Abstract
Centromeres are special structures of eukaryotic chromosomes that hold sister chromatid together and ensure proper chromosome segregation during cell division. Centromeres consist of repeated sequences, which have hindered the study of centromere mitotic recombination and its consequences for centromeric function. We use a chromosome orientation fluorescence in situ hybridization technique to visualize and quantify recombination events at mouse centromeres. We show that centromere mitotic recombination occurs in normal cells to a higher frequency than telomere recombination and to a much higher frequency than chromosome-arm recombination. Furthermore, we show that centromere mitotic recombination is increased in cells lacking the Dnmt3a and Dnmt3b DNA methyltransferases, suggesting that the epigenetic state of centromeric heterochromatin controls recombination events at these regions. Increased centromere recombination in Dnmt3a,3b-deficient cells is accompanied by changes in the length of centromere repeats, suggesting that prevention of illicit centromere recombination is important to maintain centromere integrity in the mouse.
Introduction
Centromeres are essential for the correct segregation and inheritance of genetic information by ensuring that each daughter cell receives a copy of each chromosome during cell division (Pidoux and Allshire, 2000). Mammalian centromeres are composed of long arrays of tandemly repeated DNA sequences (Sunkel and Coelho, 1995). The mouse genome contains at least two types of repetitive elements at centromeres, the major satellite repeats (6 Mb of 234-bp repeats) and the minor satellite repeats (∼600 kb of 120-bp repeats, varying from 500 kb to 1.2 Mb depending on the chromosome; Kipling et al., 1991, 1994; Guenatri et al., 2004, Kuznetsova et al., 2005). The major satellite repeats are located pericentrically, whereas the minor satellite repeats coincide with the centric constriction (Wong and Rattner, 1988). The centromere protein B (CENP-B) is found at minor satellite DNA and alphoid DNA in mouse and human cells, respectively (Horz and Altenburger, 1981; Earnshaw et al., 1989; Kipling and Warburton, 1997).
In addition, centromeres contain epigenetic marks characteristic of compacted heterochromatin domains, including histone lysine trimethylation and DNA hypermethylation (Lehnertz et al., 2003; Maison and Almouzni, 2004). Maintenance of centromeric heterochromatin has been proposed to be important for centromere function. In particular, loss of DNA methylation at pericentromeric regions caused by abrogation of the DNA methyltransferase enzymes Dnmt1 or Dnmt3a and Dnmt3b (Okano et al., 1998, 1999; Chen et al., 2004) results in defective centromere function (Chen et al., 2004; Dodge et al., 2005), although the mechanisms responsible for this are still largely unknown.
Similar to centromeres, telomeres (or the terminal ends of chromosomes) are also repeated elements, which in all vertebrates consist of tandem repeats of the TTAGGG sequence and associated proteins (Blackburn, 2001). Telomeres have an essential role in chromosome end protection and chromosomal stability. A proper length of telomeric repeats is maintained by the enzyme telomerase (Blackburn, 2001), as well as by the recombination-based alternative lengthening of telomeres or ALT mechanism (Muntoni and Reddel, 2005). Similarly to centric and pericentric chromatin, telomeres, as well as the adjacent subtelomeric regions, are enriched in epigenetic marks characteristic of constitutive heterochromatin, including histone trimethylation marks and subtelomeric DNA hypermethylation (Brock et al., 1999; Gonzalo et al., 2006; Blasco, 2007). Furthermore, these marks act as independent negative regulators of telomere length and recombination (Gonzalo et al., 2006; Blasco, 2007; Benetti et al., 2007). In particular, in the absence of Dnmts, telomeres become dramatically elongated concomitant with increased telomere recombination and in the absence of loss of histone trimethylation marks (Gonzalo et al., 2006).
Although mitotic centromere recombination has been previously described in yeast (Liebman et al., 1988), very little is known on the regulation of mammalian centromeric mitotic recombination and how this may impact on centromeric function (Warburton and Willard, 1992; Warburton et al., 1993). In this paper, we have established a technique based on chromosome orientation FISH (CO-FISH; Bailey et al., 1996), which allows measuring recombination frequencies specifically at centromeric repeats (cen-CO-FISH). Using this technique, we found that centromeric repeats are highly recombinogenic compared with the rest of the genome. In addition, we describe a role for DNA methylation in preventing centromeric mitotic recombination, as well as in controlling the length of centromeric repeats. Together, these results highlight a role for the epigenetic status of centromeric heterochromatin, and in particular of DNA methylation, in regulating centromere recombination and centromere length in mouse chromosome, which, if disrupted, may conceivably contribute to genomic instability.
Results and Discussion
A FISH-based technique to measure centromere recombination
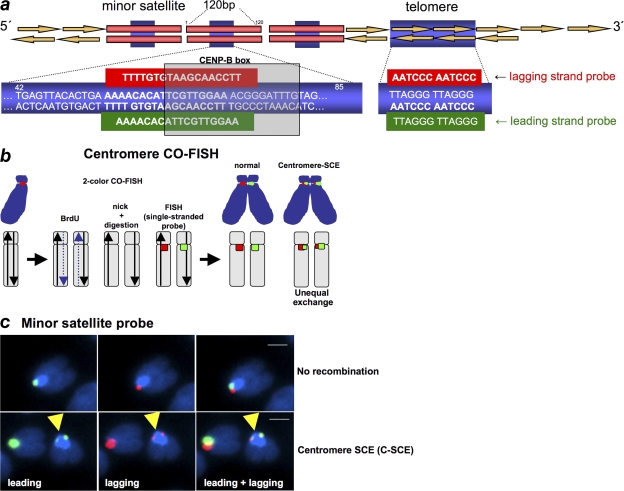
To study recombination events specifically at the centromeric regions, we have adapted the previously described two-color CO-FISH to centromeric repeats (cen-CO-FISH; Fig. 1; Materials and methods). In particular, cells that have undergone a single round of DNA replication in the presence of BrdU are used to prepare chromosome spreads. After HOECHST staining, the bromo-substituted strand is removed by nicking DNA with UV light, followed by digestion with exonuclease III (Bailey et al., 1996). Metaphases are then hybridized with strand-specific peptide nucleic acid (PNA) probes against centromeric repeats. In particular, mouse minor satellite is composed of DNA repeats of a 120-bp sequence, which contains the binding site for the CENP-B protein, the so-called CENP-B box (bp 62–78; Fig. 1 a; Wong and Rattner, 1988). To perform cen-CO-FISH, single-stranded PNA probes were designed to hybridize with the lagging and leading strands of the mouse CENP-B box sequence (Fig. 1, a and b; Materials and methods; Chen et al., 1999). In the absence of recombination events at minor satellite repeats, only one chromatid per chromosome will show a signal after hybridization with either the lagging or the leading probes (Fig. 1, b and c). In contrast, if recombination occurs within minor satellite sequences, labeling will split between the sister chromatids, giving rise to two signals of unequal intensity (Fig. 1, b and c). We refer to these events as centromere sister chromatid exchange (C-SCE) events. A C-SCE event is considered positive only when the exchange is simultaneously detected with the leading and lagging centromeric probes (Fig. 1 c). As controls for the C-SCE CO-FISH technique, we determined that the frequency of C-SCE events is similar for different slides of the same genotype processed in parallel, suggesting a similar efficiency of ExoIII digestions (Fig. S1 a, available at http://www.jcb.org/cgi/content/full/jcb.200803042/DC1). Furthermore, the fact that a significant percentage of metaphases (>10%) did not show any C-SCE event indicates that C-SCE events are true recombination events and not consequences of the possible existence of inverted repeats at some chromosome centromeric regions (Fig. S1 b).
Figure 1.
CO-FISH to measure recombination rates at minor satellite sequences. (a) A scheme of mouse minor satellite repeats and telomere repeats is shown indicating the position of the CENP-B box (bp 62–78). PNA probes are also indicated. (b) A diagram explaining the CO-FISH procedure is shown. Cells are allowed to replicate once in the presence of BrdU, giving rise to chromosomes with one BrdU-containing chromatid (dashed line). The BrdU-containing DNA strand is digested and single-stranded minor satellite probes against the lagging (red) or the leading (green) strand are hybridized to the remaining non–BrdU-labeled strand. Cen-CO-FISH labels centromeric minor satellite sequences from the lagging or leading strand. An SCE within minor satellite sequences (C-SCE) will lead to two unequal signals per chromosome after hybridization with either the leading or lagging single-stranded probes. (c) Representative Cen-CO-FISH images showing no recombination (top) or a C-SCE after hybridization with the leading and lagging minor satellite PNA probes (bottom). Bars, 1 μm.
Mammalian centromeres are highly recombinogenic
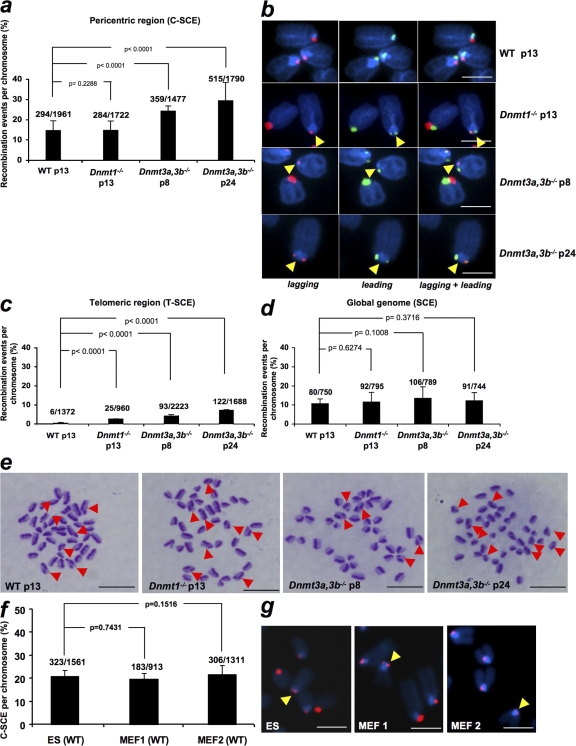
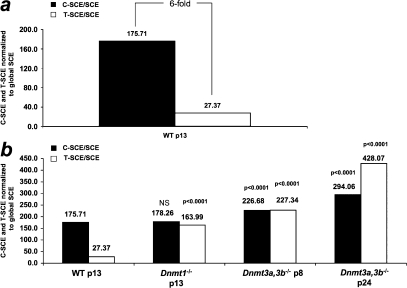
To determine the frequency of mitotic recombination at mammalian centromeres, we first measured C-SCE events per chromosome in wild-type mouse embryonic stem (ES) cells (Fig. 2, a and b) and compared these values to the frequency of SCE events at telomeres (T-SCE), which were previously determined by us using CO-FISH in the same cells (Fig. 2 c; Gonzalo et al., 2006), as well as to the frequency of global SCE events determined by a fluorescence-plus-giemsa protocol (Fig. 2, d and e; Materials and methods). Next, we normalized C-SCE, T-SCE, and SCE values by the approximate length of their respective DNA in kilobases (Tables I–III; Materials and methods) and represented C-SCE and T-SCE frequencies relative to SCE frequencies (Fig. 3 a; Materials and methods). Notably, the frequency of C-SCE events was sixfold higher than that of T-SCE events in wild-type ES cells (Fig. 3 a). The differences in mitotic recombination at these two highly repeated regions may be the result of their different DNA sequence and/or their distinct chromatin organization. In this regard, telomeres are bound by the so-called shelterin complex, which safeguards telomeres from DNA repair activities (de Lange, 2005). In contrast, centromeres are not bound by shelterin and have been described to have a more efficient DNA repair than the rest of the genome (Rief and Löbrich, 2002). Interestingly, when calculating the C-SCE/SCE and T-SCE/SCE ratios, centromeres and telomeres appeared to be dramatically more recombinogenic (∼175-fold and 27-fold, respectively) than the rest of the genome (Fig. 3 a). Although we cannot rule out that the different nature of the CO-FISH (T-SCE and C-SCE) and fluorescence-plus-giemsa (SCE) techniques could be underestimating the frequency of global SCE events, our results are in agreement with a recent study showing a 20-fold increase in T-SCE events compared with global SCE events (Bailey et al., 2004). These findings suggest that repeated regions are hot spots of recombination compared with the rest of the genome and that mechanisms must exist that prevent illicit DNA recombination at these regions to preserve their integrity.
Figure 2.
Recombination events per chromosome at the indicated chromosomal regions in wild-type and Dnmt-deficient ES cells. (a) Error bars correspond to two independent experiments (n = 2). The total number of C-SCE detected out of the total number of chromosomes analyzed per genotype is also indicated on top of each bar. (b) Representative examples of C-SCE in ES cells of the indicated passage and genotype. Yellow arrows point to the C-SCE event. Bars, 3 μm. (c) T-SCE data were obtained from Gonzalo et al. (2006). (d) Quantification of global SCE events in wild-type and Dnmt-deficient mouse ES cells at the indicated passage number. One culture of each genotype was used for the analysis. The total number of SCE events out of the total number of chromosomes analyzed is indicated on top of each bar. (e) Representative images of SCE events. The arrows indicate chromosomes showing SCE events. Bars, 10 μm. (f) Quantification of C-SCE frequencies in mouse wild-type ES cells and two independent wild-type MEF cells. No significant differences were observed between the indicated cells. Numbers above bars represent total C-SCE events out of the total number of chromosomes counted. (g) Representative CO-FISH images after labeling lagging (red) strand centromeres are shown. Yellow arrows indicate C-SCE events. Bars, 5 μm. Error bars represent standard error.
Table I. Recombination frequencies as determined by SCE events at minor satellite repeats in the indicated genotypes.
| Recombination frequency per chromosome |
Mean kb per minor satellite |
Minor satellite DNA per mouse chromosome |
C-SCE per kb | |
|---|---|---|---|---|
| Dnmt wt p13 | 0.149 | 600 | 1,200 | 1.24 × 10−4 |
| Dnmt1−/− p13 | 0.164 | 600 | 1,200 | 1.36 × 10−4 |
| Dnmt3a,3b−/−p8 | 0.243 | 600 | 1,200 | 2.02 × 10−4 |
| Dnmt3a,3b−/−p24 | 0.287 | 600 | 1,200 | 2.39 × 10−4 |
p, passage number.
Table II. Recombination frequencies as determined by SCE events at telomeric repeats in the indicated genotypes.
| Recombination frequency per chromosomea |
Mean kb per telomere | Telomeric DNA per mouse chromosome |
T-SCE per kb | |
|---|---|---|---|---|
| Dnmt wt p13 | 0.004 | 51.7 | 206.8 | 1.93 × 10−5 |
| Dnmt1−/− p13 | 0.026 | 51.7 | 206.8 | 1.25 × 10−4 |
| Dnmt3a,3b−/−p8 | 0.042 | 51.7 | 206.8 | 2.03 × 10−4 |
| Dnmt3a,3b−/−p24 | 0.072 | 51.7 | 206.8 | 3.48 × 10−4 |
p, passage number.
Values obtained from Gonzalo et al. (2006).
Table III. Global recombination frequencies as determined by SCE events in the indicated genotypes.
| Recombination frequency per chromosome |
Mouse genome (kb) | DNA per mouse chromosomea | SCE per kb | |
|---|---|---|---|---|
| Dnmt wt p13 | 0.106 | 3,000,000 | 150,000 | 7.06 × 10−7 |
| Dnmt1−/− p13 | 0.115 | 3,000,000 | 150,000 | 7.66 × 10−7 |
| Dnmt3a,3b−/−p8 | 0.134 | 3,000,000 | 150,000 | 8.93 × 10−7 |
| Dnmt3a,3b−/−p24 | 0.122 | 3,000,000 | 150,000 | 8.13 × 10−7 |
p, passage number.
The 150,000 value corresponds to the size of the mouse genome divided by the number of chromosomes multiplied by two diploid cells (300,000 kb/40 × 2 = 150 kb).
Figure 3.
Centromeres are more recombinogenic than telomeres. (a and b) Direct comparison of recombination events at centromeres and telomeres per kilobase relative to global SCE events in kilobase (Tables I–III) in wild-type and Dnmt-deficient ES cells. Note that centromeres are sixfold more recombinogenic than telomeres. The T-SCE/SCE and C-SCE/SCE ratios also indicate that both telomeres and centromeres are more recombinogenic than the global genome. C-SCE and T-SCE, but not SCE (see Fig. 2d), are further increased in the absence of Dnmts. Statistical significance comparisons with the respective wild-type value are indicated on top of each bar.
DNA methylation is a negative regulator of centromeric recombination
We recently described that abrogation of Dnmt1, or simultaneous deletion of Dnmt3a and 3b, results in increased recombination at mouse telomeres (Gonzalo et al., 2006), suggesting a role for DNA methylation in preventing illicit recombination at highly repetitive sequences in the genome. Mouse ES cells deficient for Dnmt1 or both Dnmt3a and 3b show decreased DNA methylation at both major and minor centromeric repeats in the absence of changes in histone heterochromatic marks (Chen et al., 1999, 2004; Kuznetsova et al., 2005; Gonzalo et al., 2006). In this paper, we set to determine whether decreased DNA methylation in Dnmt-deficient cells results in increased recombination rates at mouse minor satellite repeats as determined by cen-CO-FISH (Materials and methods). Interestingly, Dnmt3a,3b-deficient ES cells presented significantly increased frequencies of C-SCE per chromosome compared with wild-type controls (Fig. 2, a and b, χ2 test, P < 0.0001; and Fig. 3 b, C-SCE frequencies corrected by global SCE frequencies), whereas cells deficient for Dnmt1 showed a similar C-SCE frequency to wild-type controls (Fig. 2 a, χ2 test, P = 0.2288, not significant; and Fig. 3 b, C-SCE/SCE ratios). These results demonstrate that abrogation of the Dnmt3a,3b enzymes, which are responsible for de novo DNA methylation, leads to increased recombination at centromeric repeats in mammalian cells. As previously described (Gonzalo et al., 2006), telomere recombination (T-SCE) was also increased in Dnmt-deficient cells (Fig. 2 c, χ2 test, P < 0.0001 for all comparisons; and Fig. 3 b, T-SCE/SCE ratios). However, although Dnmt1 deficiency significantly increased recombination at telomeric repeats, this was not the case at minor satellite repeats, indicating slightly different requirements for Dnmt1 activity at these two repeated regions.
Next, we addressed whether increased recombination at pericentric and telomeric repeats in Dnmt3a,3b-deficient cells was specific to these repeated regions or, instead, was accompanied by a global increase in SCEs throughout the genome. To this end, we analyzed the frequency of global SCE in ES cells deficient for either Dnmt1 or Dnmt3a,3b compared with wild-type controls. Interestingly, no significant differences in global SCE frequencies were found between genotypes (Fig. 2, d and e). Finally, we did not detect significant differences in C-CSE frequencies when comparing pluripotent mouse ES cells to differentiated mouse embryonic fibroblasts (MEFs; Fig. 2, f and g), suggesting that centromere recombination is not significantly decreased when associated with differentiation. Collectively, these results support the notion that DNA methylation has an important role in preventing mitotic recombination specifically at highly repetitive regions but does not have a major impact on global genomic recombination.
DNA methylation of centromeric repeats influences centromere length
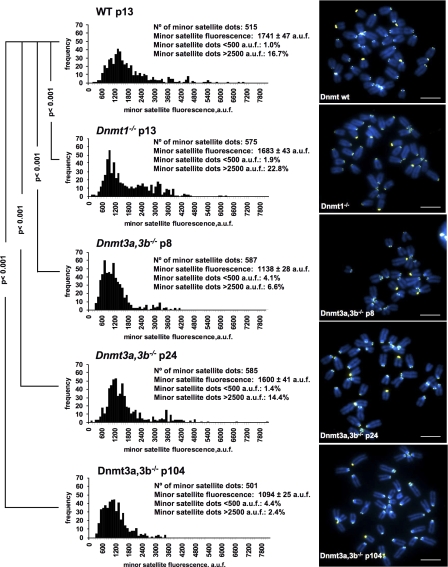
Increased recombination frequencies at telomeric sequences are associated with activation of telomerase-independent alternative pathways for telomere maintenance or alternative lengthening of telomeres (Bailey et al., 2004; Bechter et al., 2004). In particular, in the absence of either Dnmt1 or Dnmt3a,3b activity, there is a dramatic telomere elongation concomitant with increased telomere recombination frequencies in these cells (Gonzalo et al., 2006). To evaluate whether increased recombination at Dnmt3a,3b-deficient centromeres was also associated with changes in the length of centromeric repeats, we performed centromere quantitative FISH (Q-FISH) on metaphase spreads from wild-type and Dnmt-deficient ES cells using a minor satellite-specific PNA probe (Materials and methods). Interestingly, mouse ES cells deficient for either Dnmt1 or Dnmt3a,3b, showed decreased minor satellite fluorescence compared with wild-type controls. These differences were highly significant for both Dnmt1- and Dnmt3a,3b-deficient cells (Fig. 4, P < 0.001 for every comparison). Furthermore, the decrease in minor satellite fluorescence was accompanied by higher frequencies of centromeres showing very low minor satellite fluorescence (<500 arbitrary units of fluorescence; Fig. 4), which is in agreement with shorter centromeres in Dnmt3a,3b-deficient cells. The fluctuations of centromere fluorescence with increasing passages of Dnmt3a,3b-deficient may be an additional indication of centromeric instability associated with DNA hypomethylation and increased recombination (Fig. 4). Interestingly, abnormal size of centromeric fragments as detected by Southern blotting after digestion with MspI was previously reported by us with increasing passages of Dnmt3a,3b-deficient cells (see Fig. S2 a in Gonzalo et al., 2006). Collectively, these results suggest that repression of centromeric recombination by DNA methylation is an important mechanism for maintaining centromere integrity. In particular, increased mitotic recombination at centromeric sequences as the result of loss of DNA methylation may lead to altered length of centromere repeats, which in turn may affect the binding of important centromeric proteins, such as CENP-B, resulting in genomic instability. In this regard, although CENP-B is not essential for mouse development (Hudson et al., 1998; Perez-Castro et al., 1998), it has been recently described to be necessary for de novo centromere formation by recruiting other CENPs as well as epigenetic factors (Okada et al., 2007).
Figure 4.
Quantification of minor satellite length in wild-type and Dnmt-deficient ES cells. Minor satellite fluorescence distribution in arbitrary units of fluorescence (a.u.f) of wild-type (wt), Dnmt1−/−, and Dnmt3a,3b−/− ES cells at the indicated passage number as determined by Q-FISH using a mouse minor satellite PNA probe (left). Representative images of metaphase spreads of the indicated genotypes are also shown (right). A nonparametric ANOVA test, the Kruskall-wallis test (P < 0.0001), indicated that the observed differences in the centromere fluorescence were not because of coincidences or random sampling. Furthermore, a Dunn's post test indicated that the differences were significant (P < 0.001) for each pair of comparisons. The p-values in the figure refer to comparisons of the different genotypes with wild-type cells. Bars, 7 μm.
Speculation
In this paper, we describe a FISH-based technique that allows the measurement of SCE events specifically at mammalian centromeric repeats. Our results suggest that centromeres are highly recombinogenic compared with chromosome arms as well as with telomeric repeats. In addition, we demonstrate that loss of DNA methylation leads to increased centromere recombination, suggesting that the epigenetic status of centromeric heterochromatin is important to maintain centromere integrity. Importantly, defective DNA methylation did not affect global recombination frequencies, suggesting a specific role of this epigenetic mark repressing illicit recombination at repeated elements but not elsewhere in the genome. We further show that loss of DNA methylation leads to abnormal centromere length, which conceivably could alter the binding of centromeric proteins leading to abnormal centromeric function (Okada et al., 2007). Interestingly, the CENP-B box contains CpG residues, which are potential targets of the Dnmt enzymes and whose methylation status has been previously reported to affect the binding of the CENP-B protein (Mitchell et al., 1996; Tanaka et al., 2005). Furthermore, a recent study shows that PTEN-dependent expression of the centromeric protein CENP-C can lead to centromere breaks and genomic instability (Shen et al., 2007). Collectively, these findings support a model where the loss of DNA methylation at centromeric repeats may favor illicit DNA recombination, which in turn could favor genomic instability.
Materials and Methods
Cell culture
Wild-type J1 and Dnmt1−/− mutant ES cells or Dnmt3a,3b−/− double mutant ES cells were generated and maintained as described previously (Gonzalo et al., 2006). Wild-type ES cells, MEF 1 (129 sv/C57BL6 background), and MEF 2 (C57BL6 background) were used.
Mouse minor satellite Q-FISH
A PNA probe was designed to hybridize with an 18-bp sequence from the following mouse minor satellite consensus sequence: Cy3-OO-TTC CAA CGA ATG TGT TTT (bp 55–72). Metaphases were prepared and hybridized with the Cy3-labeled minor satellite PNA probe, together with DAPI counterstaining of DNA, as previously described (Samper et al., 2000). Samples were mounted on Vectashield (Vector Laboratories) imaging medium and stored at 4°C in the dark for 1 d until image acquisition using a charge-coupled device camera (COHU4912; COHU, Inc.) with a resolution of 700 × 500 pixels on a fluorescence microscope (Leitz DMRB; Leica). A 100 W/2 lamp (HBO; Osram) was used as a source. The images were acquired at room temperature using the Q-FISH software (Leica) and an HCX PL APO 100× 1.4 NA old immersion objective (Leica) with the following filters: Cy3, Y3 (excitation, band pass 570/20 nm; dicroic, 565 nm; emission, band pass 610/75 nm; Leica); DAPI, A (excitation, band pass 340–380 nm; dicroic, 400 nm; emission, long pass 425 nm; Leica). We used the TFL-Telo software (gift from P. Lansdorp, Terry Fox Laboratory, Vancouver, Canada) to quantify the fluorescence intensity of minor satellite from at least 10 metaphases for each data point. All metaphases were captured the day after hybridization at room temperature, in parallel, blindly, and with the same image acquisition conditions.
Centromere and telomere CO-FISH
Confluent MEF and ES cells were subcultured in the presence of BrdU (Sigma-Aldrich) at a final concentration of 10−5 M and were then allowed to replicate their DNA once at 37°C for 24 and 12 h, respectively. Colcemid was added in a concentration of 0.2 μg/ml for MEFs and 1 μg/ml for ES cells for the last 4 and 1 h, respectively. CO-FISH was performed as previously described (Bailey et al., 1996) with some modifications. In brief, slides were hybridized with a minor satellite PNA probe labeled with Cy3 (Cy3-oo-TTC CAA CGA ATG TGT TTT), which hybridizes with the lagging DNA strand, followed by a second hybridization with a minor satellite PNA probe labeled with FITC (Flu-oo-AAA ACA CAT TCG TTG GAA), which hybridizes with the leading DNA strand. Data on telomere CO-FISH in the same cells were obtained from a previous study (Gonzalo et al., 2006). Metaphases were captured on a fluorescence microscope (Leitz DMRB) using the conditions as described in Mouse minor satellite Q-FISH, with an additional filter for FITC detection, L5 (excitation, BP 480/40 nm; dicroic, 505 nm; emission, BP 527/30 nm; Leica).
Differential staining technique for SCE determinations
Genomic SCEs were visualized using an adapted fluorescence-plus-giemsa protocol (Perry and Wolf, 1974). Metaphases were prepared as described for the Q-FISH (Samper et al., 2000) and captured using a bright field microscope (AX70; Olympus), using a 60× immersion objective. Images were captured using a color camera (DP70; Olympus) and ImagePro Plus high-end image acquisition and analysis software (Media Cybernetics, Inc.). All the captures were performed at room temperature. Metaphases were analyzed for harlequin staining. Each color switch was scored as one SCE.
Calculation of recombination frequency per kilobase of DNA
To determine the recombination frequencies per kilobase, the mean length of mouse ES cell telomeres was considered to be 51.7 kb per telomere (as determined by us using Q-FISH), hence the total length of telomeres per chromosome was 4 × 51.7 = 206.8 kb (Table II). The length of minor satellite repeats was considered to be ∼600 kb (Guenatri et al., 2004), hence the total length of minor satellite repeats per chromosome was 2 × 600 = 1,200 kb (Table I). The length of the mouse genome is 3 × 106 kb, therefore we estimated that the mean length of a mouse chromosome is 150,000 kb (Table III). For direct comparison of recombination events at centromeres and telomeres, T-SCE and C-SCE per kilobase were expressed relative to global SCE events per kilobase (Fig. 3).
Statistical analysis
A non-parametric ANOVA test, the Kruskall-wallis test, and the Dunn's post test were used to calculate the statistical significance for each individual comparison of minor satellite fluorescence (Fig. 4, centromere Q-FISH). To calculate statistical significance of changes in SCE, T-SCE, and C-SCE frequency, we used the χ2 test. The two-sided p-values were obtained from 2 × 2 contingency table analyzed by χ2 test (including Yates' continuity correction). InStat v.2.03 (GraphPad Software, Inc.) was used for the calculations. In all cases, differences are significant for P < 0.05, very significant for P < 0.01, highly significant for P < 0.001, and extremely significant for P < 0.0001.
Online supplemental material
Fig. S1 shows controls for the centromeric CO-FISH technique. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200803042/DC1.
Supplementary Material
Acknowledgments
M.A. Blasco's laboratory is funded by the MCyT (SAF2005-00277 and GEN2001-4856-C13-08), the Regional Government of Madrid (GR/SAL/0597/2004), the European Union (TELOSENS FIGH-CT-2002-00217, INTACT LSHC-CT-2003-506803, ZINCAGE FOOD-CT-2003-506850, RISC-RAD FI6R-CT-2003-508842, and MOL CANCER MED LSHC-CT-2004-502943), the Josef Steiner Cancer Research Award (2003), and The Spanish Association Against Cancer (AECC).
Abbreviations used in this paper: CENP-B, centromere protein B; CO-FISH, chromosome orientation FISH; C-SCE, centromere sister chromatid exchange; ES, embryonic stem; MEF, mouse embryonic fibroblast; PNA, peptide nucleic acid; Q-FISH, quantitative FISH.
References
- Bailey, S.M., E.H. Goodwin, J. Meyne, and M.N. Cornforth. 1996. CO-FISH reveals inversions associated with isochromosome formation. Mutagenesis. 11:139–144. [DOI] [PubMed] [Google Scholar]
- Bailey, S.M., M.A. Brenneman, and E.H. Goodwin. 2004. Frequent recombination in telomeric DNA may extend the proliferative life of telomerase-negative cells. Nucleic Acids Res. 32:3743–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechter, O.E., J.W. Shay, and W.E. Wright. 2004. The frequency of homologous recombination in human ALT cells. Cell Cycle. 3:547–549. [PubMed] [Google Scholar]
- Benetti, R., S. Gonzalo, I. Jaco, G. Schotta, P. Klatt, T. Jenuwein, and M.A. Blasco. 2007. Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J. Cell Biol. 178:925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn, E.H. 2001. Switching and signalling at the telomere. Cell. 106:661–673. [DOI] [PubMed] [Google Scholar]
- Blasco, M.A. 2007. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 8:299–309. [DOI] [PubMed] [Google Scholar]
- Brock, G.J., J. Charlton, and A. Bird. 1999. Densely methylated sequences that are preferentially localized at telomere-proximal regions of human chromosomes. Gene. 240:269–277. [DOI] [PubMed] [Google Scholar]
- Chen, C., Y.K. Hong, S.D. Ontiveros, M. Egholm, and W.M. Strauss. 1999. Single base discrimination of CENP-B repeats on mouse and human Chromosomes with PNA-FISH. Mamm. Genome. 10:13–18. [DOI] [PubMed] [Google Scholar]
- Chen, T., N. Tsujimoto, and E. Li. 2004. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol. Cell. Biol. 24:9048–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange, T. 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19:2100–2110. [DOI] [PubMed] [Google Scholar]
- Dodge, J.E., M. Okano, F. Dick, N. Tsujimoto, T. Chen, S. Wang, Y. Ueda, N. Dyson, and E. Li. 2005. Inactivation of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability, and spontaneous immortalization. J. Biol. Chem. 280:17986–17991. [DOI] [PubMed] [Google Scholar]
- Earnshaw, W.C., H. Ratrie III, and G. Stetten. 1989. Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma. 98:1–12. [DOI] [PubMed] [Google Scholar]
- Gonzalo, S., I. Jaco, M.F. Fraga, T. Chen, E. Li, M. Esteller, and M.A. Blasco. 2006. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat. Cell Biol. 8:416–424. [DOI] [PubMed] [Google Scholar]
- Guenatri, M., D. Bailly, C. Maison, and G. Almouzni. 2004. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 166:493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horz, W., and W. Altenburger. 1981. Nucleotide sequence of mouse satellite DNA. Nucleic Acids Res. 9:683–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, D.F., K.J. Fowler, E. Earle, R. Saffery, P. Kalitsis, H. Trowell, J. Hill, N.G. Wreford, D.M. de Kretser, M.R. Cancilla, et al. 1998. Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J. Cell Biol. 141:309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipling, D., and P.E. Warburton. 1997. Centromeres, CENP-B and Tigger too. Trends Genet. 13:141–145. [DOI] [PubMed] [Google Scholar]
- Kipling, D., H.E. Ackford, B.A. Taylor, and H.J. Cooke. 1991. Mouse minor satellite DNA genetically maps to the centromere and is physically linked to the proximal telomere. Genomics. 11:235–241. [DOI] [PubMed] [Google Scholar]
- Kipling, D., H.E. Wilson, A.R. Mitchell, B.A. Taylor, and H.J. Cooke. 1994. Mouse centromere mapping using oligonucleotide probes that detect variants of the minor satellite. Chromosoma. 103:46–55. [DOI] [PubMed] [Google Scholar]
- Kuznetsova, I.S., A.N. Prusov, N.I. Enukashvily, and O.I. Podgornaya. 2005. New types of mouse centromeric satellite DNAs. Chromosome Res. 13:9–25. [DOI] [PubMed] [Google Scholar]
- Lehnertz, B., Y. Ueda, A.A. Derijck, U. Braunschweig, L. Perez-Burgos, S. Kubicek, T. Chen, E. Li, T. Jenuwein, and A.H. Peters. 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13:1192–1200. [DOI] [PubMed] [Google Scholar]
- Liebman, S.W., L.S. Symington, and T.D. Petes. 1988. Mitotic recombination within the centromere of a yeast chromosome. Science. 241:1074–1077. [DOI] [PubMed] [Google Scholar]
- Maison, C., and G. Almouzni. 2004. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 5:296–304. [DOI] [PubMed] [Google Scholar]
- Mitchell, A.R., P. Jeppesen, L. Nicol, H. Morrison, and D. Kipling. 1996. Epigenetic control of mammalian centromere protein binding: does DNA methylation have a role? J. Cell Sci. 109:2199–2206. [DOI] [PubMed] [Google Scholar]
- Muntoni, A., and R.R. Reddel. 2005. The first molecular details of ALT in human tumor cells. Hum. Mol. Genet. 14:R191–R196. [DOI] [PubMed] [Google Scholar]
- Okada, T., J. Ohzeki, M. Nakano, K. Yoda, W.R. Brinkley, V. Larionov, and H. Masumoto. 2007. CENP-B controls centromere formation depending on the chromatin context. Cell. 131:1287–1300. [DOI] [PubMed] [Google Scholar]
- Okano, M., S. Xie, and E. Li. 1998. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 19:219–220. [DOI] [PubMed] [Google Scholar]
- Okano, M., D.W. Bell, D.A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 99:247–257. [DOI] [PubMed] [Google Scholar]
- Perez-Castro, A.V., F.L. Shamanski, J.J. Meneses, T.L. Lovato, K.G. Vogel, R.K. Moyzis, and R. Pedersen. 1998. Centromeric protein B null mice are viable with no apparent abnormalities. Dev. Biol. 201:135–143. [DOI] [PubMed] [Google Scholar]
- Perry, P., and S. Wolf. 1974. New Giemsa method for the differential staining of sister chromatids. Nature. 251:156–158. [DOI] [PubMed] [Google Scholar]
- Pidoux, A.L., and R.C. Allshire. 2000. Centromeres: getting a grip of chromosomes. Curr. Opin. Cell Biol. 12:308–319. [DOI] [PubMed] [Google Scholar]
- Rief, N., and M. Löbrich. 2002. Efficient rejoining of radiation-induced DNA double-strand breaks in centromeric DNA of human cells. J. Biol. Chem. 277:20572–20582. [DOI] [PubMed] [Google Scholar]
- Samper, E., F.A. Goytisolo, P. Slijepcevic, P.P. Van Buul, and M.A. Blasco. 2000. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 1:244–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, W.H., A.S. Balajee, J. Wang, H. Wu, C. Eng, P.P. Pandolfi, and Y. Yin. 2007. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 128:157–170. [DOI] [PubMed] [Google Scholar]
- Sunkel, C.E., and P.A. Coelho. 1995. The elusive centromere: sequence divergence and functional conservation. Curr. Opin. Genet. Dev. 5:756–767. [DOI] [PubMed] [Google Scholar]
- Tanaka, Y., H. Kurumizaka, and S. Yokoyama. 2005. CpG methylation of the CENP-B box reduces human CENP-B binding. FEBS J. 272:282–289. [DOI] [PubMed] [Google Scholar]
- Warburton, P.E., and H.F. Willard. 1992. PCR amplification of tandemly repeated DNA: analysis of intra- and interchromosomal sequence variation and homologous unequal crossing-over in human alpha satellite DNA. Nucleic Acids Res. 20:6033–6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton, P.E., J.S. Waye, and H.F. Willard. 1993. Nonrandom localization of recombination events in human alpha satellite repeat unit variants: implications for higher-order structural characteristics within centromeric heterochromatin. Mol. Cell. Biol. 13:6520–6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, A.K., and J.B. Rattner. 1988. Sequence organization and cytological localization of the minor satellite of mouse. Nucleic Acids Res. 16:11645–11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.