interactions that are distinctly preferential rather than promiscuous. As interactions among HLH proteins provide a mechanism for their specificity, we discuss the possible function of this interaction network in Drosophila. Additionally, we describe distinct features of members of the E(spl) family that were previously considered redundant.
MATERIALS AND METHODS
Constructs.
Two different versions of the two-hybrid system were used, the Brent system (46) and the Hollenberg system (47, 48), both of which test for interactions by tethering the interacting complex to DNA via the DNA-binding domain of the bacterial repressor LexA. Both versions use lacZ reporters driven by eight tandem LexA operators: the Brent system reporter is on a high copy plasmid, pSH18-34 (URA3), whereas the Hollenberg reporter is integrated into the URA3 gene of a special yeast strain (see below). Fusions of coding regions of interest to the LexA DNA-binding domain are referred to as the “LexA constructs,” e.g., LexA-m5. pEG202 (HIS3) and pBTM116 (TRP1) were used as the LexA fusion vectors. Two different activation domains were used, B42 on vector pJG4–5 (TRP1) or VP16 on vector pVP16 (LEU2) to generate fusion constructs, e.g., B42-m5, VP16-sc.
Fusions of the E(spl) genes and ato were produced by ligating PCR amplification products (verified by sequencing) into pJG4-5 as 5′ EcoRI-3′ XhoI fragments containing the entire ORF. LexA fusion constructs were produced by transferring the coding EcoRI–XhoI cassette from the B42 fusion construct to the pEG202 vector. gro was cloned into pEG202 using a cDNA fragment that lacks the 14 N-terminal codons.
The VP16 fusions were constructed as in-frame insertions of PCR-amplified BamHI fragments containing the entire ORF, whereas the LexA (pBTM116) fusions were constructed using appropriate PCR primers as EcoRI/BamHI or EcoRI/BglII insertions where the EcoRI site abuts the ATG start site. Deletion constructs LexA–m3 bHLH and LexA–m3 bHLH-O contain amino acids 1–77 and 1–143 of m3 respectively. VP16-Sc bHLH contains amino acids and 1–178 of Sc. VP16-ScS>D was made using the following 3′ PCR primer, which bears a point mutation: 5′-cgtcggatccgtcactgctcctgccatagatcgatgtagt-3′. The mutant protein ends in DYIDLWQEQ instead of DYISLWQEQ.
Sequences of other PCR primers and construction details are available upon request.
Yeast Strains and Interaction Assays.
The yeast strains used with the Brent system are as follows: EGY 48 (Mata, trp1, his3, ura3, and (lexAop)6-Leu 2) (49) and FT4 (Matα, trp1, his3, ura3, and leu2). LexA fusions and reporter plasmid were transformed into EGY48, and B42 fusions were transformed into FT4. In the Hollenberg system the strains were: AMR70 [Matα his3Δ200 lys2-801am trp1-901 leu2-3, 112 URA3::(lexAop)8-lacZ] and L40 [Mata his3Δ200 trp1–901 leu2–3, 112 ade2 lys2–801am URA3::(lexAop)8-lacZ LYS2::(lexAop)4-HIS3] (48). The mating protocol and β-galactosidase liquid assays are described in ref. 50. Diploids bearing all three constructs were first tested on 5-bromo-4-chloro-3-indolylβ-d-galactoside (X-Gal) indicator plates.
Western Blot Analysis.
Extracts of yeast strains were analyzed on SDS polyacrylamide gels and blotted onto nitrocellulose. Rabbit anti-LexA antibody was a gift from Roger Brent (Harvard Medical School, Boston). Mouse anti-HA epitope tag (anti-influenza virus hemagglutinin epitope tag) mAb was from Boehringer Mannheim. Horseradish peroxidase-conjugated anti-rabbit and anti-mouse antibodies were from Jackson ImmunoResearch. Diaminobenzidine (Sigma) was used to develop the filters.
RESULTS
Criteria for Two-Hybrid System Interactions.
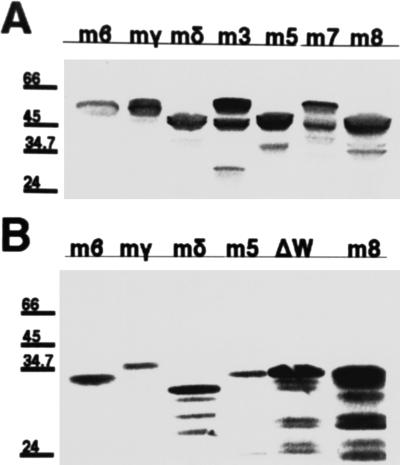
The proteins tested in this study were as follows: (i) three proneural bHLH proteins, Ac, Sc and Ato, as well as the structurally related neural precursor/proneural protein Ase; (ii) the ubiquitous bHLH E-protein Da; (iii) the negative HLH regulator Emc; (iv) a number of bHLH repressors, namely the seven members of the E(spl)-complex (M8, M7, M5, M3, Mβ, Mγ and Mδ), along with the structurally related Hairy; (v) the ubiquitous WD40 nuclear protein Gro; and (vi) Runt, a non-HLH nuclear factor involved in later steps of neural fate specification, as well as in early embryonic segmentation along with Hairy, and bearing the variant C-terminal tetrapeptide WRPY instead of the WRPW found in the members of the Hairy/E(spl) class (51, 52). These proteins were fused to the LexA DNA-binding domain and/or to an acidic activator domain (see Materials and Methods). Western blot analysis confirmed the ability of a number of our fusion constructs to direct the synthesis of proteins of the expected lengths (Fig. 2). Because known interactions between proteins tested were reproduced in the two-hybrid system (see below), it appears that our fusion proteins are properly modified and folded in yeast so as to retain dimerization activity. We did encounter a small number of cases where the fusion protein had no activity, made evident by its inability to interact with any of the reciprocal constructs tested. These fusions (LexA-runt, VP16-l′sc, B42-m7, and B42-m3) were not examined further.
Figure 2.
Western blot analysis of fusion constructs. (A) Extracts from LexA construct bearing cells probed with an anti-LexA antibody. (B) B42 extracts probed with an anti-HA epitope tag antibody. ΔW: B42-m5ΔW. Sizes of markers in kDa are indicated on the side of each blot. Proteins of approximately expected sizes were observed as the major bands.
As a consequence of vector incompatibility between the Brent and Hollenberg two-hybrid systems (see Materials and Methods), some LexA fusions could only be tested against the VP16 fusions, whereas the remaining could be tested against all acidic domain fusions. The interactions determined using the Hollenberg system (in AMR70 × L40 diploids) are shown in Table 1; those detected with the Brent system (in EGY48 × FT4 diploids) are shown in Tables 2 and 3. Interactions were assayed as the level of β-galactosidase activity obtained from a yeast strain bearing all three constructs: LexAop-lacZ reporter and the two fusion constructs, LexA and B42/VP16. As expected with a quantitative enzyme assay, a continuous range of activities was obtained. The actual values are displayed in Tables 1, 2, and 3, however, for the convenience of discussing the results, we set a threshold for what we consider a significant interaction. The two different systems gave differing ranges of β-galactosidase activities, which are partly due to the fact that the Brent system places the reporter gene on a high copy plasmid, whereas the Hollenberg version uses a chromosomally integrated reporter. In what follows, we set 1.5 units as the minimum level for interaction in Table 1. In Tables 2 and 3 the corresponding threshold is set to 100 units for B42 fusions and 200 units for fusions of the stronger VP16 activation domain. The lowest of these thresholds was determined by comparing behavior of yeast patches on X-Gal plates with liquid assay levels—only patches that would turn light blue by 12 h after plating were considered positive.
Table 1.
Interaction pairs tested using the Hollenberg two-hybrid system
| LexA fusion | VP16 fusion
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| VP16 | Runt | M3 | Sc | Sc bHLH | Sc S>D | Ac | Ase | Da | Gro | |
| Hairy | 0.2 | 0.1 | 0.2 | 0.1 | 0.2 | 0.1 | 0.2 | 0.1 | 0.2 | 36.2 ± 1.6 |
| M3 | 0.4 | 0.5 | 0.7 | 3.8 ± 1.3 | 6.1 ± 0.3 | 1 ± 0.2 | 25 ± 1.3 | 19 ± 0.4 | 1.4 ± 0.1 | 101.3 ± 20.6 |
| M3 bHLH | 0.2 | 0.2 | 1.2 | 2.2 ± 0.5 | 6.8 ± 0.3 | 0.6 | 7.2 ± 0.0 | 10.2 ± 1.5 | 0.4 | w |
| M3 bHLH-O | 0.2 | 0.1 | 0.2 | 2.5 ± 0.6 | 4.2 ± 0.5 | 0.4 | 3.1 ± 0.3 | 9.4 ± 0.1 | 0.9 | w |
| M8 | 0.5 | 0.2 | 0.2 | 0.2 | 1.4 ± 0.1 | 0.2 | 0.3 | 0.5 | 22 ± 0.6 | 232.2 ± 22.5 |
| Emc | 0.6 | 0.1 | 0.1 | 141 ± 16 | 135 ± 1.4 | 100 ± 31 | 2.5 ± 0.0 | 0.6 | 147 ± 6.1 | 0.1 |
| Gro | 0.1 | 16 ± 4 | 0.5 | NT | NT | NT | NT | NT | NT | 0.1 |
β-galactosidase levels normalized to the number of cells are shown. Standard deviations are indicated by ±. NT: not tested. w: not tested by liquid assay/white patch on X-Gal. An unfused VP16 domain was used to control for fortuitous activation ability of the LexA fusions. Above-threshold values are set in bold. The VP16-M3 fusion shows no above-threshold interactions in this system; however, we detected interactions using the more sensitive Brent system (see Table 3).
Table 2.
B-42 fusions tested using the Brent two-hybrid system
| LexA fusion | B42 fusion
|
|||||||
|---|---|---|---|---|---|---|---|---|
| alone* | Mβ | Mγ | M5 | M5ΔW | M8 | Mδ | Ato | |
| Mβ | 4 | 71 ± 24 | 64 ± 2 | 75 ± 18 | 93 ± 19 | 42 ± 1 | 92 ± 19 | 17 ± 6 |
| Mγ | 1 | 33 ± 10 | 35 ± 3 | 201 ± 3 | 110 ± 2 | 68 ± 30 | 33 ± 7 | 8 ± 3 |
| M7 | 6 | 50 ± 16 | 29 ± 13 | 213 ± 43 | 71 ± 12 | 55 ± 5 | 31 ± 2 | 28 ± 4 |
| M5 | 0 | 85 ± 44 | 162 ± 94 | 63 ± 34 | 45 ± 25 | 10 ± 6 | 183 ± 115 | 2 ± 1 |
| M5ΔW | 0 | 84 ± 24 | 124 ± 9 | 23 ± 10 | 26 ± 5 | 19 ± 2 | 109 ± 10 | 4 ± 1 |
| M8 | 1 | 88 ± 5 | 125 ± 61 | 20 ± 2 | 18 ± 6 | 10 ± 5 | 22 ± 4 | 4 ± 3 |
| M3 | 0 | 23 ± 9 | 16 ± 8 | 18 ± 2 | 24 ± 2 | 4 ± 2 | 161 ± 73 | 37 ± 12 |
| Mδ | 1 | 78 ± 25 | 25 ± 9 | 69 ± 3 | 45 ± 13 | 16 ± 10 | 37 ± 24 | 5 ± 1 |
| Gro | 0 | 77 ± 3 | 98 ± 17 | 179 ± 66 | 0 | 25 ± 3 | 242 ± 128 | 0 |
The numbers are β-galactosidase units × 10−1. β-galactosidase activity was normalized to the number of cells. Standard deviations are indicated by ±. Above-threshold values are set in bold.
Activity tested in the haploid strain bearing only the LexA fusion and the reporter plasmid.
Table 3.
VP16 fusions tested using the Brent two-hybrid system
| LexA fusion | VP16 fusion
|
|||||
|---|---|---|---|---|---|---|
| Ac | Sc | Ase | Da | M3 | Gro | |
| Mβ | 227 ± 88 | 176 ± 53 | 97 ± 23 | 157 ± 40 | 18 ± 3 | 141 ± 6 |
| Mγ | 128 ± 17 | 178 ± 75 | 78 ± 11 | 74 ± 26 | 3 ± 2 | 150 ± 14 |
| M7 | 150 ± 44 | 57 ± 10 | 147 ± 23 | 151 ± 20 | 14 ± 1 | 160 ± 23 |
| M5 | 10 ± 1 | 2 ± 1 | 6 ± 5 | 56 ± 39 | 8 ± 3 | 156 ± 19 |
| M5ΔW | NT | NT | NT | 42 ± 14 | NT | 0 |
| M8 | 10 ± 7 | 1 | 17 ± 11 | 200 ± 36 | 4 ± 1 | 209 ± 51 |
| M3 | 182 ± 51 | 150 ± 28 | 82 ± 22 | 7 ± 4 | 20 ± 4 | 131 ± 19 |
| Mδ | 8 ± 4 | 7 ± 4 | 9 ± 5 | 2 ± 1 | 28 ± 4 | 178 ± 75 |
| Gro | 0 | 1 | 1 | 1 | 28 ± 1 | 10 ± 1 |
The numbers are β-galactosidase units × 10−1. β-galactosidase activity was normalized to the number of cells. Standard deviations are indicated by ±. Above-threshold values are set in bold.
NT: not tested.
Two additional conditions were imposed in identifying potentially significant interactions: (i) the LexA construct must not activate transcription in the absence of an activation domain construct and (ii) each construct must be able to activate transcription with at least one reciprocal construct, but not with all reciprocal constructs. As is evident from Tables 1, 2, and 3, each row and column contain at least one instance of interaction. A few LexA and B42 fusions display interactions with the entire panel of reciprocal constructs; however, we have documented instances of noninteraction for each of these fusions with reciprocal constructs not presented here.
Proneural Proteins and Da Interact with Emc.
Combinations between LexA-Emc and VP16 fusions of ASC proteins and Da were tested to confirm that the assay reproduced known protein–protein interactions within the HLH family (Table 1). Indeed, Emc gave strong interactions with both Sc and Da and an interaction above background with Ac. No interaction between Emc and Ase was observed, despite the structural similarity between Ase and Ac/Sc (53). Unlike Ac and Sc, which in the adult PNS compete with Emc to define the precise pattern of macrochaetae precursors, Ase is only expressed in the neural precursor cell once this cell has been determined as a consequence of high Ac/Sc activity (54–56). The inability of Ase to be inactivated by Emc would help to stably maintain the neural precursor fate despite the continuing presence of Emc protein (26).
Proneural Proteins and Da Interact with a Subset of E(spl) Proteins.
We next tested the potential involvement of E(spl) bHLH proteins and Hairy in the well-established interacting network of Da-Ac-Sc-Emc. LexA fusions of all seven E(spl) proteins were tested against the VP16 fusions of Ac, Sc, Ase, and Da, as well as B42-Ato (Tables 2 and 3). Hairy was tested against only the VP16 fusions (Table 1). E(spl) proteins displayed interactions with Da and all proneural proteins. In contrast, Hairy did not interact with any of the activator bHLH proteins tested.
Activator bHLH-E(spl) heterodimer combinations did not form indiscriminately. Specifically, LexA-Mδ was unable to bind any of the activator proteins tested (Tables 2 and 3). In contrast, LexA-M7, -Mβ, and -Mγ displayed dimerization capability with all activator bHLH proteins (except Mγ-Ato; Table 2). The remaining E(spl) proteins exhibited selectivity with LexA-m5 and LexA-m8, interacting only with Da, whereas LexA-M3 interacted with Ac, Sc, Ase, and Ato, but not with Da (Tables 2 and 3). All interactions were quite strong, with the exception of M7-Sc, M5-Da, and Mβ-Ato, which were at the lower end of the range.
To determine which of the functional domains of an E(spl) protein are involved in the dimerization with bHLH activators, we constructed two truncated versions of M3; one, LexA-m3bHLH-O, ending after the Orange domain (see Fig. 1) and the other, LexA-m3bHLH, ending after the bHLH domain. Both of these were able to dimerize with Ac, Sc, and Ase (Table 1), the interaction with Ac being weaker compared with full-length LexA-M3. VP16-ScbHLH, a C-terminal truncation of Sc, ending after the bHLH domain does not significantly affect interaction with M3. Interestingly, a point mutation in the C-terminal domain of Sc converting a conserved (among Ac, Sc, and L′sc) serine (S340) to aspartic acid, VP16-ScS > D, results in a severe reduction in the strength of its interaction with M3, without affecting the interaction with LexA-Emc (Table 1) or a LexA-Da (bHLH domain) construct (data not shown). As this serine is potentially phosphorylated, and the aspartic acid mimics the negative charge of the phosphate moiety, it suggests that phosphorylation of that residue may regulate the affinity of Ac, Sc, and L′sc for E(spl) proteins.
Interactions Among E(spl) bHLH Proteins.
All seven E(spl) bHLH proteins were fused to LexA, whereas, of the B42 fusions, only five were functional: Mβ, Mγ, M5, M8, and Mδ. Virtually all combinations among E(spl) proteins produced detectable lacZ activation—the single exception being LexA-M3 with B42-M8 (Table 2). Furthermore, in most cases, the strength of the interaction was not affected by orientation, i.e. which partner was fused to LexA and which to B42. One construct, B42-M8, produced consistently lower β-galactosidase levels than the reciprocal LexA-M8. We believe that the B42 fusion produces a less functional M8 moiety that nonetheless retains enough function to display interactions with the LexA fusions.
From inspection of Tables 2 and 3 we note that preferences exist for and against specific E(spl) pairs. The strongest interactions (>1000 β-galactosidase activity units) were Mγ–M5, Mγ–M8, Mδ–M5, Mδ–M3, and M5–M7. At the other end of the spectrum, weak interactions (<200 units) were observed for M3–Mγ, M3–M5, M3–M8 (no interaction), Mδ–M8, M5–M8, and M8–M8. The existence of a wide range of β-galactosidase values obtained points to the fact that the reporter/acidic fusion combination used has sufficient capacity to discriminate interactions according to their relative affinities.
As our B42-M3 fusion was inactive, we tested a VP16-M3 fusion, which gave transcriptional activation (Table 3), albeit weakly (e.g., compare M3–Mδ interactions in the two orientations). Nonetheless, it reproduced the trends observed for LexA-M3 (Table 2), including the noninteraction with M8, and further detected M3 homodimerization.
Interactions Involving Non-HLH Proteins.
The E(spl)-complex contains a number of transcription units interspersed with the seven bHLH-encoding ones. Of these non-HLH genes, groucho has been shown to encode a ubiquitous nuclear protein involved in a number of processes, including neuroblast specification (38, 39, 41, 57). Two-hybrid analysis has shown that the E(spl) bHLH proteins and Hairy interact with Groucho (41). We have confirmed the fact that Gro can strongly bind Hairy and the E(spl) bHLH proteins regardless of fusion orientation. In contrast, neither Da nor the proneural proteins were observed to do so (Tables 1, 2, and 3).
The C-terminal tetrapeptide common to E(spl) and Hairy is necessary and sufficient for Groucho binding (41, 58). We have confirmed this using our C-terminally truncated LexA-M3bHLH and M3bHLH-O constructs, both of which lack the tetrapeptide and accordingly show no interaction with Groucho. We have further shown that a nonsense point mutation in the last codon of M5 (M5ΔW) abolishes its interaction with Gro, but continues to retain its ability to interact with all E(spl) proteins as efficiently as wild-type M5 (Table 2). Runt bears the variant C terminus WRPY, and has the ability to associate with Gro (Table 1). Despite its interaction with Gro, Runt appears not to interact with any of the other proteins tested, namely Hairy, E(spl) M3 and M8, and Emc (Table 1). This is consistent with the apparent noninvolvement of Runt in the initial segregation of neural precursors, although Runt is involved in subsequent steps of neural development, as well as in embryo segmentation along with Hairy (52).
DISCUSSION
Summary of Two-Hybrid Interactions.
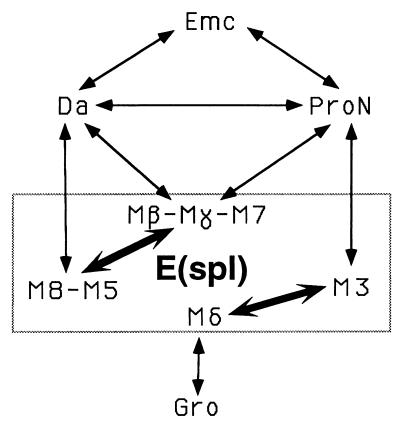
Using the yeast two-hybrid system, we have confirmed previously documented bHLH interactions (1, 6, 10, 25, 41) and demonstrated that E(spl) proteins can associate with each other and with proneural proteins (Ac, Sc, Ato), as well as with Ase and Da (summarized in Fig. 3).
Figure 3.
Summary of interactions. Interactions are indicated by arrows connecting the proteins involved. Proteins grouped together display qualitatively identical interactions with other proteins in this study. ProN, proneural proteins, typified by Ac and Sc. All E(spl) proteins (in box) interact with each other and with Gro. Thick arrows in the box indicate the strongest interactions within the E(spl) family.
The existence of some E(spl) homodimers has been previously demonstrated by the in vitro ability of individual E(spl) proteins to bind specific DNA sites (18, 19). Here, we show that virtually all E(spl) homo- or heterodimers can form in yeast, with marked preferences for certain combinations (Fig. 3). It is possible that in fly cells these heterodimers are favored over homodimers, a plausible scenario because the expression domains of the seven E(spl) bHLH proteins overlap extensively. In our assay, Hairy could only be tested against E(spl) M3 and displayed no interaction—it is formally possible that it may interact with another E(spl) protein.
In addition to associating with each other, E(spl) proteins display robust interactions with the bHLH activators of the proneural class, as well as with Da. Based on their partner preferences, we can divide the seven E(spl) proteins into subgroups (Fig. 3). If these interactions reflect some aspect of their functions in vivo, we can account for the conservation of the seven E(spl) proteins between Drosophila melanogaster and D. hydei (40).
Our deletion analysis of M3 and Sc is consistent with their interaction being mediated by their respective bHLH domains. The dependence of the E(spl)–activator HLH interactions on the HLH domain is nicely reflected in the fact that the functional grouping of the E(spl) proteins correlates well with the amino acid sequences of their bHLH domains (13, 14), e.g., M5 and M8 have highly similar bHLH regions, different from those of the M7/Mβ/Mγ group, which also display high intragroup similarity.
The Hairy/E(spl) class bHLH proteins known in Drosophila to date include the seven E(spl) proteins, Hairy, and Deadpan (Dpn), a protein involved in neural precursor cell differentiation (but not in the precursor selection process), as well as X/A ratio calculation (12, 59). Dpn has also been shown to interact with Da, but not with Sc, using a yeast two-hybrid assay (60). Unlike E(spl) and Dpn, LexA-Hairy displays no interactions with either Da or ASC proteins. We earlier observed mutually antagonistic effects of early overexpression of hairy and sc on sex determination that were consistent with a model in which the two proteins interact (20, 43). In view of our present data, this could be accounted for if Hairy acts as a repressor and Sc as an activator of the target gene Sxl independently of one another. It is still possible that Hairy and Sc do interact in a complex, but the yeast system may be unable to reproduce the in vivo dimerization abilities of Hairy. We do not believe this would be due to misfolding of LexA-Hairy, as it interacts with Groucho as well as with novel proteins identified in a two-hybrid screen (G.P. and S.M.P., unpublished data). It is conceivable that endogenous yeast proteins mask some dimerization domain of Hairy or that Hairy actively represses the yeast reporter construct. Further work will be needed to distinguish among various alternative explanations.
Recently, Gigliani et al. (73) reported pairwise interactions between the HLH domains of E(spl) and Ac/Sc proteins. Their assay was based on the ability of λ repressor fusion proteins to confer immunity to λ. Even though a wide range of interactions was observed, there were significant differences from our results. Most conspicuous was the inability of Mβ, Mγ, and Mδ to form heterodimers with either E(spl) or Ac/Sc proteins in the bacterial assay. Also, the interactions reported between E(spl) proteins and Ac/Sc led to the categorization of M5 and M8 together with M7, which is contrary to our classification. The differences are most likely due to the different systems used to assay interactions and the use of truncated vs. full-length proteins. In vivo evidence of association among the different proteins in question is needed to decide which of the two systems better approximates the biologically relevant interactions.
Implications on the Function of Hairy/E(spl) and Proneural Proteins.
One cannot extrapolate from results obtained in a heterologous system to the situation in the parent organism with certainty. The interactions we report must be considered as potential and their in vivo relevance has to be confirmed by future genetic experiments. The occurrence of these interactions in Drosophila will depend on a number of parameters, such as presence and concentrations of the two partners, posttranslational modifications, presence of competitor proteins, and relative interaction affinities. Of these parameters, the only one that may be gleaned by the two-hybrid system is interaction affinity, reflected by the level of transcriptional activation in yeast (49). In our case, the measurements were generally congruent when we tested an interacting pair in the two possible orientations. We therefore conclude that our results are a gross indicator of respective interaction affinities.
Expression of proneural genes in embryos and imaginal discs occurs in fields of cells that overlap extensively with the expression domains of emc and E(spl) (26, 61, 62). Even ase, which is usually expressed in neural precursors, is expressed in the proneural field of the wing margin sensilla, where it should have the opportunity to interact with E(spl) proteins (54). gro and da are ubiquitously expressed (38, 63). hairy expression, on the other hand, is absent from the embryonic central nervous system primordium (64), but may occur transiently in imaginal PNS precursors (27, 65). As these proteins can coexist in Drosophila cells, we can use our two-hybrid data to extend current models for their function, which can subsequently be directly tested in Drosophila.
Proneural genes are under complex transcriptional control. After their initial expression in proneural clusters, ac and sc probably rely on autoactivation for persistent expression in the cell that will become the neural precursor (25, 34). This autoactivation requires class A E-boxes (EA, CAGNTG) that bind Da/proneural heterodimers. Hairy/E(spl) proteins inhibit this autoactivation, but none of them have been reported to bind EA sites. In the case of Hairy, this inhibition has been shown to be absolutely dependent on the presence of a nearby Class C site (EC, CACGCG), that is bound by Hairy or E(spl) homodimers (18, 19). A similar situation exists for the early promoter of Sxl: it can be activated by Da/Sc binding to an EA box and repressed by Dpn binding to a nearby EC box (66). DNA-bound activator and repressor proteins could exert their effects without interacting with each other; alternatively, a repressor could act only on specific activators to abolish their transcriptional activation upon repressor-activator contact. Indeed, E(spl) proteins could use the latter mechanism given the facts that (i) they have strong affinities for specific activator proteins and (ii) their target sites are found in the vicinity of EA activator binding sites in the ac, Sxl, and other promoters (18, 19, 66, 67). In agreement with this model is the finding that E(spl) M8 and M5 repress Da-activated transcription from an E(spl)m8-CAT fusion reporter in cultured cells, whereas basal transcription (without Da) is unaffected by their presence (68). If indeed Hairy does not interact with Da and proneural proteins, it could achieve repression by contacting other factors, perhaps the basal transcription machinery itself or factors affecting chromatin.
In this postulated DNA-bound Da/proneural/E(spl) higher order complex, repression could be regulated by the affinity of the repressor for the activator rather than the occupancy of their respective target sites. The Sc-S > D mutant suggests that this affinity could be regulated by phosphorylation (Table 1), i.e. phosphorylation of Sc could relieve it from the negative effects of a nearby bound E(spl). Because a truncated Sc protein (Sc bHLH) retains the ability to interact with M3, it is possible that the C-terminal tail itself does not contact M3, rather, that its phosphorylation could affect some intramolecular conformational change that specifically abolishes Sc-E(spl) interaction.
The strong interactions observed between E(spl) proteins and proneural proteins might lead one to hypothesize that E(spl) proteins act like Emc, i.e. by sequestering HLH activators. A suggestion that this is indeed possible comes from in vivo data that detect residual activities of E(spl) proteins with mutated basic domains (69, 70). Still, the fact that these activities are weak confirms that these proteins do not normally function like Emc, but, rather, require intact basic domains. Indeed, in tissue culture transfection assays, E(spl) proteins repress transcription only in the presence of a cognate DNA target site (18, 19, 37, 68). Repression of promoters driven solely by EA boxes has not been reported, nor has addition of E(spl), Hairy, or Dpn been able to cause any perturbation to Da/Ac (Sc)–EA DNA complexes in vitro (10, 66).
A number of vertebrate proteins bearing the domains characteristic of Hairy/E(spl) proteins have been isolated. They appear to be functionally similar to the related Drosophila proteins. At least one has been shown to be transcribed as a response to Notch signaling (71) and to antagonize bHLH activators (15). In terms of their potential interactions with other HLH proteins, such proteins exhibit a variety of behaviors. HES-1 is able to abolish the formation of activator HLH–EA complexes in vitro, by interacting with the E-protein E47 and/or with activators such as MyoD (15). Consequently, in transcriptional assays, it represses E47/activator-induced transcription. In contrast, HES-2 seems unable to repress E47-activated transcription, whereas it does repress the basal activity of a β-actin promoter, when bound upstream of it (72). Studies of the interaction abilities of vertebrate HES proteins using the two-hybrid system or other methods should shed light on the mechanisms of action of these factors.
Acknowledgments
We thank Sarah Bray, Amy Chen, Fotis Kafatos, Dave Turner, and the Delidakis and Parkhurst labs for comments on the manuscript. We also thank Roger Brent, Russ Finley, Stan Hollenberg, Dave Turner, Anne Vojtek, and Tony Zervos for providing materials and advice for the two-hybrid system, and Rena Skoula and George Apidianakis for help in plasmid construction and yeast transformation. This work was supported by a EU SCIENCE Program grant and a Human Frontier Science Program Research grant (to C.D.), and by a National Institutes of Health grant and Leukemia Society of America Scholar Award (to S.M.P.). S.M.P. is a Pew Scholar in the Biomedical Sciences.
ABBREVIATIONS
- HLH
helix-loop-helix
- bHLH
basic HLH
- Emc
extramacrochaetae
- E(spl)
Enhancer of split
- ASC
achaete–scute complex
- X-Gal
5-bromo-4-chloro-3-indolylβ-d-galactoside
- PNS
peripheral nervous system
References
- 1.Murre C, Schonleber McCaw P, Baltimore D. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 2.Murre C, Schonleber McCaw P, Vaessin H, Caudy M, Jan L Y, Jan Y N, Hauschka S D, Lassar A B, Weintraub H, Baltimore D. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 3.Weintraub H, Davis R, Tapscott S, Thayer M, Krause M, Benezra R, Blackwell T K, Turner D, Rupp R, Hollenberg S, Zhuang Y, Lassar A. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- 4.Lassar A B, Davis R L, Wright W E, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- 5.Campuzano S, Modolell J. Trends Genet. 1992;8:202–208. doi: 10.1016/0168-9525(92)90234-u. [DOI] [PubMed] [Google Scholar]
- 6.Cabrera C, V, Alonso M C. EMBO J. 1991;10:2965–2973. doi: 10.1002/j.1460-2075.1991.tb07847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benezra R, Davis R L, Lockshon D, Turner D L, Weintraub H. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 8.Ellis H M, Spann D R, Posakony J W. Cell. 1990;61:27–38. doi: 10.1016/0092-8674(90)90212-w. [DOI] [PubMed] [Google Scholar]
- 9.Garrell J, Modolell J. Cell. 1990;61:39–48. doi: 10.1016/0092-8674(90)90213-x. [DOI] [PubMed] [Google Scholar]
- 10.Van Doren M, Ellis H M, Posakony J W. Development (Cambridge, UK) 1991;113:245–255. doi: 10.1242/dev.113.1.245. [DOI] [PubMed] [Google Scholar]
- 11.Rushlow C A, Hogan A, Pinchin S M, Howe K M, Lardelli M T, Ish-Horowicz D. EMBO J. 1989;8:3095–3103. doi: 10.1002/j.1460-2075.1989.tb08461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bier E, Vässin H, Younger-Shepherd S, Jan L Y, Jan Y N. Genes Dev. 1992;6:2137–2151. doi: 10.1101/gad.6.11.2137. [DOI] [PubMed] [Google Scholar]
- 13.Klämbt C, Knust E, Tietze K, Campos-Ortega J A. EMBO J. 1989;8:203–210. doi: 10.1002/j.1460-2075.1989.tb03365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delidakis C, Artavanis-Tsakonas S. Proc Natl Acad Sci USA. 1992;89:8731–8735. doi: 10.1073/pnas.89.18.8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sasai Y, Kageyama R, Tagawa Y, Shigemoto R, Nakanishi S. Genes Dev. 1992;6:2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- 16.Feder J N, Li L, Jan L Y, Jan Y N. Genomics. 1994;20:56–61. doi: 10.1006/geno.1994.1126. [DOI] [PubMed] [Google Scholar]
- 17.Tietze K, Oellers N, Knust E. Proc Natl Acad Sci USA. 1992;89:6152–6156. doi: 10.1073/pnas.89.13.6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohsako S, Hyer J, Panganiban G, Oliver I, Caudy M. Genes Dev. 1994;8:2743–2755. doi: 10.1101/gad.8.22.2743. [DOI] [PubMed] [Google Scholar]
- 19.Van Doren M, Bailey A M, Esnayra J, Ede K, Posakony J W. Genes Dev. 1994;8:2729–2742. doi: 10.1101/gad.8.22.2729. [DOI] [PubMed] [Google Scholar]
- 20.Dawson S R, Turner D L, Weintraub H, Parkhurst S M. Mol Cell Biol. 1995;15:6923–6931. doi: 10.1128/mcb.15.12.6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wainwright S M, Ish-Horowicz D. Mol Cell Biol. 1992;12:2475–2483. doi: 10.1128/mcb.12.6.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarman A P, Grau Y, Jan L Y, Jan Y N. Cell. 1993;73:1307–1321. doi: 10.1016/0092-8674(93)90358-w. [DOI] [PubMed] [Google Scholar]
- 23.Moscoso del Prado J, García-Bellido A. Roux’s Arch Dev Biol. 1984;193:242–245. doi: 10.1007/BF01260345. [DOI] [PubMed] [Google Scholar]
- 24.Caudy M, Vässin H, Brand M, Tuma R, Jan L Y, Jan Y N. Cell. 1988;55:1061–1067. doi: 10.1016/0092-8674(88)90250-4. [DOI] [PubMed] [Google Scholar]
- 25.Van Doren M, Powell P A, Pasternak D, Singson A, Posakony J W. Genes Dev. 1992;6:2592–2605. doi: 10.1101/gad.6.12b.2592. [DOI] [PubMed] [Google Scholar]
- 26.Cubas P, Modolell J. EMBO J. 1992;11:3385–3393. doi: 10.1002/j.1460-2075.1992.tb05417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skeath J B, Carroll S B. Genes Dev. 1991;5:984–995. doi: 10.1101/gad.5.6.984. [DOI] [PubMed] [Google Scholar]
- 28.Cubas P, de Celis J-F, Campuzano S, Modolell J. Genes Dev. 1991;5:996–1008. doi: 10.1101/gad.5.6.996. [DOI] [PubMed] [Google Scholar]
- 29.Stern C. Am Sci. 1954;42:213–247. [Google Scholar]
- 30.Ghysen A, Dambly-Chaudiére C. Genes Dev. 1988;2:495–501. doi: 10.1101/gad.2.5.495. [DOI] [PubMed] [Google Scholar]
- 31.Gómez-Skarmeta J-L, Diez del Corral R, de la Calle-Mustienes E, Ferrés-Marcó D, Modolell J. Cell. 1996;85:95–105. doi: 10.1016/s0092-8674(00)81085-5. [DOI] [PubMed] [Google Scholar]
- 32.Orenic T V, Held L I, Paddock S W, Carroll S B. Development (Cambridge, UK) 1993;118:9–20. doi: 10.1242/dev.118.1.9. [DOI] [PubMed] [Google Scholar]
- 33.Skeath J B, Carroll S B. Development (Cambridge, UK) 1992;114:939–946. doi: 10.1242/dev.114.4.939. [DOI] [PubMed] [Google Scholar]
- 34.Ruiz-Gómez M, Ghysen A. EMBO J. 1993;12:1121–1130. doi: 10.1002/j.1460-2075.1993.tb05753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin-Bermudo M D, Carmena A, Jinénez F. Development (Cambridge, UK) 1995;121:219–224. doi: 10.1242/dev.121.1.219. [DOI] [PubMed] [Google Scholar]
- 36.Jennings B, Preiss A, Delidakis C, Bray S. Development (Cambridge, UK) 1994;120:3537–3548. doi: 10.1242/dev.120.12.3537. [DOI] [PubMed] [Google Scholar]
- 37.Heitzler P, Bourouis M, Ruel L, Carteret C, Simpson P. Development (Cambridge, UK) 1996;122:161–171. doi: 10.1242/dev.122.1.161. [DOI] [PubMed] [Google Scholar]
- 38.Delidakis C, Preiss A, Hartley D A, Artavanis-Tsakonas S. Genetics. 1991;129:803–823. doi: 10.1093/genetics/129.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schrons H, Knust E, Campos-Ortega J A. Genetics. 1992;132:481–503. doi: 10.1093/genetics/132.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maier D, Marte B M, Schäfer W, Yu Y, Preiss A. Proc Natl Acad Sci USA. 1993;90:5464–5468. doi: 10.1073/pnas.90.12.5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paroush Z, Finley R L F J, Kidd T, Wainwright S M, Ingham P, W, Brent R, Ish-Horowicz D. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 42.Torres M, Sanchez L. EMBO J. 1989;8:3079–3086. doi: 10.1002/j.1460-2075.1989.tb08459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parkhurst S M, Bopp D, Ish-Horowicz D. Cell. 1990;63:1179–1191. doi: 10.1016/0092-8674(90)90414-a. [DOI] [PubMed] [Google Scholar]
- 44.Cline T W. Trends Genet. 1993;9:385–390. doi: 10.1016/0168-9525(93)90138-8. [DOI] [PubMed] [Google Scholar]
- 45.Fields S, Song O. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 46.Gyuris J, Golemis E A, Chertkov H, Brent R. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 47.Vojtek A B, Cooper J A. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 48.Hollenberg S M, Sternglanz R, Cheng P F, Weintraub H. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Estojak J, Brent R, Golemis E A. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith G A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1995. [Google Scholar]
- 51.Kania M A, Bonner A S, Duffy J B, Gergen J P. Genes Dev. 1990;4:1701–1713. doi: 10.1101/gad.4.10.1701. [DOI] [PubMed] [Google Scholar]
- 52.Duffy J B, Kania M A, Gergen J P. Development (Cambridge, UK) 1991;113:1223–1230. doi: 10.1242/dev.113.4.1223. [DOI] [PubMed] [Google Scholar]
- 53.Alonso M C, Cabrera C V. EMBO J. 1988;7:2585–2591. doi: 10.1002/j.1460-2075.1988.tb03108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dominguez M, Campuzano S. EMBO J. 1993;12:2049–2060. doi: 10.1002/j.1460-2075.1993.tb05854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brand M, Jarman A P, Jan L Y, Jan Y N. Development (Cambridge, UK) 1993;119:1–17. doi: 10.1242/dev.119.Supplement.1. [DOI] [PubMed] [Google Scholar]
- 56.Jarman A P, Brand M, Jan L Y, Jan Y N. Development (Cambridge, UK) 1993;119:19–29. doi: 10.1242/dev.119.Supplement.19. [DOI] [PubMed] [Google Scholar]
- 57.Hartley D A, Preiss A, Artavanis-Tsakonas S. Cell. 1988;55:785–795. doi: 10.1016/0092-8674(88)90134-1. [DOI] [PubMed] [Google Scholar]
- 58.Fisher A L, Ohsako S, Caudy M. Mol Cell Biol. 1996;16:2670–2677. doi: 10.1128/mcb.16.6.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Younger-Shepherd S, Vaessin H, Bier E, Jan L Y, Jan Y N. Cell. 1992;70:911–922. doi: 10.1016/0092-8674(92)90242-5. [DOI] [PubMed] [Google Scholar]
- 60.Liu Y, Belote J M. Mol Gen Genet. 1995;248:182–189. doi: 10.1007/BF02190799. [DOI] [PubMed] [Google Scholar]
- 61.Knust E, Schrons H, Grawe F, Campos-Ortega J A. Genetics. 1992;132:505–518. doi: 10.1093/genetics/132.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jennings B, de Celis J, Delidakis C, Preiss A, Bray S. Development (Cambridge, UK) 1995;121:3745–3752. [Google Scholar]
- 63.Vaessin H, Brand M, Jan L Y, Jan Y N. Development (Cambridge, UK) 1994;120:935–945. doi: 10.1242/dev.120.4.935. [DOI] [PubMed] [Google Scholar]
- 64.Carroll S B, Laughon A, Thalley B S. Genes Dev. 1988;2:883–890. doi: 10.1101/gad.2.7.883. [DOI] [PubMed] [Google Scholar]
- 65.Carroll S B, Whyte J S. Genes Dev. 1989;3:905–916. [Google Scholar]
- 66.Hoshijima K, Kohyama A, Watakabe I, Inoue K, Sakamoto H, Shimura Y. Nucleic Acids Res. 1995;23:3441–3448. doi: 10.1093/nar/23.17.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singson A, Leviten M W, Bang A G, Hua X H, Posakony J W. Genes Dev. 1994;8:2058–2071. doi: 10.1101/gad.8.17.2058. [DOI] [PubMed] [Google Scholar]
- 68.Oellers N, Dehio M, Knust E. Mol Gen Genet. 1994;244:465–473. doi: 10.1007/BF00583897. [DOI] [PubMed] [Google Scholar]
- 69.Tietze K, Schrons H, Campos-Ortega J A, Knust E. Roux’s Arch Dev Biol. 1993;203:10–17. doi: 10.1007/BF00539885. [DOI] [PubMed] [Google Scholar]
- 70.Nakao K, Campos-Ortega J A. Neuron. 1996;16:275–286. doi: 10.1016/s0896-6273(00)80046-x. [DOI] [PubMed] [Google Scholar]
- 71.Jarriault S, Brou C, Logeat F, Schroeter E H, Kopan R, Israel A. Nature (London) 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 72.Ishibashi M, Sasai Y, Nakanishi S, Kageyama R. Eur J Biochem. 1993;215:645–652. doi: 10.1111/j.1432-1033.1993.tb18075.x. [DOI] [PubMed] [Google Scholar]
- 73.Gigliani F, Longo F, Gaddini L, Battaglia P A. Mol Gen Genet. 1996;251:628–634. doi: 10.1007/BF02174111. [DOI] [PubMed] [Google Scholar]