Abstract
Changes in the morphology of a dendritic spine require remodeling of its actin-based cytoskeleton. Biochemical mechanisms underlying actin remodeling have been studied extensively, but little is known about the physical organization of the actin-binding proteins that mediate remodeling in spines. Long-term potentiation-inducing stimuli trigger expansion of the spine head, suggesting local extension and branching of actin filaments. Because filament branching requires the Arp2/3 complex, we used quantitative immunoelectron microscopy to elucidate the organization of ARPC-2 (Arp2/3 complex subunit 2), an essential component of the complex. Our data from CA1 hippocampus indicate that Arp2/3 concentrates within spines in a previously unrecognized torroidal domain, apparently specialized to mediate actin filament branching.
Keywords: actin, cytoskeleton, hippocampus, LTP, pyramidal cell, synaptic plasticity
Introduction
Spines, actin-rich protrusions that emerge from the dendritic shaft of excitatory neurons in the mammalian forebrain, are the primary locus of synaptic contacts. Spines can change their size and shape in response to synaptic activity (Lippman and Dunaevsky, 2005). Recent in vitro evidence shows that long-term potentiation (LTP) induction protocols lead to spine enlargement (Kopec et al., 2006; Park et al., 2006; Harvey and Svoboda, 2007); moreover, the synapses on these enlarged spines express more AMPA-type glutamate receptors (Matsuzaki et al., 2001; Kasai et al., 2003). That similar conditions obtain in vivo is suggested by ultrastructural evidence that larger spines have larger postsynaptic densities (PSDs) (Harris and Stevens, 1989), and that larger PSDs host more AMPA receptors (Kharazia and Weinberg, 1999; Takumi et al., 1999; Nusser, 2000). Therefore, spine size is tightly linked to synaptic strength, implying machinery within the spine that coordinates modifications in synaptic efficacy and spine size. Changes in spine morphology require remodeling of the actin filaments comprising the “spinoskeleton” (Chen et al., 2007; Lynch et al., 2007). That actin remodeling itself may play a causal role in LTP (Fifkova and Delay, 1982) is supported by evidence that spines enlarge before AMPA receptors accumulate at the spine surface (Kopec et al., 2006), and more directly, by evidence that blocking actin remodeling pharmacologically interferes with LTP (Fukazawa et al., 2003).
The actin cytoskeleton is reorganized dynamically via an elaborate network of biochemical cascades. Filament branching requires the actin-related protein 2/3 (Arp2/3) complex (Goley and Welch, 2006; Pollard, 2007). After activation, this evolutionarily conserved supramolecular assembly initiates a new “daughter” actin filament, branching from the side of an existing “mother” filament. The Arp2/3 complex plays a central role in a wide range of basic processes throughout the eukaryotes, including cell migration, phagocytosis, exocytosis, and endocytosis (Bretschneider et al., 2004; Hurst et al., 2004; Toshima et al., 2005; Zuo et al., 2006; Cai et al., 2007; Galletta et al., 2008). Recently, Arp2/3 was shown also to play a key role in regulating the morphology of dendritic spines (Kim et al., 2006). Recent work suggests considerable organization within hippocampal spines (Blanpied et al., 2002; Cooney et al., 2002; Rácz and Weinberg, 2004, 2006; Rácz et al., 2004; Park et al., 2006), leading us to wonder whether compartmentalization of Arp2/3 within the spine might help to maintain this internal organization. Using immunoelectron microscopy, we show here that Arp2/3 complex subunit 2 (ARPC-2; an essential component of the Arp2/3 complex) (Mullins et al., 1997; Gournier et al., 2001) in the spine head concentrates in a restricted domain likely to play a special role in actin remodeling.
Materials and Methods
Experiments were performed on adult Sprague Dawley rats; housing and experimental procedures complied with Institutional Animal Care and Use Committee guidelines. Animals anesthetized with pentobarbital (60 mg/kg, i.p.) were perfused intracardially with saline, followed by a mixture of depolymerized paraformaldehyde (PFA) (4%), glutaraldehyde (0.2–0.5%), and picric acid (1%) in 0.1 m phosphate buffer, pH 7.4 (for EM), or 4% PFA (for LM). Fifty micrometer coronal Vibratome sections were processed for immunohistochemistry (Rácz et al., 2004; Rácz and Weinberg, 2004, 2006).
The primary antibody was a polyclonal rabbit anti-ARPC-2 (also called p34 in mammals, and arc35 in yeast) generated against a peptide corresponding to amino acids 285–298 of human ARPC-2 (Upstate Biotechnology #07-227). This widely used antibody has been well characterized (Di Nardo et al., 2005; Kempiak et al., 2005; Hotulainen and Lappalainen, 2006). Western blot analysis confirmed that the antibody recognizes a single band in rat brain homogenate, migrating at ∼34 kDa (supplemental Fig. 1, available at www.jneurosci.org as supplemental material).
Electron micrographs of 125 randomly selected immunolabeled spine profiles, including synapses, were taken from proximal and middle regions of CA1 stratum radiatum from three animals. Membrane perimeters, profile areas, and distances were measured using NIH ImageJ (v1.38; see http://rsb.info.nih.gov/ij). To determine “axo-dendritic” position of immunogold particles, we defined the lateral edges of the PSD for a random sample of clearly defined synapses and measured the distance from each gold particle lying within this synaptic region to the postsynaptic membrane (supplemental Fig. 2, inset, available at www.jneurosci.org as supplemental material) (Valtschanoff and Weinberg, 2001; Rácz and Weinberg, 2004).
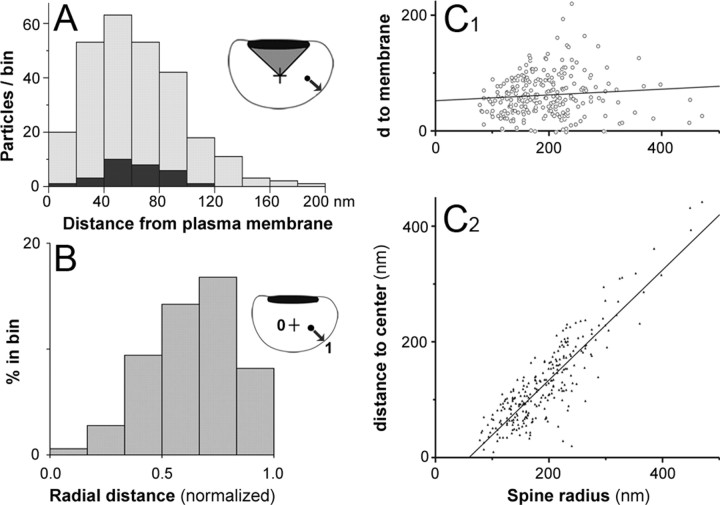
Using the same micrographs, we measured distances of gold particles from the closest point on the spine plasma membrane and from the geometric center of the spine profile (defined as the bisection of a line from the PSD center to the furthest point on the spine profile) to determine “radial position” (see Fig. 3 A, inset).
Figure 3.
Radial organization of labeling. A, Stacked histogram shows labeling in relationship to the spine membrane; data from the PSD wedge (inset) are shown in black. Immunogold particles concentrated 20–100 nm from the membrane. B, Histogram shows distribution of normalized positions (0 corresponds to a particle at the spine center; 1.0 to a particle at the plasma membrane) (for details, see Materials and Methods). Particles concentrated between 0.4 and 0.8 normalized radial units within the spine head (see inset; black dot represents gold particle, and the arrow represents distance from the particle to the membrane). C1, Scatterplot: radius of the spine head is unrelated to distance to the plasma membrane; the line shows least-squares linear regression fit (r = 0.09). C2, In contrast, note the striking relationship between spine radius and distance to spine center (r = 0.87).
Normalized radial position, rN (the fraction of the distance from the center to the plasma membrane), was computed according to the following equation:
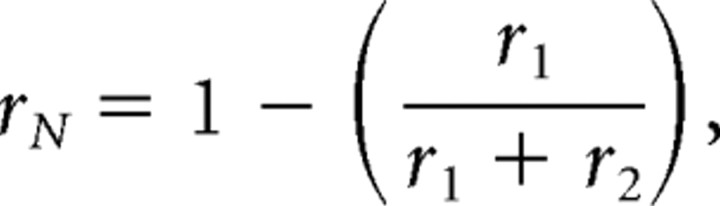 |
where r 1 is the radial distance from the membrane to the particle, and r 2 is the radial distance of the particle from the geometric center of the spine. Therefore, 1.0 corresponds to a particle at the plasma membrane, and 0 corresponds to a particle at the spine center. Only particles lying within the spine were considered.
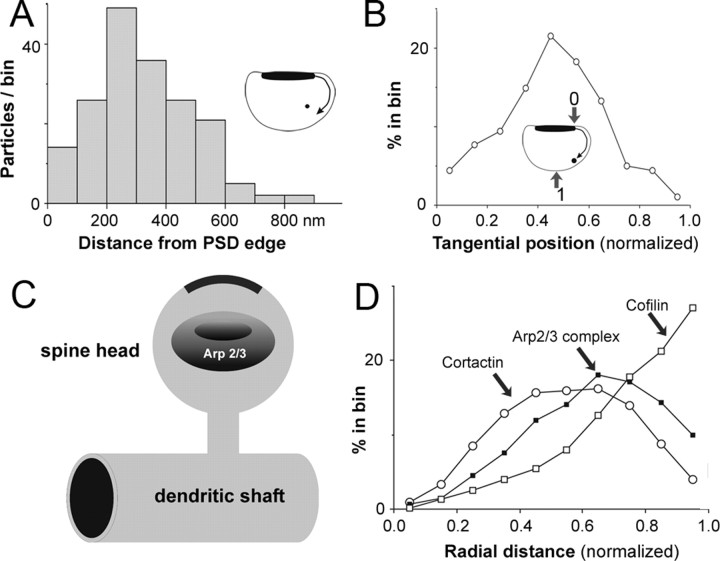
To investigate “tangential” distribution of ARPC-2 within the spinoplasm, we identified the point on the plasma membrane closest to each particle and determined the distance from there to the PSD edge, measured along the membrane (see Fig. 4 A, inset). To compensate for differences in spine sizes, we normalized particle positions, such that 0 corresponded to the PSD edge and 1.0 to a point equidistant from both edges of the PSD (Fig. 4 B, inset).
Figure 4.
Organization of ABPs in spines. A, Tangential distribution of ARPC-2 (see inset and Materials and Methods). ARPC-2 was sparse near the synapse, peaking 200–400 nm from the PSD. B, Normalized particle positions: 0 corresponds to PSD edge, and 1.0 corresponds to the point equidistant from both edges of the PSD (inset). Particles concentrate ∼0.4–0.6 U away from the PSD. C, Schematic representation of the Arp2/3 complex-enriched domain. D, Graph showing spatial distribution of cofilin (white squares), the Arp2/3 complex (black squares), and cortactin (white circles) within spines. Cofilin concentrates closest to the plasma membrane [postembedding immunogold data from Rácz and Weinberg (2006)], Arp2/3 at an intermediate position, and cortactin peaks farthest from the membrane [preembedding (Rácz and Weinberg, 2004)].
Data were collected in Microsoft Excel for additional analysis; Kaleidagraph (Synergy Software) and CricketGraph (Cricket Software) were used to compute statistics and generate graphs. We detected no significant differences in data from different animals (p > 0.2; two-sided t tests) and therefore pooled the data. Line graphs were smoothed digitally, using a three-point weighted running average. For all but the end points, y j(smoothed) = [y (j − 1) + 2yj + y (j + 1)]/4. For the end points, y 1(smoothed) = (2y 1 + y 2)/3, and y n(smoothed) = [y (n − 1) + 2yn]/3.
Results
Immunostaining for the Arp2/3 complex was widespread throughout the gray matter of the brain. Staining was prominent in areas with numerous spiny neurons. In the hippocampus, staining was remarkably strong in the CA1 subregion (Fig. 1 A). ARPC-2 immunostaining in CA1 concentrated in the cell bodies and dendrites of pyramidal neurons, sparing the nucleus (Fig. 1 B,C). Numerous intensely stained puncta were visible at high magnification (Fig. 1 D), raising the suspicion that ARPC-2 might be associated with spines.
Figure 1.
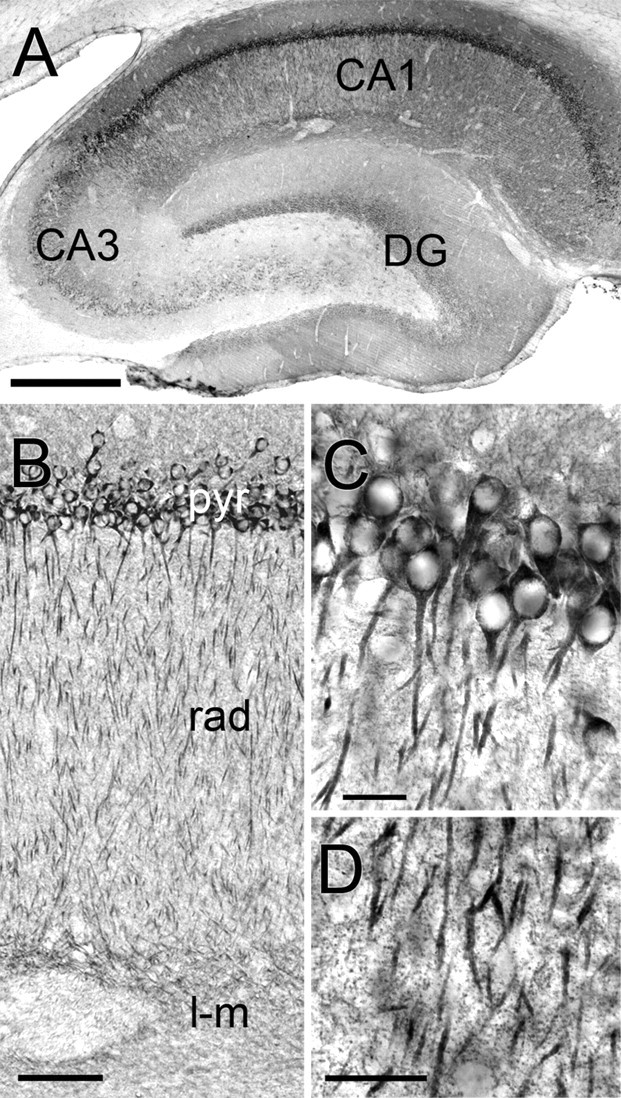
Immunoperoxidase staining for ARPC-2 in hippocampus. A, Low-magnification view; staining is strongest in CA1. B, Higher-magnification view shows laminar staining. pyr, Pyramidal layer; rad, stratum radiatum; l-m, stratum lacunosum-moleculare. C, Pyramidal cell layer of CA1. Staining is prominent in perikarya and apical dendrites but spares nuclei. D, High magnification reveals immunopositive puncta in the neuropil of stratum radiatum, likely corresponding to dendritic spines. Scale bars: A, 1 mm; B, 250 μm; C, D, 50 μm.
To obtain a clearer understanding of its subcellular organization, we performed immunogold labeling for ARPC-2, using pre-embedding methods to optimize sensitivity. After labeling with silver-enhanced 1.4 nm gold, numerous electron-dense particles were seen in dendrites and spines, and occasionally in axon terminals. Particles were seen throughout the spine, seemingly more common away from the plasma membrane (Fig. 2 A–D). Particles in spines and dendritic shafts were often associated with filamentous structures resembling F-actin, especially at branch points (Fig. 2 C,D). Particles were also associated with vesicular structures occasionally visible within spines or in shafts near the spine neck (Fig. 2 E,F).
Figure 2.
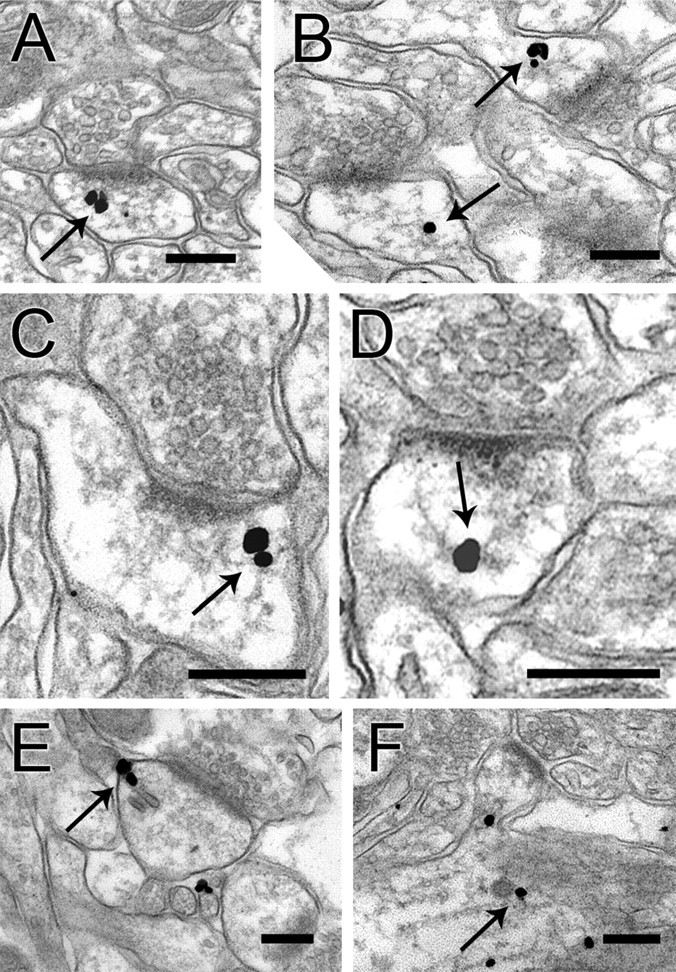
Preembedding immunogold labeling in dendritic spines. A, B, Immunogold particles (arrows) within the spine cytoplasm, typically lying away from the membrane. C, D, Particles coding for ARPC-2 were often associated with thin filaments, in the vicinity of branch points (arrows). E, Labeling associated with a likely endosome in a spine profile. F, Gold particles associated with filamentous structures and a coated vesicle (arrow) within a dendritic shaft. Another gold particle is associated with a vesicle lying in the neck of a spine from this shaft. Scale bars, 250 nm.
We performed quantitative analysis to determine whether the Arp2/3 complex is restricted to specific spine domains. Because the domain defined by the synapse may exhibit distinct properties, we compared gold particles lying within the wedge defined by the lateral edges of the PSD and the geometric center of the spine with particles lying outside of this area (Fig. 3 A). Finding no obvious differences, we pooled the two samples for subsequent analysis. ARPC-2 concentrated away from the plasma membrane but also lay away from the geometric center of the spine, peaking between 20 and 100 nm from the plasma membrane (Fig. 3 A). Data were generally consistent among animals (means of 66.7 ± 3.2, 62.6 ± 3.7, and 59.9 ± 4.4 nm). Pooling data from the three animals, the label was an average of 63.3 ± 2.1 nm from the plasma membrane (n = 263 particles). These data suggest that the Arp2/3 complex concentrates in a shell between the plasma membrane and the center of the spine.
Gold particles were uncommon >100 nm from the membrane (Fig. 3 A). However, these data come from spine profiles of different radii (range, 80–470 nm; median, 177 nm). To pool the data more effectively, we normalized radial position (see Materials and Methods). Labeling again concentrated in a radial zone close to the plasma membrane (Fig. 3 B). Further adjustment to correct for unequal bin areas did not change our conclusion that Arp2/3 concentrates in an intermediate zone between the center and the plasma membrane (supplemental Fig. 3A, available at www.jneurosci.org as supplemental material).
We examined the distance of particles from the membrane, and from the spine center, as a function of spine radius (Fig. 3 C). Unexpectedly, we found no relationship between spine size and distance to the membrane (r 2 < 0.01) but a very strong relationship between spine size and distance to the center (r 2 > 0.76; p < 0.001). We conclude that the antigen tends to lie at a fixed distance from the plasma membrane, independent of spine size.
We then asked whether the Arp2/3 complex has a preferred “angular” location with respect to the axis defined by the PSD. We found that ARPC-2 was sparse in the wedge defined by the synaptic specialization, instead concentrating in a region 200–400 nm from the PSD edge, measured along the membrane (Fig. 4 A). Because this tangential distance was correlated with the total perimeter of the spine (data not shown), we normalized particle positions such that 0 corresponded to the PSD edge and 1.0 to the point equidistant along the perimeter of the spine from both edges of the PSD (Fig. 4 B, inset). We considered only particles lying away from the spine center (normalized radial distance > 0.4) (Fig. 3 B, supplemental Fig. 3B, available at www.jneurosci.org as supplemental material), because the true tangential position of the few particles lying in the center is inherently poorly defined. Pooling these data, we found that ARPC-2 concentrated in a region 0.4–0.6 normalized units from the PSD (Fig. 4 B) (mean, 0.44 ± 0.02; n = 189 particles). We conclude that in three dimensions, ARPC-2 concentrates within a donut-shaped zone lying halfway between the plasma membrane and the center and halfway between the PSD and the spine neck (Fig. 4 C).
Discussion
Methodological constraints
The Arp2/3 complex contains ARPC-2 and six other proteins assembled into a tightly linked biochemical package (Robinson et al., 2001), suggesting that ARPC-2 is a reliable probe for this complex, although this has not been demonstrated directly in brain. Proteomic studies have identified Arp2/3 subunits within the PSD (Li et al., 2004; Peng et al., 2004). Our results also suggest the Arp2/3 complex is present (although sparse) within the PSD. However, because pre-embedding techniques may fail to detect antigens within the PSD, because of limited access of antibodies (Lorincz et al., 2002; Rácz and Weinberg, 2004), pre-embedding may underestimate the fraction of antigen lying within the PSD.
The intrinsic noise of the immunogold technique can be reduced by averaging data from a large unbiased sample. This approach revealed unexpected features of the spatial organization of the Arp2/3 complex within the spine head. By focusing on average values, we may have failed to detect heterogeneity among spines. However, because the averaging process should attenuate any nonrandom organization detected, the true distribution of Arp2/3 in vivo likely peaks even more sharply within the domain we identified. An error is introduced by examining random single sections to estimate antigen distribution, rather than serial sections. That this error is relatively modest is suggested by the strong correlation between spine size and PSD length (r = 0.68) (supplemental Fig. 4, available at www.jneurosci.org as supplemental material), similar to, although less impressive than, the r = 0.88 from fully reconstructed CA1 spines (Harris and Stevens, 1989). Therefore, our method should provide a good qualitative estimate of protein organization but is likely to underestimate the true extent of ARPC-2 compartmentalization.
The techniques used here provide an averaged “snapshot” of a dynamic process, which reflects the most common distribution of the Arp2/3 complex in spines, but misses transients. However, even in very dynamic systems like the leading edges of migrating cells, the spatial location of Arp2/3 must be highly restricted to maintain typical actin treadmilling (Svitkina and Borisy, 1999; Atilgan et al., 2005; Shao et al., 2006). Therefore, the restricted spatial organization within the spinoplasm seen here likely represents the focus of activity of the Arp2/3 complex.
Actin dynamics and the spinoskeleton
Activity-dependent changes in spine morphology are tightly linked to long-term synaptic plasticity (Fifkova and Delay, 1982; Matus, 2000; Knott et al., 2006; Chen et al., 2007; Harvey and Svoboda, 2007). Underlying these shape changes is a complex interplay of molecular cascades triggered by synaptic input, leading ultimately to reorganization of the actin cytoskeleton (for review, see Carlisle and Kennedy, 2005; Calabrese et al., 2006; Schubert and Dotti, 2007; Honkura et al., 2008). Recent studies suggest that Arp2/3 plays a crucial role in synaptic plasticity: disruption of WAVE-1 (Wiskott-Aldrich syndrome protein family member 1), an upstream activator of the Arp2/3 complex, impairs learning and memory (Soderling et al., 2003, 2007). Moreover, loss of WAVE-1 also leads to abnormal spine morphology (Kim et al., 2006).
During morphogenesis, filopodia are thought to transform into mature, mushroom-shaped spines (Ethell and Pasquale, 2005) as a result of the protrusive force created by actin polymerization (Tada and Sheng, 2006). A linear bundle of actin reported in the spine, densest in the neck, may represent the primordial axis of the original filopodial extension (Fifkova and Delay, 1982; Smart and Halpain, 2000). Honkura et al. (2008) found that a dynamic pool of actin treadmills at the periphery, whereas a stable actin pool resides at the base of the spine. In this context, evidence that Arp2/3 plays a key role in the expansion of the head associated with mature dendritic spines might be predicted (Kim et al., 2006). The data reported here show that the Arp2/3 complex is restricted primarily to a torroidal domain within the spinoplasm, presumably representing a zone specialized for branch-point creation (Kiehart and Franke, 2002; Goley and Welch, 2006). Growth of filaments from this domain may generate the forces necessary to cause protrusion of the spine membrane during activity-dependent spine morphing, as observed in model systems (Pollard, 2007).
Endocytosis and the Arp2/3 complex in spines
Specialized endosomal compartments have been reported in hippocampal spines (Blanpied et al., 2002; Cooney et al., 2002). We reported previously that three key endocytic proteins localize in a lateral domain of dendritic spines (tangential distance, ∼0.4–0.6 normalized units), corresponding to the functionally defined “endocytic zone,” a region that plays a special role in the activity-dependent endocytosis necessary for synaptic trafficking of receptors (Perez-Otano and Ehlers, 2004; Rácz et al., 2004; Lu et al., 2007). Recent evidence from non-neuronal systems implicates the actin cytoskeleton in regulated endocytosis (for review, see Kaksonen et al., 2006; Smythe and Ayscough, 2006; Galletta et al., 2008); moreover, Arp2/3 complex-mediated actin polymerization is a prerequisite for clathrin-coated vesicle formation and movement (Merrifield et al., 2004; Martin et al., 2006). Recent in vitro experiments show that proper regulation of Arp2/3 is important for NMDA receptor-mediated endocytosis of AMPA receptors (Rocca et al., 2008). The present in vivo results support this finding, raising the intriguing possibility that this Arp2/3 complex-rich domain may contribute to efficient regulation of postsynaptic endocytosis.
Organization of actin-binding proteins within the spine
Actin-binding proteins (ABPs) within the spine exhibit remarkable spatial segregation. Previous work found that cofilin concentrates at the plasma membrane (Rácz and Weinberg, 2006; Zhou et al., 2007), where filament growth is likely restricted by its depolymerizing activity. By activating cofilin, long-term depression-evoking stimuli lead to spine shrinkage (Zhou et al., 2004, 2007); conversely, stimuli that trigger LTP also inactivate cofilin, permitting stable spine enlargement (Meng et al., 2003; Lang et al., 2004). In contrast, cortactin concentrates in the spine core, and its loss decreases spine density (Hering and Sheng, 2003), consistent with the suggestion that the core is stabilized by cortactin (Rácz and Weinberg, 2004). The present data indicate that the Arp2/3 complex concentrates in an annulus within the spinoplasm, presumably representing a zone specialized for actin branching. Together, these data (summarized in Fig. 4 D) suggest a previously unrecognized compartmentalization of actin regulation in dendritic spines.
Footnotes
This work was supported by Hungarian Scientific Research Fund Grant OTKA PF60416 and Veterinary Faculty Grant NKB15719 (B.R.), and by National Institutes of Health Grant NS-35527 (R.J.W.). B.R. performed the microscopy, analyzed data, and wrote this paper. R.J.W. provided support and helped with data analysis and writing. We thank Michael Ehlers, James Bear, and Scott Soderling for comments on this manuscript; Kristen Phend for specimen preparation; and Susan Burette for Western blotting.
References
- Atilgan E, Wirtz D, Sun SX. Morphology of the lamellipodium and organization of actin filaments at the leading edge of crawling cells. Biophys J. 2005;89:3589–3602. doi: 10.1529/biophysj.105.065383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpied TA, Scott DB, Ehlers MD. Dynamics and regulation of clathrin coats at specialized endocytic zones of dendrites and spines. Neuron. 2002;36:435–449. doi: 10.1016/s0896-6273(02)00979-0. [DOI] [PubMed] [Google Scholar]
- Bretschneider T, Diez S, Anderson K, Heuser J, Clarke M, Muller-Taubenberger A, Kohler J, Gerisch G. Dynamic actin patterns and Arp2/3 assembly at the substrate-attached surface of motile cells. Curr Biol. 2004;14:1–10. doi: 10.1016/j.cub.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Cai L, Marshall TW, Uetrecht AC, Schafer DA, Bear JE. Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell. 2007;128:915–929. doi: 10.1016/j.cell.2007.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese B, Wilson MS, Halpain S. Development and regulation of dendritic spine synapses. Physiology (Bethesda) 2006;21:38–47. doi: 10.1152/physiol.00042.2005. [DOI] [PubMed] [Google Scholar]
- Carlisle HJ, Kennedy MB. Spine architecture and synaptic plasticity. Trends Neurosci. 2005;28:182–187. doi: 10.1016/j.tins.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Chen LY, Rex CS, Casale MS, Gall CM, Lynch G. Changes in synaptic morphology accompany actin signaling during LTP. J Neurosci. 2007;27:5363–5372. doi: 10.1523/JNEUROSCI.0164-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney JR, Hurlburt JL, Selig DK, Harris KM, Fiala JC. Endosomal compartments serve multiple hippocampal dendritic spines from a widespread rather than a local store of recycling membrane. J Neurosci. 2002;22:2215–2224. doi: 10.1523/JNEUROSCI.22-06-02215.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nardo A, Cicchetti G, Falet H, Hartwig JH, Stossel TP, Kwiatkowski DJ. Arp2/3 complex-deficient mouse fibroblasts are viable and have normal leading-edge actin structure and function. Proc Natl Acad Sci USA. 2005;102:16263–16268. doi: 10.1073/pnas.0508228102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethell IM, Pasquale EB. Molecular mechanisms of dendritic spine development and remodeling. Prog Neurobiol. 2005;75:161–205. doi: 10.1016/j.pneurobio.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Fifkova E, Delay RJ. Cytoplasmic actin in neuronal processes as a possible mediator of synaptic plasticity. J Cell Biol. 1982;95:345–350. doi: 10.1083/jcb.95.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa Y, Saitoh Y, Ozawa F, Ohta Y, Mizuno K, Inokuchi K. Hippocampal LTP is accompanied by enhanced F-actin content within the dendritic spine that is essential for late LTP maintenance in vivo. Neuron. 2003;38:447–460. doi: 10.1016/s0896-6273(03)00206-x. [DOI] [PubMed] [Google Scholar]
- Galletta BJ, Chuang DY, Cooper JA. Distinct roles for Arp2/3 regulators in actin assembly and endocytosis. PLoS Biol. 2008;6:e1. doi: 10.1371/journal.pbio.0060001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- Gournier H, Goley ED, Niederstrasser H, Trinh T, Welch MD. Reconstitution of human Arp2/3 complex reveals critical roles of individual subunits in complex structure and activity. Mol Cell. 2001;8:1041–1052. doi: 10.1016/s1097-2765(01)00393-8. [DOI] [PubMed] [Google Scholar]
- Harris KM, Stevens JK. Dendritic spines of CA 1 pyramidal cells in the rat hippocampus: serial electron microscopy with reference to their biophysical characteristics. J Neurosci. 1989;9:2982–2997. doi: 10.1523/JNEUROSCI.09-08-02982.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey CD, Svoboda K. Locally dynamic synaptic learning rules in pyramidal neuron dendrites. Nature. 2007;450:1195–1200. doi: 10.1038/nature06416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hering H, Sheng M. Activity-dependent redistribution and essential role of cortactin in dendritic spine morphogenesis. J Neurosci. 2003;23:11759–11769. doi: 10.1523/JNEUROSCI.23-37-11759.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkura N, Matsuzaki M, Noguchi J, Ellis-Davies GC, Kasai H. The subspine organization of actin fibers regulates the structure and plasticity of dendritic spines. Neuron. 2008;57:719–729. doi: 10.1016/j.neuron.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Hotulainen P, Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J Cell Biol. 2006;173:383–394. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst IR, Zuo J, Jiang J, Holliday LS. Actin-related protein 2/3 complex is required for actin ring formation. J Bone Miner Res. 2004;19:499–506. doi: 10.1359/JBMR.0301238. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Toret CP, Drubin DG. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2006;7:404–414. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- Kasai H, Matsuzaki M, Noguchi J, Yasumatsu N, Nakahara H. Structure-stability-function relationships of dendritic spines. Trends Neurosci. 2003;26:360–368. doi: 10.1016/S0166-2236(03)00162-0. [DOI] [PubMed] [Google Scholar]
- Kempiak SJ, Yamaguchi H, Sarmiento C, Sidani M, Ghosh M, Eddy RJ, Desmarais V, Way M, Condeelis J, Segall JE. A neural Wiskott-Aldrich Syndrome protein-mediated pathway for localized activation of actin polymerization that is regulated by cortactin. J Biol Chem. 2005;280:5836–5842. doi: 10.1074/jbc.M410713200. [DOI] [PubMed] [Google Scholar]
- Kharazia VN, Weinberg RJ. Immunogold localization of AMPA and NMDA receptors in somatic sensory cortex of albino rat. J Comp Neurol. 1999;412:292–302. doi: 10.1002/(sici)1096-9861(19990920)412:2<292::aid-cne8>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Kiehart DP, Franke JD. Actin dynamics: the Arp2/3 complex branches out. Curr Biol. 2002;12:R557–R559. doi: 10.1016/s0960-9822(02)01053-9. [DOI] [PubMed] [Google Scholar]
- Kim Y, Sung JY, Ceglia I, Lee KW, Ahn JH, Halford JM, Kim AM, Kwak SP, Park JB, Ho Ryu S, Schenck A, Bardoni B, Scott JD, Nairn AC, Greengard P. Phosphorylation of WAVE1 regulates actin polymerization and dendritic spine morphology. Nature. 2006;442:814–817. doi: 10.1038/nature04976. [DOI] [PubMed] [Google Scholar]
- Knott GW, Holtmaat A, Wilbrecht L, Welker E, Svoboda K. Spine growth precedes synapse formation in the adult neocortex in vivo. Nat Neurosci. 2006;9:1117–1124. doi: 10.1038/nn1747. [DOI] [PubMed] [Google Scholar]
- Kopec CD, Li B, Wei W, Boehm J, Malinow R. Glutamate receptor exocytosis and spine enlargement during chemically induced long-term potentiation. J Neurosci. 2006;26:2000–2009. doi: 10.1523/JNEUROSCI.3918-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang C, Barco A, Zablow L, Kandel ER, Siegelbaum SA, Zakharenko SS. Transient expansion of synaptically connected dendritic spines upon induction of hippocampal long-term potentiation. Proc Natl Acad Sci USA. 2004;101:16665–16670. doi: 10.1073/pnas.0407581101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li KW, Hornshaw MP, Van Der Schors RC, Watson R, Tate S, Casetta B, Jimenez CR, Gouwenberg Y, Gundelfinger ED, Smalla KH, Smit AB. Proteomics analysis of rat brain postsynaptic density. Implications of the diverse protein functional groups for the integration of synaptic physiology. J Biol Chem. 2004;279:987–1002. doi: 10.1074/jbc.M303116200. [DOI] [PubMed] [Google Scholar]
- Lippman J, Dunaevsky A. Dendritic spine morphogenesis and plasticity. J Neurobiol. 2005;64:47–57. doi: 10.1002/neu.20149. [DOI] [PubMed] [Google Scholar]
- Lorincz A, Notomi T, Tamas G, Shigemoto R, Nusser Z. Polarized and compartment-dependent distribution of HCN1 in pyramidal cell dendrites. Nat Neurosci. 2002;5:1185–1193. doi: 10.1038/nn962. [DOI] [PubMed] [Google Scholar]
- Lu J, Helton TD, Blanpied TA, Rácz B, Newpher TM, Weinberg RJ, Ehlers MD. Postsynaptic positioning of endocytic zones and AMPA receptor cycling by physical coupling of dynamin-3 to Homer. Neuron. 2007;55:874–889. doi: 10.1016/j.neuron.2007.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch G, Rex CS, Gall CM. LTP consolidation: substrates, explanatory power, and functional significance. Neuropharmacology. 2007;52:12–23. doi: 10.1016/j.neuropharm.2006.07.027. [DOI] [PubMed] [Google Scholar]
- Martin AC, Welch MD, Drubin DG. Arp2/3 ATP hydrolysis-catalysed branch dissociation is critical for endocytic force generation. Nat Cell Biol. 2006;8:826–833. doi: 10.1038/ncb1443. [DOI] [PubMed] [Google Scholar]
- Matsuzaki M, Ellis-Davies GC, Nemoto T, Miyashita Y, Iino M, Kasai H. Dendritic spine geometry is critical for AMPA receptor expression in hippocampal CA1 pyramidal neurons. Nat Neurosci. 2001;4:1086–1092. doi: 10.1038/nn736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus A. Actin-based plasticity in dendritic spines. Science. 2000;290:754–758. doi: 10.1126/science.290.5492.754. [DOI] [PubMed] [Google Scholar]
- Meng Y, Zhang Y, Tregoubov V, Falls DL, Jia Z. Regulation of spine morphology and synaptic function by LIMK and the actin cytoskeleton. Rev Neurosci. 2003;14:233–240. doi: 10.1515/revneuro.2003.14.3.233. [DOI] [PubMed] [Google Scholar]
- Merrifield CJ, Qualmann B, Kessels MM, Almers W. Neural Wiskott Aldrich Syndrome Protein (N-WASP) and the Arp2/3 complex are recruited to sites of clathrin-mediated endocytosis in cultured fibroblasts. Eur J Cell Biol. 2004;83:13–18. doi: 10.1078/0171-9335-00356. [DOI] [PubMed] [Google Scholar]
- Mullins RD, Stafford WF, Pollard TD. Structure, subunit topology, and actin-binding activity of the Arp2/3 complex from Acanthamoeba. J Cell Biol. 1997;136:331–343. doi: 10.1083/jcb.136.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusser Z. AMPA and NMDA receptors: similarities and differences in their synaptic distribution. Curr Opin Neurobiol. 2000;10:337–341. doi: 10.1016/s0959-4388(00)00086-6. [DOI] [PubMed] [Google Scholar]
- Park M, Salgado JM, Ostroff L, Helton TD, Robinson CG, Harris KM, Ehlers MD. Plasticity-induced growth of dendritic spines by exocytic trafficking from recycling endosomes. Neuron. 2006;52:817–830. doi: 10.1016/j.neuron.2006.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Kim MJ, Cheng D, Duong DM, Gygi SP, Sheng M. Semiquantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. J Biol Chem. 2004;279:21003–21011. doi: 10.1074/jbc.M400103200. [DOI] [PubMed] [Google Scholar]
- Perez-Otano I, Ehlers MD. Learning from NMDA receptor trafficking: clues to the development and maturation of glutamatergic synapses. Neurosignals. 2004;13:175–189. doi: 10.1159/000077524. [DOI] [PubMed] [Google Scholar]
- Pollard TD. Regulation of actin filament assembly by arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- Rácz B, Weinberg RJ. The subcellular organization of cortactin in hippocampus. J Neurosci. 2004;24:10310–10317. doi: 10.1523/JNEUROSCI.2080-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rácz B, Weinberg RJ. Spatial organization of cofilin in dendritic spines. Neuroscience. 2006;138:447–456. doi: 10.1016/j.neuroscience.2005.11.025. [DOI] [PubMed] [Google Scholar]
- Rácz B, Blanpied TA, Ehlers MD, Weinberg RJ. Lateral organization of endocytic machinery in dendritic spines. Nat Neurosci. 2004;7:917–918. doi: 10.1038/nn1303. [DOI] [PubMed] [Google Scholar]
- Robinson RC, Turbedsky K, Kaiser DA, Marchand JB, Higgs HN, Choe S, Pollard TD. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- Rocca DL, Martin S, Jenkins EL, Hanley JG. Inhibition of Arp2/3-mediated actin polymerization by PICK1 regulates neuronal morphology and AMPA receptor endocytosis. Nat Cell Biol. 2008;10:259–271. doi: 10.1038/ncb1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert V, Dotti CG. Transmitting on actin: synaptic control of dendritic architecture. J Cell Sci. 2007;120:205–212. doi: 10.1242/jcs.03337. [DOI] [PubMed] [Google Scholar]
- Shao D, Forge A, Munro PM, Bailly M. Arp2/3 complex-mediated actin polymerisation occurs on specific pre-existing networks in cells and requires spatial restriction to sustain functional lamellipod extension. Cell Motil Cytoskel. 2006;63:395–414. doi: 10.1002/cm.20131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart FM, Halpain S. Regulation of dendritic spine stability. Hippocampus. 2000;10:542–554. doi: 10.1002/1098-1063(2000)10:5<542::AID-HIPO4>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Smythe E, Ayscough KR. Actin regulation in endocytosis. J Cell Sci. 2006;119:4589–4598. doi: 10.1242/jcs.03247. [DOI] [PubMed] [Google Scholar]
- Soderling SH, Langeberg LK, Soderling JA, Davee SM, Simerly R, Raber J, Scott JD. Loss of WAVE-1 causes sensorimotor retardation and reduced learning and memory in mice. Proc Natl Acad Sci USA. 2003;100:1723–1728. doi: 10.1073/pnas.0438033100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderling SH, Guire ES, Kaech S, White J, Zhang F, Schutz K, Langeberg LK, Banker G, Raber J, Scott JD. A WAVE-1 and WRP signaling complex regulates spine density, synaptic plasticity, and memory. J Neurosci. 2007;27:355–365. doi: 10.1523/JNEUROSCI.3209-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T, Sheng M. Molecular mechanisms of dendritic spine morphogenesis. Curr Opin Neurobiol. 2006;16:95–101. doi: 10.1016/j.conb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Takumi Y, Ramirez-Leon V, Laake P, Rinvik E, Ottersen OP. Different modes of expression of AMPA and NMDA receptors in hippocampal synapses. Nat Neurosci. 1999;2:618–624. doi: 10.1038/10172. [DOI] [PubMed] [Google Scholar]
- Toshima J, Toshima JY, Martin AC, Drubin DG. Phosphoregulation of Arp2/3-dependent actin assembly during receptor-mediated endocytosis. Nat Cell Biol. 2005;7:246–254. doi: 10.1038/ncb1229. [DOI] [PubMed] [Google Scholar]
- Valtschanoff JG, Weinberg RJ. Laminar organization of the NMDA receptor complex within the postsynaptic density. J Neurosci. 2001;21:1211–1217. doi: 10.1523/JNEUROSCI.21-04-01211.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Martinez SJ, Haber M, Jones EV, Bouvier D, Doucet G, Corera AT, Fon EA, Zisch AH, Murai KK. EphA4 signaling regulates phospholipase Cgamma1 activation, cofilin membrane association, and dendritic spine morphology. J Neurosci. 2007;27:5127–5138. doi: 10.1523/JNEUROSCI.1170-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Homma KJ, Poo MM. Shrinkage of dendritic spines associated with long-term depression of hippocampal synapses. Neuron. 2004;44:749–757. doi: 10.1016/j.neuron.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Zuo X, Zhang J, Zhang Y, Hsu SC, Zhou D, Guo W. Exo70 interacts with the Arp2/3 complex and regulates cell migration. Nat Cell Biol. 2006;8:1383–1388. doi: 10.1038/ncb1505. [DOI] [PubMed] [Google Scholar]