Abstract
Ribosome biogenesis takes place successively in the nucleolar, nucleoplasmic, and cytoplasmic compartments. Numerous nonribosomal factors transiently associate with the nascent ribosomes, but the mechanisms driving ribosome formation are mostly unknown. Here, we show that an energy-consuming enzyme, the AAA-type (ATPases associated with various cellular activities) ATPase Rix7, restructures a novel pre-60S particle at the transition from the nucleolus to nucleoplasm. Rix7 interacts genetically with Nsa1 and is targeted to the Nsa1-defined preribosomal particle. In vivo, Nsa1 cannot dissociate from pre-60S particles in rix7 mutants, causing nucleolar Nsa1 to escape to the cytoplasm, where it remains associated with aberrant 60S subunits. Altogether, our data suggest that Rix7 is required for the release of Nsa1 from a discrete preribosomal particle, thereby triggering the progression of 60S ribosome biogenesis.
Introduction
The biogenesis of ribosomes is a fundamental process that utilizes a substantial amount of the cell's energy resources (Warner, 1999). Most of our current knowledge concerning this highly dynamic multistep process comes from studies with the yeast Saccharomyces cerevisiae. The combined use of proteomic, genetic, and cell biological methods has revealed that a multitude of protein trans-acting factors (>150) are required for the assembly and maturation of preribosomal particles as they travel from the nucleolus to the cytoplasm (Fatica and Tollervey, 2002; Fromont-Racine et al., 2003; Tschochner and Hurt, 2003; de la Cruz et al., 2004; Granneman and Baserga, 2004). Among these are energy-consuming enzymes such as GTPases, protein kinases, ATP-dependent RNA helicases, and AAA-type (ATPases associated with various cellular activities) ATPases, which suggests that the energy derived from nucleotide hydrolysis confers directionality to ribosome assembly. Therefore, to unravel the mechanisms of ribosome formation, it is essential to identify the substrate proteins of these energy-consuming enzymes.
Recent advances using the tandem affinity purification (TAP) method (Rigaut et al., 1999) have allowed refinement of the ribosome assembly pathway that was outlined upon identification of 90S, 66S, and 43S preribosomal particles in the 1970s (Trapman et al., 1975). These proteomic approaches revealed for the first time the protein and ribosomal RNA (rRNA) composition of several distinct and successive pre-40S and pre-60S ribosomal particles (Bassler et al., 2001; Saveanu et al., 2001; Harnpicharnchai et al., 2001; Fatica et al., 2002; Nissan et al., 2002; Schäfer et al., 2003). Contrary to the relatively low compositional complexity of 43S preparticles, the early pre-60S particles contain >40 associated trans-acting factors. As a consequence, the maturation of nuclear pre-60S particles is accompanied by major compositional changes that lead to a sequential reduction of complexity and to the acquisition of export competence.
AAA-type ATPases belong to the larger AAA+ family of ubiquitous ATPases (Neuwald et al., 1999). These proteins are found in all organisms—the yeast genome alone encodes around 50 AAA+ ATPases—and they are involved in a variety of cellular processes. In common to all these proteins is the presence of a structurally conserved ATPase domain of around 250 amino acids (AAA domain) that contains, besides other characteristic features, Walker A (P-loop) and Walker B (DExx-box) motifs responsible for ATP binding and hydrolysis, respectively (Patel and Latterich, 1998). A hallmark feature of AAA+ ATPases, regardless of whether they harbor one (type I), two (type II), or six (e.g., dynein and Rea1) AAA domains, is their assembly into the functionally active “doughnut-shaped” oligomeric, mostly hexameric rings (Vale, 2000), which undergo conformational changes during the ATPase cycle (Hanson and Whiteheart, 2005). Accordingly, AAA ATPases are believed to transmit the tension of a nucleotide-dependent conformational switch to bound substrate proteins, thus promoting the unfolding of proteins, the dissociation of protein–protein interactions, or the generation of unidirectional movement along a track (Vale, 2000; Hanson and Whiteheart, 2005).
The three AAA ATPases that have been shown to be involved in ribosome biogenesis in yeast act at different stages during formation of 60S ribosomal subunits. The type II AAA ATPase Rix7 was cloned by complementation of the rix7-1 mutant, which had been isolated in a visual screen by virtue of its predominantly nucleolar accumulation of the Rpl25-GFP large subunit reporter (Gadal et al., 2001a). Rix7 localizes throughout the nucleus in exponentially growing cells, and its mutational inactivation leads to a striking destabilization of the 27SB precursor rRNA (pre-rRNA), which suggests that Rix7 is required for correct assembly, and therefore stability, of pre-60S ribosomal particles. Rea1, which is the largest yeast protein and is related to dynein heavy chain, is specifically associated with the nucleoplasmic Rix1 pre-60S particle, where it forms the tail of the tadpole-like particle (Nissan et al., 2002, 2004). Finally, Drg1 contributes to the recycling of trans-acting factors from cytoplasmic pre-60S particles (Pertschy et al., 2007).
The closest yeast homologue of Rix7 is Cdc48, which, together with its mammalian orthologue p97, has been the most intensively studied AAA ATPase over the past 15 years. The diverse cellular functions of Cdc48/p97, which include activation of a membrane-bound transcription factor, participation in the ER-associated degradation (ERAD) pathway, and control of membrane fusion, are all linked to the recognition of ubiquitinated substrates and their dissociation from unmodified binding partners (Jentsch and Rumpf, 2007). The highly homologous D1 and D2 AAA domains carry out different functions: D1 is rather rigid and promotes hexamer formation, whereas D2 contains the ATPase activity and undergoes major conformational changes during the ATPase cycle (Song et al., 2003; Wang et al., 2003; DeLaBarre and Brunger, 2005). Cdc48/p97 binds via its N-terminal domain either directly or indirectly, through substrate-recruiting cofactors, to ubiquitinated substrate proteins. Moreover, the relative position of the N-terminal domain changes depending on the nucleotide bound to the D2 domain (DeLaBarre and Brunger, 2005; Pye et al., 2006), which suggests that the movement of the substrate-bound N-terminal domain may lead to substrate release from its binding partner.
In this study, we provide evidence that the AAA ATPase Rix7 is required for the release of the pre-60S factor Nsa1 from pre-60S ribosomal particles and could thereby power the progression of 60S ribosome biogenesis.
Results
RIX7 exhibits an intimate genetic link to the pre-60S factor NSA1
Rix7 exhibits the typical domain organization of type II AAA ATPases, and notably contains two central AAA domains (termed D1 and D2) flanked by a long N-terminal and a very short C-terminal extension. The N-terminal extension of Rix7 consists of a 174–amino acid leader (hereafter referred to as the N-terminal domain) of moderate evolutionary conservation that is followed by a predicted NLS (for a schematic representation, see Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200801181/DC1). Given that the N-terminal domain of Cdc48, which is the closest yeast homologue of Rix7, is involved in cofactor binding and substrate processing (see Introduction), we sought to identify potential pre-60S ribosomal substrate proteins of Rix7 by performing synthetic lethal (sl) screens with rix7 alleles where the functional integrity of the N-terminal domain is affected. To this end, we first generated by random PCR mutagenesis and progressive N-terminal deletion, respectively, two temperature-sensitive (ts) rix7 mutants lacking the first 14 N-terminal amino acids: rix7-31 (I23T, Y117H, and S162P) and rix7Δ14N (Fig. S1, B and C). Similar to the original rix7-1 (P224L in D1) allele, these new rix7 ts mutants exhibited defects in the formation and nuclear export of 60S ribosomal subunits (Fig. S1, D and E). Moreover, examination of the pre-rRNA processing phenotype of the rix7Δ14N mutant revealed a strong decrease in the steady-state levels of the 27SB and 7S pre-rRNAs (unpublished data), as described previously for the rix7-1 allele (Gadal et al., 2001a).
Next, we performed the sl screen using the rix7-31 mutant, which we considered more suitable than the rix7Δ14N mutant because of its moderate growth defect at the semipermissive temperature of 30°C (Fig. S1 B). This sl screen yielded one sl mutant (sl30), which could be complemented by NSA1 (Fig. 1 A). Thus, this genetic finding indicated that Rix7 and Nsa1 functionally interact. Nsa1 is an essential, conserved WD repeat protein that was previously found to be associated with the Nop7-purified pre-60S particles and whose depletion caused a reduction in 60S subunits (Harnpicharnchai et al., 2001). The chromosomal NSA1 gene was recovered from strain sl30 (this allele is called nsa1-1), and DNA sequencing confirmed that NSA1 was indeed mutated (W230R). The recovered nsa1-1 allele conferred a moderate slow-growth phenotype at all tested temperatures (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200801181/DC1), and the combination with rix7-31 resulted in a synthetic enhancement of the respective single mutant phenotypes (unpublished data).
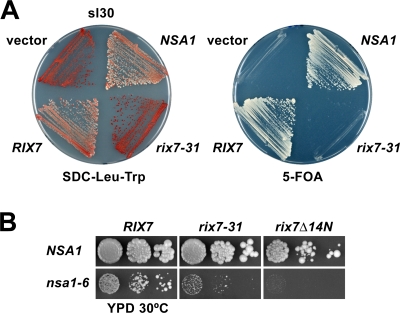
Figure 1.
Rix7 is functionally linked to the WD40-repeat protein Nsa1. (A) Identification of Nsa1 in an sl screen with the rix7-31 mutant allele. Mutant sl30 was transformed with YCplac111 (empty vector), pHAC111-RIX7, pHAC111-rix7-31, and YCplac111-NSA1. Transformants were streaked out on SDC-Leu-Trp and SDC + 5-fluoroorotic acid (5-FOA) plates, which were incubated for 5 d at 30°C. (B) The rix7-31 or rix7Δ14N mutants synthetically enhance the growth phenotype of the nsa1-6 strain. The RIX7/NSA1 double shuffle strain was transformed with plasmids that carry the indicated wild-type or mutant alleles. After 5-FOA shuffling at 23°C, cells were spotted in 10-fold serial dilution steps onto yeast peptone dextrose plates, which were incubated for 4 d at 30°C.
For further functional analyses of Nsa1, we created the nsa1-6 ts allele (L43S, L375P, and F406S) by random PCR mutagenesis (Fig. S2 A). When nsa1-6 was combined with rix7-31 or rix7Δ14N, a strong synthetic enhancement phenotype was observed (Fig. 1 B). At the semipermissive temperature of 23°C, Nsa1-6 was stably expressed and associated with pre-60S particles; however, its expression levels were decreased at 30 and 37°C (Fig. S2, B, C, and D). Polysome profile analysis indicated that nsa1-6 cells, when grown at 30°C, had significantly reduced amounts of 60S subunits paralleled by the appearance of half-mer polysomes (Fig. S2 E). Moreover, the large ribosomal subunit reporter Rpl25-EGFP accumulated in the nucleus in the nsa1-6 mutant (Fig. S2 F). To further explore the functional environment of Nsa1, we performed an sl screen with the nsa1-6 allele and found only one sl mutant, which was complemented by RIX7 (unpublished data). The recovered rix7-4 allele from this sl strain harbored one mutation in the N-terminal domain (I73F), which conferred a strong slow-growth phenotype at 30°C and a ts phenotype at 37°C (Fig. S1 B). Furthermore, the rix7-4 mutation also affected the formation and nuclear export of 60S ribosomal subunits (Fig. S1, D and E).
Notably, in an independent approach by performing a yeast two-hybrid screen using full-length Nsa1 as bait, we isolated four identical library clones expressing amino acids 133–557 of Rix7 (see Materials and methods). Further N-terminal truncation revealed that a Rix7 fragment, spanning amino acids 166–557 and lacking the N-terminal domain, was sufficient to confer a robust yeast two-hybrid interaction (Fig. S3 A, available at http://www.jcb.org/cgi/content/full/jcb.200801181/DC1). However, we were not able to reconstitute a direct interaction between Rix7 and Nsa1 in in vitro binding assays (unpublished data).
Altogether, these comprehensive genetic and two-hybrid analyses indicate an intimate functional interaction between Rix7 and the pre-60S factor Nsa1 and suggest a model in which Nsa1 could be a substrate of the AAA ATPase Rix7 (see Discussion).
Rix7 is specifically associated with a novel pre-60S ribosomal particle specified by the Nsa1-TAP bait protein
The genetic and two-hybrid data suggested that Nsa1 and Rix7 could physically interact in vivo with each other either directly or as subunits in a larger complex. Hence, we isolated Rix7-TAP by the TAP method and looked for a coenrichment of Nsa1. Notably, Nsa1 could be detected, albeit in substoichiometric amounts, in the Rix7-TAP purification by Coomassie staining (Fig. S3 B); however, this association was lost when more stringent washing conditions were applied (unpublished data), which suggests that the interaction between Nsa1 and Rix7 is transient or regulated. However, Rix7-TAP did also not purify other preribosomal factors in stoichiometric amounts, which indicates that Rix7 in general only transiently associates with preribosomal particles.
In a reciprocal experiment, we tested whether Rix7 is copurified during affinity-purification of Nsa1-TAP. The Nsa1-TAP bait protein purified a very distinct pre-60S particle of an apparent stoichiometric composition, containing a myriad of known pre-60S factors and ribosomal L-proteins (Fig. 2 A). Remarkably, the pre-60S particle specified by the Nsa1 bait did not contain early (e.g., Rrp5 and Noc1) or late (e.g., Rea1, Rix1, and Arx1) pre-60S factors but did carry Noc3, an indicator of an intermediate pre-60S particle (Fig. 2, A and B; Nissan et al., 2002). Moreover, Northern analysis revealed an almost exclusive association of the 27SB pre-rRNA with the Nsa1 pre-60S particle; correspondingly, only very little mature 25S and 5.8S rRNAs were present (see Fig. 4 B). Altogether, these data indicated that the Nsa1-purified pre-60S particle represents a late nucleolar pre-60S ribosomal particle (see also the following paragraph). However, Rix7 could not be found as a stoichiometric component in this Nsa1-TAP preparation.
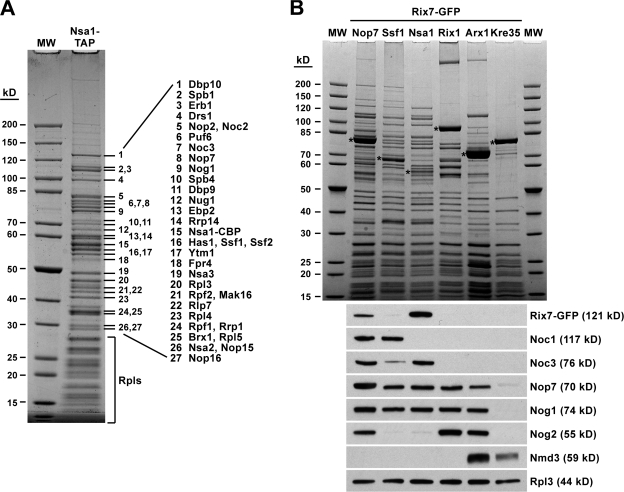
Figure 2.
Rix7 is associated with the Nsa1 pre-60S particle. (A) Protein composition of the Nsa1 pre-60S ribosomal particle. Nsa1-TAP was affinity-purified via the TAP method, and the final EGTA eluate was analyzed by SDS-PAGE and Coomassie staining. The indicated proteins (1–27) were identified by mass spectrometry. (B) Nsa1 defines a distinct pre-60S ribosomal particle that specifically carries Rix7. The indicated TAP-tagged bait proteins were affinity-purified from cells expressing Rix7-GFP. The final EGTA eluates were analyzed by SDS-PAGE and Coomassie staining (top) and Western blotting using anti-GFP, anti-Noc1, anti-Noc3, anti-Nop7, anti-Nog1, anti-Nog2, anti-Nmd3, and anti-Rpl3 antibodies (bottom). The bands marked with asterisks correspond to the purified bait proteins. MW, molecular weight protein standard; Rpls, ribosomal L proteins.
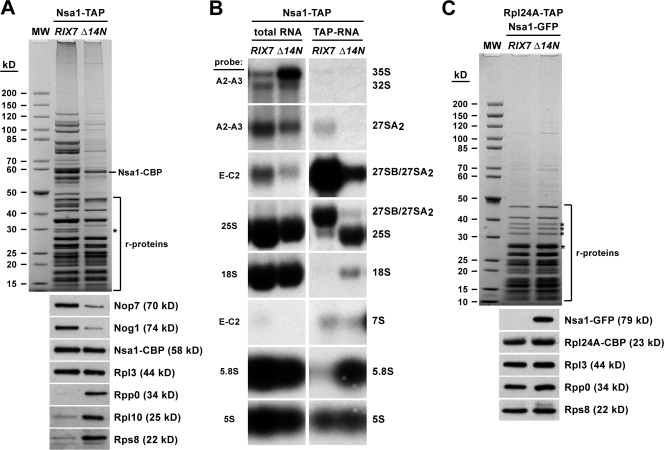
Figure 4.
Nsa1 is associated with mature 60S ribosomes in the rix7Δ14N mutant. (A) Affinity purification of Nsa1-TAP from wild-type RIX7 or rix7Δ14N mutant cells that were first grown at 23°C and then shifted for 3 h to 30°C. The final EGTA eluates were analyzed by SDS-PAGE and Coomassie staining (top) and Western blotting using anti-Nop7, anti-Nog1, anti-CBP, anti-Rpl3, anti-Rpp0, anti-Rpl10, and anti-Rps8 antibodies (bottom). The band marked with an asterisk corresponds to Rpp0. The bands corresponding to ribosomal proteins are indicated. (B) rRNA composition of Nsa1-TAP affinity-purified from wild-type RIX7 or rix7Δ14N mutant cells that were first grown at 23°C and then shifted for 3 h to 30°C. Aliquots of total RNA and of the final EGTA eluates were resolved on a 1.5% agarose-formaldehyde gel, transferred to a nylon membrane, and analyzed by Northern blotting using the indicated probes to detect pre-rRNA and mature rRNA species. (C) Affinity purification of mature 60S ribosomal subunits via Rpl24A-TAP from wild-type RIX7 or rix7Δ14N mutant cells expressing Nsa1-GFP. Cells were first grown at 23°C and then shifted for 3 h to 30°C. The final EGTA eluates were analyzed by SDS-PAGE and Coomassie staining (top) and Western blotting using anti-GFP, anti-CBP, anti-Rpl3, anti-Rpp0, and anti-Rps8 antibodies (bottom). The bands marked with asterisks contain Stm1, Rpl5, Rpp0/Asc1, and Rpl2/Rps1/Rps3/Rps4/Rps0/Rpl8/Rps6, respectively. MW, molecular weight protein standard.
To test whether Rix7 is present in the Nsa1 particle in substoichiometric amounts, Rix7 was GFP-tagged, and the Nsa1-TAP preparation was analyzed by SDS-PAGE and Western blotting using anti-GFP antibodies (Fig. 2 B). Other TAP-tagged bait proteins (Nop7, Ssf1, Rix1, Arx1, and Kre35), which are well-described markers for different (earlier and later) pre-60S particles, served as controls (Fatica et al., 2002; Nissan et al., 2002). Moreover, antibodies against several pre-60S marker proteins were used to further distinguish between early nucleolar, late nucleolar/early nucleoplasmic, and late nucleoplasmic pre-60S particles (Fig. 2 B, bottom). These biochemical analyses demonstrated that Rix7-GFP was strongly coenriched in the Nsa1 particle but absent from the other pre-60S particles except for the Nop7-TAP preparation. However, Nop7-TAP did not coenrich a discrete pre-60S particle; rather, it was associated with a larger number of different pre-60S particles (Fig. 2 B). Importantly, Rix7 was not associated with the early nucleolar Ssf1 particle and the intermediate nucleoplasmic Rix1 (Ipi1-Ipi3-Rea1) particle. These findings indicated that Nsa1 defines a novel pre-60S intermediate that can be placed between the Ssf1 particle of highly complex composition and the Rix1 particle, which is significantly reduced in its complexity but has acquired new biogenesis factors. Accordingly, a major structural rearrangement accompanied by the dissociation of many pre-60S factors might occur at the transition from the Nsa1 particle to the Rix1 particle. The observation that Rix7 is exclusively associated with the Nsa1 particle suggests that Rix7 may release specific biogenesis factors (e.g., Nsa1) from this pre-60S particle and thus serve as an energy-requiring trigger for ribosome biogenesis (see Discussion).
Rix7 is required for the release of Nsa1 from pre-60S particles
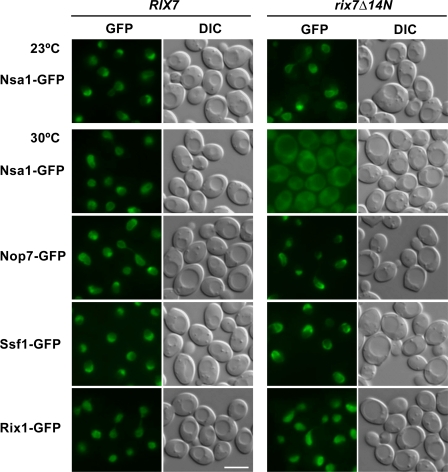
To address a possible role of Rix7 in removing Nsa1 from the Nsa1-defined pre-60S particle during ribosome biogenesis, we analyzed the intracellular localization of Nsa1-GFP in the rix7Δ14N mutant. In wild-type and rix7Δ14N mutant cells grown at 23°C, the permissive temperature for rix7Δ14N, Nsa1-GFP was localized predominantly in the nucleolus, with a fainter nucleoplasmic staining (Fig. 3). However, when rix7Δ14N cells were shifted for 3 h to the semipermissive temperature (30°C; see also Fig. S1 B for its growth), Nsa1-GFP was strongly mislocalized to the cytoplasm, whereas other GFP-labeled pre-60S markers (i.e., Nop7, Ssf1, and Rix1) remained concentrated inside the nucleus (Fig. 3). This finding suggested that Nsa1 is not efficiently removed from late nucleolar pre-60S particles when Rix7 is nonfunctional and hence escapes to the cytoplasm, possibly in association with aberrant preribosomal particles.
Figure 3.
Nsa1-GFP mislocalizes to the cytoplasm in the rix7Δ14N mutant. The subcellular localization of the indicated GFP-tagged proteins was assessed by fluorescence microscopy in wild-type RIX7 and rix7Δ14N mutant cells that were first grown in yeast peptone dextrose at 23°C and then shifted for 3 h to 30°C. Location of Nsa1-GFP was also determined in RIX7 and rix7Δ14N cells that were grown at the permissive temperature of 23°C (top). Bar, 5 μm.
Nsa1 remains associated with mature 60S subunits upon inactivation of Rix7
To demonstrate that cytoplasmic Nsa1 remained bound to mature 60S subunits, biochemical studies were performed. Nsa1-TAP was affinity-purified from rix7Δ14N mutant cells shifted to the semipermissive temperature (30°C), and the composition of the purified material was analyzed by SDS-PAGE and Coomassie staining or Western blotting using antibodies against preribosomal factors and ribosomal proteins. This analysis showed that Nsa1-TAP isolated from the rix7Δ14N mutant contained significantly reduced amounts of pre-60S factors, but the quantity of associated ribosomal proteins was apparently not altered (Fig. 4 A). However, among the ribosomal proteins were also Rpl10 and Rpp0, which have been suggested to associate only with mature 60S subunits in the cytoplasm (Zinker and Warner, 1976; Kruiswijk et al., 1978), and the small subunit protein Rps8 (Fig. 4 A). In contrast, these cytoplasmic 60S ribosomal subunit proteins and Rps8 were absent when Nsa1-TAP was affinity-purified from wild-type cells (Fig. 4 A). In agreement with an association with mature 60S subunits, Northern analysis revealed that mature rRNA species were specifically purified with the Nsa1-TAP bait in the rix7Δ14N mutant but not when Nsa1-TAP was isolated from wild-type RIX7 cells (Fig. 4 B). Additionally, Nsa1-GFP was copurified in a reciprocal approach with Rpl24A-TAP, another ribosomal protein exclusively contained within mature 60S subunits (Kruiswijk et al., 1978), from rix7Δ14N mutant cells (Fig. 4 C).
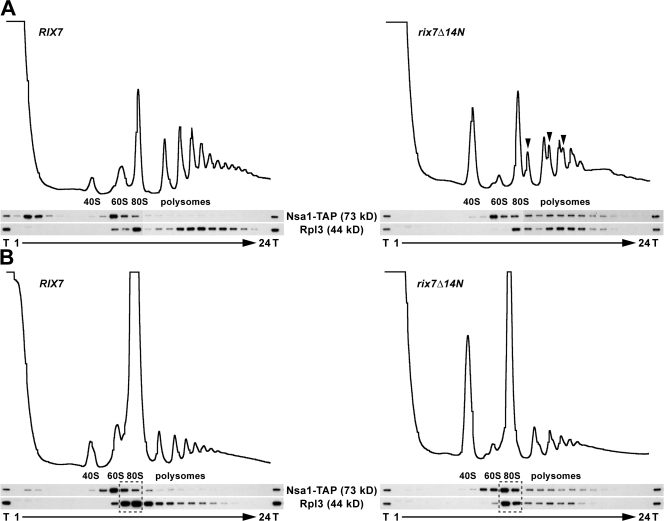
To determine if Nsa1-containing 60S subunits were present in translating ribosomes, polysomal profiles of whole cell lysates were analyzed by sucrose gradient centrifugation. Contrary to the wild-type situation, a significant pool of Nsa1-TAP was detected in the polysomal fractions of rix7Δ14N mutant cells (Fig. 5 A). Moreover, under polysome runoff conditions (omission of cycloheximide), Nsa1 was significantly redistributed from the polysomal to the 80S fractions, which indicated that Nsa1 was associated with translating ribosomes (Fig. 5 B).
Figure 5.
Nsa1 is associated with translating ribosomes in the rix7Δ14N mutant. Whole cell lysates derived from wild-type RIX7 and rix7Δ14N mutant strains (both expressing Nsa1-TAP), which were first grown at 23°C and then shifted for 3 h to 30°C, were analyzed by sucrose gradient centrifugation under polysome-conserving conditions (A) or polysome runoff conditions (B). The profiles were recorded at A254. The peaks of free 40S and 60S subunits, 80S ribosomes, and polysomes are indicated. Half-mers are indicated by arrowheads. Aliquots of the total cell lysate (T) and the fractions from the gradient were analyzed by Western blotting using anti-ProtA and anti-Rpl3 antibodies. The fractions boxed with broken lines indicate the 80S fractions into which the polysomes are redistributed under polysome runoff conditions.
All together, our data indicated that Rix7 is required to remove Nsa1 from pre-60S ribosomal particles. Therefore, when Rix7 is mutated, Nsa1 remains associated with pre-60S particles (see also the following paragraph) and travels to the cytoplasm, where it is retained on aberrant mature 60S subunits.
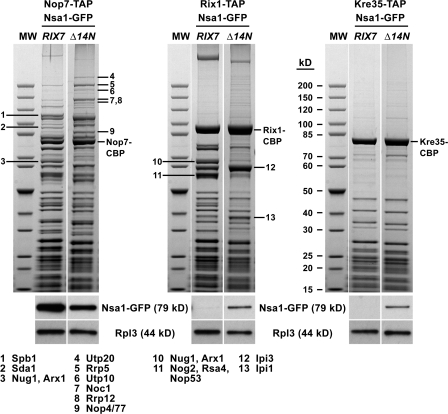
To investigate how the failure to remove Nsa1 from pre-60S subunits alters the composition of other pre-60S particles, we isolated Nop7-TAP, Rix1-TAP, and Kre35-TAP from wild-type and rix7Δ14N cells. As shown in Fig. 6, the Nop7 particle purified from rix7Δ14N cells exhibited a more complex composition and acquired very early pre-60S factors (e.g., Utp20, Rrp5, Utp10, Noc1, Rrp12, and Nop4/77) when compared with wild-type cells. Consistent with these findings, the Nop7 particle isolated from this mutant now also contained pre-rRNA species typically found in very early pre-60S intermediates (i.e., 35S, 32S, and 27SA2 pre-rRNAs; Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200801181/DC1). In contrast, the downstream Rix1 particle exhibited a simpler composition and contained reduced amounts of some of its typical pre-60S factors (e.g., Nug1, Arx1, Nog2, and Rsa4) but gained Nsa1-GFP instead (Fig. 6). Furthermore, Nsa1-GFP was also present on the cytoplasmic Kre35 particle (Fig. 6). These findings indicated that the blockage of removing Nsa1 from a pre-60S intermediate alters the composition and abundance of upstream and downstream pre-60S particles. Thus, the normal evolvement of pre-60S particles is disturbed in the rix7Δ14N mutant.
Figure 6.
Changes in the composition of “upstream” and “downstream” pre-60S ribosomal particles isolated from the rix7Δ14N mutant. Nop7-TAP, Rix1-TAP, and Kre35-TAP were affinity-purified from wild-type RIX7 or rix7Δ14N mutant cells expressing Nsa1-GFP. Cells were first grown at 23°C and then shifted for 3 h to 30°C. The final EGTA eluates were analyzed by SDS-PAGE and Coomassie staining (top) and Western blotting using anti-GFP and anti-Rpl3 antibodies (bottom). The indicated proteins (1–13) were identified by mass spectrometry. White lines indicate that intervening lanes have been spliced out. MW, molecular weight protein standard.
Discussion
Energy-consuming enzymes such as GTPases, AAA ATPases, RNA helicases, and protein kinases are thought to use their energy to drive ribosome biogenesis, but the mechanisms of their actions are largely unknown. In this study, we show that the pre-60S factor Nsa1 is functionally linked to the AAA ATPase Rix7 and that release of Nsa1 from a novel late nucleolar pre-60S particle requires the Rix7 function. In this way, Rix7 could power an energy-requiring step during ribosome biogenesis. When Rix7 is mutated, Nsa1 remains associated with pre-60S particles and travels to the cytoplasm, where it is retained on aberrant mature 60S subunits.
Nsa1 is associated with a unique pre-60S particle that differs from the pre-60S particles characterized so far. Comparison of the protein composition of the Nsa1-purified pre-60S intermediate with other well-characterized pre-60S particles suggests that Nsa1 coenriches a discrete, late nucleolar pre-60S particle. Moreover, Nsa1 appears to be tightly bound to this biogenesis intermediate because it was not significantly released by treatment with increasing concentrations of salt (unpublished data). Therefore, it is possible that dissociation of Nsa1 from this pre-60S particle may require the expenditure of energy in vivo. In line with a specific role of Rix7 in this process, Rix7 is exclusively associated with the Nsa1-defined pre-60S particle but not with other pre-60S intermediates, which suggests that it may transiently act upon Nsa1 when associated with this pre-60S particle. Moreover, Nsa1 is sharply released when the pre-60S particles enter the nucleoplasm, as indicated by its absence from the nucleoplasmic Rix1 particle. Considering that we observed a robust yeast two-hybrid interaction between full-length Nsa1 and a fragment of Rix7, our data suggest that Nsa1 could be a preribosomal substrate of Rix7. Alternatively, the Rix7-triggered release of Nsa1 from the pre-60S particle may be an indirect consequence of Rix7 acting on other pre-60S factors, with one of these being its direct substrate. However, we did not observe any significant coenrichment of other trans-acting factors in the Nsa1 TAP from the rix7Δ14N mutant background, which suggests that Nsa1 may be indeed a direct substrate of Rix7.
Nsa1 defines a distinct nucleolar pre-60S particle, which we could place between the early nucleolar Ssf1-defined and the later nucleoplasmic Rix1-defined pre-60S particle, which also carries another AAA ATPase, Rea1 (Nissan et al., 2002). Interestingly, upon transition of pre-60S particles from the nucleolus to the nucleoplasm, a major compositional change occurs, which is indicated by the release of a myriad of pre-60S factors. Accordingly, the “upstream” Ssf1 particle contains a large number of early trans-acting factors, whereas the “downstream” Rix1 particle lacks most of these pre-60S factors but has acquired new biogenesis factors including Rea1, Rix1, Ipi1, Ipi3, and Nog2. It is conceivable that the Rix7-dependent release of Nsa1 from a late nucleolar pre-60S particle may be a trigger to induce the successive evolution of the pre-60S particle in the nucleoplasm. In case of an inefficient release of Nsa1 upon mutational inactivation of Rix7, the generated pre-60S particles either do not further evolve and are degraded in the nucleus or escape as aberrant pre-60S subunits to the cytoplasm. Moreover, a fraction of the pre-60S particles appear to also be retained in the nucleolus, as suggested by the observation that the protein composition of Nop7-purified pre-60S particles—which, in wild-type cells, is a mixture of several pre-60S intermediates—is shifted toward one of the early pre-60S particles. The pool of pre-60S particles that escaped from disassembly and nucleolar retention contains Nsa1, which accumulates over time on cytoplasmic 60S subunits. The occurrence of Nsa1 in polyribosomes indicates that the Nsa1-containing 60S subunits are competent to engage in translation; however, it is possible that they may not be fully functional in all aspects of the translation process.
The discovery of the pre-60S factor Nsa1 as a potential substrate protein of Rix7 raises many new questions. How would the release of Nsa1 be timed and triggered? Is the N-terminal domain of Rix7, which is genetically linked to Nsa1, also involved in Nsa1 recognition and its subsequent dissociation from pre-60S subunits? Considering the significant homology between Rix7/NVL and Cdc48/p97 over the D1 and D2 domains, we speculate that their mechanisms of substrate binding and ATP hydrolysis–dependent release could be similar. Hence, the N-terminal domain of Rix7 might also contribute to the recognition and processing of a modified substrate, which might consist of ubiquitinated or sumoylated Nsa1. Indeed, Rix7 and Nsa1 have previously been genetically connected to the SUMO pathway (Panse et al., 2006), and, moreover, Nsa1 is also polyubiquitinated (unpublished data). Thus, the mechanistic details of how Rix7, possibly by directly acting on Nsa1, restructures the Nsa1-defined pre-60S ribosomal particle remains a challenging question for future studies.
Materials and methods
Yeast strains and yeast genetic methods
The S. cerevisiae strains used in this study are listed in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200801181/DC1); all strains are derivatives of W303 (Thomas and Rothstein, 1989) or DS1-2b (Nissan et al., 2002). For the yeast two-hybrid analysis, the reporter strain PJ69-4A was used (James et al., 1996). Deletion disruption and C-terminal tagging at the genomic locus were performed as described previously (Longtine et al., 1998; Puig et al., 2001; Janke et al., 2004). Preparation of media, yeast transformation, and genetic manipulations were performed according to established procedures.
Plasmid constructs and random PCR mutagenesis
All recombinant DNA techniques were performed according to established procedures using Escherichia coli DH5α for cloning and plasmid propagation. All cloned DNA fragments generated by PCR amplification were verified by sequencing. Plasmids used in this study are listed in Table S2 (available at http://www.jcb.org/cgi/content/full/jcb.200801181/DC1). Random PCR mutagenesis of RIX7 and NSA1 was performed essentially as described previously (Kressler et al., 1999).
sl screens
sl screens were performed as described previously (Kressler et al., 1999). The sl screen with rix7-31 was performed at 30°C using strain Y4138, and it yielded one sl candidate (sl30). The sl screen with nsa1-6 was performed at 23°C using strain Y3900, and it yielded one sl candidate (sl38).
Yeast two-hybrid screens and interaction analysis
For the yeast two-hybrid interaction analysis, the plasmids expressing the bait proteins, fused to the GAL4 DNA-binding domain (G4BD), and the prey proteins, fused to the GAL4 activation domain (G4AD), were cotransformed into the reporter strain PJ69-4A (James et al., 1996). Yeast two-hybrid interactions were documented by spotting representative transformants in 10-fold serial dilution steps on synthetic dextrose complete (SDC)-Trp-Leu, SDC-Trp-Leu-His (HIS3 reporter), and SDC-Trp-Leu-Ade (ADE2 reporter) plates. Growth on SDC-Trp-Leu-His plates is indicative of a weak interaction, whereas only relatively strong interactions permit growth on SDC-Trp-Leu-Ade plates. As a positive control, the combination of plasmids pVA3-1 and pTD1-1 (both from Clontech Laboratories, Inc.) was used; these plasmids also served as negative controls when combined with plasmids expressing the prey and bait proteins under investigation, respectively.
For the yeast two-hybrid screen with full-length Nsa1, PJ69-4A was first transformed with YEplac195-NSA1-G4BD and then with a yeast two-hybrid library constructed in pMPM137 (N-terminal G4AD; CEN, TRP1, and ADH1 promoter; a gift of M.P. Mayer, Universität Heidelberg, Heidelberg, Germany). From a total of ∼1,340,000 transformants, four identical library plasmids containing a fragment of RIX7, coding for amino acids 133–557, were recovered.
Fluorescence microscopy and Rpl25-EGFP reporter assay
The Rpl25-EGFP reporter assay to monitor large ribosomal subunit export was performed as described previously (Gadal et al., 2001b). Cells were examined by fluorescence microscopy using an microscope (Imager Z1) equipped with a 100×/63× NA 1.4 Plan-Apochromat oil immersion lens and using DICIII, HE-EGFP, DAPI, or HECy3 filters (all from Carl Zeiss, Inc.). Images were acquired with an AxioCam MRm camera and AxioVision 4.3 software (Carl Zeiss, Inc.).
Sucrose gradient analysis and fractionation
Cell extracts for polysome profile analyses were prepared as described previously (Kressler et al., 1997). For the polysome runoff experiments, cycloheximide was omitted. Sucrose gradients were analyzed and fractionated using an UA-6 system (Teledyne ISCO) with continuous monitoring at A254. For Western analysis of sucrose gradient fractions, five A260 units of cell extract were layered onto 7–50% sucrose gradients, and the gradients were centrifuged at 38,000 rpm in a rotor (SW40; Beckman Coulter) for 4 h and 30 min. Fractions of 500 μl were collected and processed as described previously (Kressler et al., 1999). The precipitated fractions were resuspended in 100 μl of 3× sample loading buffer, and 5 μl each fraction was separated on NuPAGE SDS 4–12% gradient polyacrylamide gels (Invitrogen) and analyzed by Western blotting. The position of the 40S, 60S, 80S and polysomes was determined from the UV profile of the sucrose gradient.
Preparation of total yeast protein extracts and Western analysis
Total yeast protein extracts were prepared as described previously (Yaffe and Schatz, 1984). Cultures were grown to an OD600 of around 0.8 and protein extracts were prepared from an equivalent of one OD600 of cells. Western blot analysis was performed according to standard protocols. The following primary antibodies were used in this study: mouse monoclonal anti-GFP (1:3,000; Roche) and anti-Rpl3 (1:5,000; J.R. Warner, Albert Einstein College of Medicine, New York, NY); rabbit polyclonal anti-CBP (1:15,000; Open Biosystems), anti-Arc1 (1:20,000; Simos et al., 1996), anti-Nmd3 (1:5,000; A.W. Johnson, University of Texas, Austin, TX), anti-Noc1 (1:500; H. Tschochner, Universität Regensburg, Regensburg, Germany), anti-Noc3 (1:200; H. Tschochner), anti-Nog1 (1:10,000; M. Fromont-Racine, Institut Pasteur, Paris, France), anti-Nog2 (1:10,000; M. Fromont-Racine), anti-Nop7 (1:40,000; B. Stillman, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY), anti-Rpl10 (1:2,000; B.L. Trumpower, Dartmouth Medical School, Hanover, NH), anti-Rpp0 (1:2,000; J.P.G. Ballesta, Universidad Autonoma, Madrid, Spain), and anti-Rps8 (1:1,000; H. Tschochner). Secondary goat anti–rabbit or anti–mouse (both from Bio-Rad Laboratories, Inc.) horseradish peroxidase–conjugated antibodies were used at a dilution of 1:7,500. For detection of TAP-tagged proteins, the peroxidase anti-peroxidase complex antibody was used at a dilution of 1:15,000 (Dako). Proteins were visualized using enhanced chemiluminescence detection kits (Immobilon Western [Millipore]; and Amersham ECL [GE Healthcare]).
TAP and mass spectrometry
TAP purifications of TAP-tagged bait proteins were, unless otherwise indicated, performed in a buffer containing 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1.5 mM MgCl2, 5% glycerol, and 0.1% NP-40 essentially as described previously (Puig et al., 2001; Nissan et al., 2002). For tobacco etch virus protease cleavage, DTT was added to a final concentration of 1 mM to the buffer. Elution from calmodulin–Sepharose beads was performed in the presence of 5 mM EGTA. The EGTA eluates were precipitated by the addition of TCA to a final concentration of 10% and dissolved in SDS sample buffer. The samples were then separated on NuPAGE SDS 4–12% gradient polyacrylamide gels (Invitrogen) and stained with colloidal Coomassie (Sigma-Aldrich). Mass spectrometric identification of the proteins contained in Coomassie-stained bands was performed as described previously (Bassler et al., 2001; Nissan et al., 2002).
rRNA composition of pre-60S particles
To minimize RNA degradation during TAP purification, tobacco etch virus protease was preincubated with RiboLock RNase inhibitor (Fermentas). Total RNA and rRNA was extracted from total extracts and EGTA eluates, respectively, as described previously (de la Cruz et al., 1998). Aliquots of total RNA (∼5 μg) and the EGTA eluate (∼15%) were resolved on a 1.5% agarose-formaldehyde gel and transferred to a nylon membrane for Northern hybridization. The same filter was hybridized consecutively with the following probes detecting pre-rRNA and mature rRNA species: A2-A3, 5′-TGTTACCTCTGGGCCC-3′; E-C2, 5′-GGCCAGCAATTTCAAGTTA-3′; 25S, 5′-CTCCGCTTATTGATATGC-3′; 18S, 5′-CATGGCTTAATCTTTGAGAC-3′; 5.8S, 5′-GCGTTCTTCATCGATGC-3′; and 5S, 5′-GGTCACCCACTACACTACTCGG-3′.
Online supplemental material
Fig. S1 contains the basic phenotypic characterization of the N-terminal rix7 mutants used in this study. Fig. S2 contains the basic phenotypic characterization of the nsa1 mutants used in this study. Fig. S3 shows that full-length Nsa1 and a fragment of Rix7 interact in the yeast two-hybrid assay, and that Nsa1 can be copurified with Rix7-TAP. Fig. S4 reveals the rRNA composition of the Nop7 pre-60S particle isolated from wild-type RIX7 or rix7Δ14N mutant cells. Table S1 indicates the yeast strains used in this study. Table S2 indicates the plasmids used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200801181/DC1.
Acknowledgments
We are indebted to the following colleagues for their kind gift of antibodies: J.P.G. Ballesta (anti-Rpp0), M. Fromont-Racine (anti-Nog1 and anti-Nog2), A.W. Johnson (anti-Nmd3), B. Stillman (anti-Nop7), B.L. Trumpower (anti-Rpl10), H. Tschochner (anti-Noc1, anti-Noc3, and anti-Rps8), and J.R. Warner (anti-Rpl3). We would especially like to thank M.P. Mayer for providing the yeast two-hybrid library. We also acknowledge the excellent technical assistance of P. Ihrig and S. Merker under the supervision of J. Lechner. We thank G. Bange, J. Bassler, A. Köhler, S. Lutz, and V. Panse for fruitful discussions and T. Nissan for providing the RIX7-TAP strain.
D. Kressler was the recipient of an European Molecular Biology Organization long-term fellowship (ALTF381-2003) and a Swiss National Science Foundation fellowship (PA00A--101445). This work was supported by grants from the Deutsche Forschungsgemeinschaft (Hu363/10-3 and the Gottfried Wilhelm Leibniz Program) to E. Hurt.
Abbreviations used in this paper: AAA, ATPases associated with various cellular activities; pre-rRNA, precursor rRNA; rRNA, ribosomal RNA; SDC, synthetic dextrose complete; sl, synthetic lethal; TAP, tandem affinity purification; ts, temperature sensitive.
References
- Bassler, J., P. Grandi, O. Gadal, T. Leßmann, E. Petfalski, D. Tollervey, J. Lechner, and E. Hurt. 2001. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell. 8:517–529. [DOI] [PubMed] [Google Scholar]
- DeLaBarre, B., and A.T. Brunger. 2005. Nucleotide dependent motion and mechanism of action of p97/VCP. J. Mol. Biol. 347:437–452. [DOI] [PubMed] [Google Scholar]
- de la Cruz, J., D. Kressler, M. Rojo, D. Tollervey, and P. Linder. 1998. Spb4p, an essential putative RNA helicase, is required for a late step in the assembly of 60S ribosomal subunits in Saccharomyces cerevisiae.RNA. 4:1268–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz, J., D. Kressler, and P. Linder. 2004. Ribosomal subunit assembly. In The Nucleolus. M.O.J. Olson, editor. Landes Bioscience, Georgetown, TX. 258–285.
- Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313–318. [DOI] [PubMed] [Google Scholar]
- Fatica, A., A.D. Cronshaw, M. Dlakic, and D. Tollervey. 2002. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell. 9:341–351. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine, M., B. Senger, C. Saveanu, and F. Fasiolo. 2003. Ribosome assembly in eukaryotes. Gene. 313:17–42. [DOI] [PubMed] [Google Scholar]
- Gadal, O., D. Strauss, J. Braspenning, D. Hoepfner, E. Petfalski, P. Philippsen, D. Tollervey, and E. Hurt. 2001. a. A nuclear AAA-type ATPase (Rix7p) is required for biogenesis and nuclear export of 60S ribosomal subunits. EMBO J. 20:3695–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal, O., D. Strauss, J. Kessl, B. Trumpower, D. Tollervey, and E. Hurt. 2001. b. Nuclear export of 60S ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 21:3405–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman, S., and S.J. Baserga. 2004. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 296:43–50. [DOI] [PubMed] [Google Scholar]
- Hanson, P.I., and S.W. Whiteheart. 2005. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6:519–529. [DOI] [PubMed] [Google Scholar]
- Harnpicharnchai, P., J. Jakovljevic, E. Horsey, T. Miles, J. Roman, M. Rout, D. Meagher, B. Imai, Y. Guo, C.J. Brame, et al. 2001. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell. 8:505–515. [DOI] [PubMed] [Google Scholar]
- James, P., J. Halladay, and E.A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 144:1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke, C., M.M. Magiera, N. Rathfelder, C. Taxis, S. Reber, H. Maekawa, A. Moreno-Borchart, G. Doenges, E. Schwob, E. Schiebel, and M. Knop. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962. [DOI] [PubMed] [Google Scholar]
- Jentsch, S., and S. Rumpf. 2007. Cdc48 (p97): a “molecular gearbox” in the ubiquitin pathway? Trends Biochem. Sci. 32:6–11. [DOI] [PubMed] [Google Scholar]
- Kressler, D., J. de la Cruz, M. Rojo, and P. Linder. 1997. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae.Mol. Cell. Biol. 17:7283–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler, D., M. Doère, M. Rojo, and P. Linder. 1999. Synthetic lethality with conditional dbp6 alleles identifies Rsa1p, a nucleoplasmic protein involved in the assembly of 60S ribosomal subunits. Mol. Cell. Biol. 19:8633–8645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruiswijk, T., R.J. Planta, and J.M. Krop. 1978. The course of the assembly of ribosomal subunits in yeast. Biochim. Biophys. Acta. 517:378–389. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., A. McKenzie III, D.J. Demarini, N.G. Shah, A. Wach, A. Brachat, P. Philippsen, and J.R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae.Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Neuwald, A.F., L. Aravind, J.L. Spouge, and E.V. Koonin. 1999. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9:27–43. [PubMed] [Google Scholar]
- Nissan, T.A., J. Bassler, E. Petfalski, D. Tollervey, and E. Hurt. 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21:5539–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan, T.A., K. Galani, B. Maco, D. Tollervey, U. Aebi, and E. Hurt. 2004. A pre-ribosome with a tadpole-like structure functions in ATP-dependent maturation of 60S subunits. Mol. Cell. 15:295–301. [DOI] [PubMed] [Google Scholar]
- Panse, V.G., D. Kressler, A. Pauli, E. Petfalski, M. Gnädig, D. Tollervey, and E. Hurt. 2006. Formation and nuclear export of preribosomes are functionally linked to the small-ubiquitin-related modifier pathway. Traffic. 7:1311–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, S., and M. Latterich. 1998. The AAA team: related ATPases with diverse functions. Trends Cell Biol. 8:65–71. [PubMed] [Google Scholar]
- Pertschy, B., C. Saveanu, G. Zisser, A. Lebreton, M. Tengg, A. Jacquier, E. Liebminger, B. Nobis, L. Kappel, I. van der Klei, et al. 2007. Cytoplasmic recycling of 60S preribosomal factors depends on the AAA protein Drg1. Mol. Cell. Biol. 27:6581–6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Séraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 24:218–229. [DOI] [PubMed] [Google Scholar]
- Pye, V.E., I. Dreveny, L.C. Briggs, C. Sands, F. Beuron, X. Zhang, and P.S. Freemont. 2006. Going through the motions: the ATPase cycle of p97. J. Struct. Biol. 156:12–28. [DOI] [PubMed] [Google Scholar]
- Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Séraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030–1032. [DOI] [PubMed] [Google Scholar]
- Saveanu, C., D. Bienvenu, A. Namane, P.E. Gleizes, N. Gas, A. Jacquier, and M. Fromont-Racine. 2001. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 20:6475–6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer, T., D. Strauss, E. Petfalski, D. Tollervey, and E. Hurt. 2003. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 22:1370–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simos, G., A. Segref, F. Fasiolo, K. Hellmuth, A. Shevchenko, M. Mann, and E.C. Hurt. 1996. The yeast protein Arc1p binds to tRNA and functions as a cofactor for the methionyl- and glutamyl-tRNA synthetases. EMBO J. 15:5437–5448. [PMC free article] [PubMed] [Google Scholar]
- Song, C., Q. Wang, and C.C. Li. 2003. ATPase activity of p97-valosin-containing protein (VCP). D2 mediates the major enzyme activity, and D1 contributes to the heat-induced activity. J. Biol. Chem. 278:3648–3655. [DOI] [PubMed] [Google Scholar]
- Thomas, B.J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell. 56:619–630. [DOI] [PubMed] [Google Scholar]
- Trapman, J., J. Retèl, and R.J. Planta. 1975. Ribosomal precursor particles from yeast. Exp. Cell Res. 90:95–104. [DOI] [PubMed] [Google Scholar]
- Tschochner, H., and E. Hurt. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13:255–263. [DOI] [PubMed] [Google Scholar]
- Vale, R.D. 2000. AAA proteins. Lords of the ring. J. Cell Biol. 150:F13–F19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., C. Song, X. Yang, and C.C. Li. 2003. D1 ring is stable and nucleotide-independent, whereas D2 ring undergoes major conformational changes during the ATPase cycle of p97-VCP. J. Biol. Chem. 278:32784–32793. [DOI] [PubMed] [Google Scholar]
- Warner, J.R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24:437–440. [DOI] [PubMed] [Google Scholar]
- Yaffe, M.P., and G. Schatz. 1984. Two nuclear mutations that block mitochondrial protein import in yeast. Proc. Natl. Acad. Sci. USA. 81:4819–4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinker, S., and J.R. Warner. 1976. The ribosomal proteins of Saccharomyces cerevisiae. Phosphorylated and exchangeable proteins. J. Biol. Chem. 251:1799–1807. [PubMed] [Google Scholar]