Abstract
The processing of Arabidopsis thaliana microRNAs (miRNAs) from longer primary transcripts (pri-miRNAs) requires the activity of several proteins, including DICER-LIKE1 (DCL1), the double-stranded RNA-binding protein HYPONASTIC LEAVES1 (HYL1), and the zinc finger protein SERRATE (SE). It has been noted before that the morphological appearance of weak se mutants is reminiscent of plants with mutations in ABH1/CBP80 and CBP20, which encode the two subunits of the nuclear cap-binding complex. We report that, like SE, the cap-binding complex is necessary for proper processing of pri-miRNAs. Inactivation of either ABH1/CBP80 or CBP20 results in decreased levels of mature miRNAs accompanied by apparent stabilization of pri-miRNAs. Whole-genome tiling array analyses reveal that se, abh1/cbp80, and cbp20 mutants also share similar splicing defects, leading to the accumulation of many partially spliced transcripts. This is unlikely to be an indirect consequence of improper miRNA processing or other mRNA turnover pathways, because introns retained in se, abh1/cbp80, and cbp20 mutants are not affected by mutations in other genes required for miRNA processing or for nonsense-mediated mRNA decay. Taken together, our results uncover dual roles in splicing and miRNA processing that distinguish SE and the cap-binding complex from specialized miRNA processing factors such as DCL1 and HYL1.
Posttranscriptional gene regulation by microRNAs (miRNAs) is essential for the development and function of multicellular eukaryotes (1). In plants, miRNAs are processed from primary transcripts that contain partially complementary foldbacks of variable lengths (pri-miRNAs). Pri-miRNAs are similar to messenger RNAs (mRNAs) in being transcribed by DNA-dependent RNA polymerase II (pol II) and carrying a seven-methyl guanosine (m7G) cap at the 5′ end and a polyadenosine (polyA) tail at the 3′ end (1). Pri-miRNAs are processed to yield 20- to 22-nt-long mature miRNAs by an RNAseIII-like domain containing protein called DICER-LIKE1 (DCL1) (2–4). DCL1 interacts with the double-stranded (ds)RNA-binding protein HYPONASTIC LEAVES1 (HYL1) and the zinc finger protein SERRATE (SE) to ensure proper processing of pri-miRNAs (5–13). In common with dcl1 mutants, pri-miRNA levels are increased in plants that lack HYL1 or SE activity, whereas the amounts of mature miRNAs are reduced (5–9). All three proteins are found in nuclear processing centers, called D-bodies or SmD3/SmB nuclear bodies (12, 13).
Mutants deficient in miRNA biogenesis suffer from a large range of morphological defects. Plants with null alleles of DCL1 or SE die as embryos, and even moderate reduction of DCL1 activity leads to a broad spectrum of developmental abnormalities (8, 14, 15). The weak se-1 allele causes only mild defects, including an alteration of phyllotaxis and the name-sake serrated leaves (16, 17). It has been noted before that the se-1 phenotype is reminiscent of that of another mutant with impaired RNA metabolism, ABA hypersensitive 1 (abh1). Both mutants respond more strongly to the hormone abscisic acid (ABA), both have serrated leaves, and the late-flowering phenotype caused by an active allele of FRIGIDA (FRI) is also suppressed in both (18, 19). They differ, however, in other phenotypes, such as the rate of leaf production, indicating their functions only partially overlap.
ABH1/CBP80 encodes the large subunit of the nuclear cap-binding complex (CBC) (18). The CBC, which was originally identified through its role in pre-mRNA splicing in human cells, consists of two subunits, CBP20 and CBP80, that together bind to m7G-cap structures of mRNAs (20). Further analyses revealed crucial roles of the CBC in many different aspects of mRNA metabolism in human and yeast (21, 22). For example, the CBC inhibits deadenylation by interacting with a polyA-specific ribonuclease, it associates with components of the mRNA export machinery, and it regulates export of U snRNAs (23–25). The binding of the CBC to mRNAs during the pioneer round of translation is essential for mRNA quality control (26, 27). Similarly, the interaction with the CBC is important for the activity of UPF1, a well known RNA helicase with a central role in nonsense-mediated mRNA decay (NMD) (28). In yeast, the CBC associates with U1 snRNP, a component of the pre-mRNA splicing commitment complex, to ensure proper mRNA maturation (29–32).
The A. thaliana genome contains single genes for both the large and small subunits, ABH1/CBP80 and CBP20. As the human and yeast proteins, the heterodimeric CBP80/CBP20 complex binds to m7G-cap structures and is mainly localized in the nucleus (18, 33). Consistent with closely related activities, inactivation of CBP20 causes a similar serrated leaf phenotype and increased drought resistance as seen in abh1/cbp80 mutants (18, 34).
Although the effects of ABH1/CBP80 and CBP20 on plant morphology and physiology have been documented in quite some detail (18, 19, 34), it is less clear how these relate to the known biochemical functions of the CBC. The suppression of late flowering caused by an active FRI allele has been shown to be due to reduced expression of FLOWERING LOCUS C (FLC), a downstream target of FRI (19). Many factors implicated in splicing and other aspects of RNA metabolism regulate FLC expression, but whether any of them directly regulate splicing of the FLC sense transcript is still an open question (35). Alternative splicing of the FLC pre-mRNA has been observed, but the functional significance is uncertain (36, 37). Nevertheless, abh1/cbp80 mutations change the complex splicing pattern at FLC, and variants in which intron 1 is retained overaccumulate in abh1/cbp80 mutants. In addition, ABH1/CBP80 affects a natural antisense RNA at the CONSTANS locus (37).
Here, we show that, similar to SE, the A. thaliana CBC is important for proper pri-miRNA processing in Arabidopsis. Conversely, whole-genome tiling array analyses revealed that the CBC and SE have overlapping functions in pre-mRNA splicing, but that this role is not shared by other factors required for miRNA maturation. We propose that, in analogy to splicing, the CBC may provide a platform for recruitment of miRNA maturation factors, and that SE plays an important role for CBC function in both splicing and miRNA processing.
Results and Discussion
Requirement of CBC for miRNA Biogenesis.
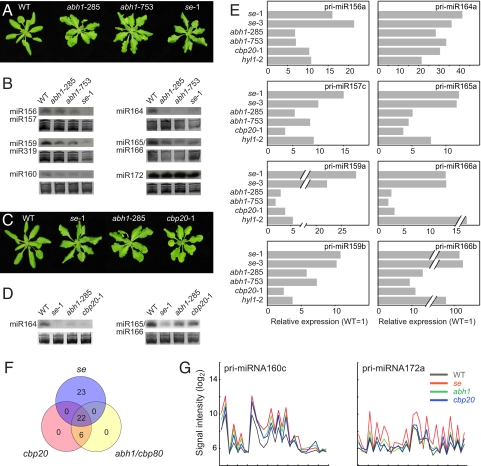
Previous studies reported similar effects of abh1/cbp80 and se-1 mutations on flowering time, leaf development, and seedling responses to the hormone ABA (17–19). We isolated two additional abh1/cbp80 T-DNA insertion alleles (abh1-285 and abh1-753). These closely resembled se-1 mutants in vegetative morphology and exhibit the same leaf serration phenotype as reported for other abh1/cbp80 alleles (Fig. 1A).
Fig. 1.
Requirement of the CBC for pri-miRNA processing. (A) Comparative gross morphology of 21-day-old wild-type (WT), abh1-285, abh1-753, and se-1 plants. (B) Detection of selected miRNAs by RNA blot in 10-day-old wild-type, abh1-285, abh1-753, and se-1 plants. Ethidium-stained gels on which tRNA and rRNA bands are visible are shown as loading control. (C) Comparative gross morphology of 21-day-old wild-type, se-1, abh1-285, and cbp20 plants. (D) Detection of selected miRNAs in 10-day-old wild-type, se-1, abh1-285, and cbp20 plants. (E) Real-time RT-PCR analysis of selected pri-miRNAs in wild-type, se-1, se-3, abh1-285, abh1-753, cbp20, and hyl1 plants. (F) Overlap of pri-miRNAs that accumulate to higher levels in abh1/cbp80-285, cbp20 and se-1 mutants. (G) As two examples for pri-miRNAs that accumulate in mutants, hybridization intensities on tiling arrays are shown for pri-miR160c and pri-miR172a. Values are averaged from three biological replicates each. Accumulation in se-1 is apparent for both pri-miRNA transcripts, but only pri-miR160c appears to be affected also by abh1 and cbp20 mutations. Tick marks indicate 100 bases each.
Because both SE and ABH1/CBP80 have pleiotropic phenotypic effects, their similar mutant phenotypes might be simply due to cross-regulation of SE mRNA accumulation by ABH1/CBP80. RNA blot analysis did not reveal obvious changes in SE mRNA levels or splicing pattern in plants with mutations in either ABH1/CBP80 or in CBP20, which encodes the other CBC subunit. Similarly, ABH1/CBP80 and CBP20 mRNAs were unaffected in se-1 mutants [supporting information (SI) Fig. S1].
The recent discovery that SE is required for miRNA processing (7–9) prompted us to test whether ABH1 affects the accumulation of mature miRNAs as well. Small RNA blots showed that levels of a subset of miRNAs examined were reduced in two different abh1/cbp80 alleles, although the effects were not always as severe as those of se-1 (Fig. 1B).
Because ABH1/CBP80 acts in concert with CBP20, we also included cbp20 mutants in our analysis. cbp20 and abh1/cbp80 mutants are known to have similar defects in morphology and ABA response (34) (Fig. 1C). Consistent with ABH1/CBP80 and CBP20 acting in the same complex, we found RNA expression levels of the encoding genes to be highly correlated, as deduced from the AtGenExpress developmental expression atlas (38) (Fig. S2). A very similar expression profile was seen for SE as well, supporting the proposal of related biochemical roles. The effects of a cbp20 mutant allele on miRNA levels were also very similar to those of abh1/cbp80 (Fig. 1 C and D). Taken together, these results indicate that the CBC is necessary for the accumulation of at least a subset of mature miRNA species.
Requirement of CBC for Pri-miRNA Processing.
Reduced miRNA levels can be due to diminished pri-miRNA levels or to less-efficient processing of the pri-miRNA transcripts into miRNAs. To distinguish between these possibilities, we analyzed the steady-state levels of pri-miRNAs by quantitative real-time PCR (qRT). In abh1/cbp80 and cbp20 mutants, pri-miRNA transcripts accumulated to higher levels than in wild type, indicating that pri-miRNAs are less effectively converted into mature miRNAs, similar to what is seen in known miRNA processing mutants, such as hyl1 (Fig. 1E) (5, 6, 10).
Interestingly, not all miRNAs were equally affected by inactivation of ABH1/CBP80 or CBP20. For example, miR156/157 and miR172 seemed to be largely insensitive to loss of CBC activity. This is in agreement with the observation that certain pri-miRNA transcripts accumulated to higher levels in se mutants than in abh1/cbp80 or cbp20 mutants (Fig. 1E). In this context, it is worth noting that, similar to the corresponding yeast mutants (29, 32), abh1/cbp80 and cbp20 null mutants are impaired in development and physiology but still viable, at least under the benign environment of the laboratory. In contrast, null mutations in DCL1 and SE cause embryonic lethality (8, 14, 15). Therefore, we conclude that the CBC is not an essential part of the miRNA processing machinery, but that it has more of a supporting role. Perhaps CBC function is not required for pri-miRNAs that are particularly strongly expressed or other proteins with cap-binding activity act redundantly with the CBC. In this respect, the CBC appears similar to HYL1, null alleles of which are viable and which also have a weaker effect on miRNA processing (Fig. 1E) (5, 6).
To obtain a global view of pri-miRNAs, we analyzed mRNA populations of se, abh1/cbp80, and cbp20 mutants with whole-genome tiling arrays. The Affymetrix Tiling 1.0R array represents a single strand of the A. thaliana genome, with the 25-mer probes spaced on average 10 nt apart (39). In se mutants, 45 of 162 analyzed pri-miRNAs showed increased hybridization signals compared with wild type (false discovery rate <2.5%). Consistent with the qRT-PCR data, a smaller number, 28, was significantly affected in abh1/cbp80 and cbp20 mutants, with 22 common to all three mutants (Fig. 1 F and G; Table S1). These findings support the conclusion that SE has a more prominent role in pri-miRNA processing than the CBC.
Accumulation of Unspliced Transcripts in abh1/cbp80, cbp20, and se Mutants.
In humans and yeast, the CBC is involved in several aspects of RNA processing and quality control, such as pre-mRNA splicing and NMD (22). It has been suggested that the A. thaliana CBC affects splicing of FLC mRNA (37), but its broader effects on splicing, if any, have been unknown. To address this question, we analyzed intron accumulation using the above-mentioned tiling arrays, which cover 80,190 introns with at least one probe. A. thaliana introns have a lower GC content than exons, and intronic probes therefore often have comparatively poor hybridization properties. Hence, for high-confidence predictions we analyzed only the 30,615 introns covered by at least three probes. These introns are >100 nt in length, and they represent 11,882 genes.
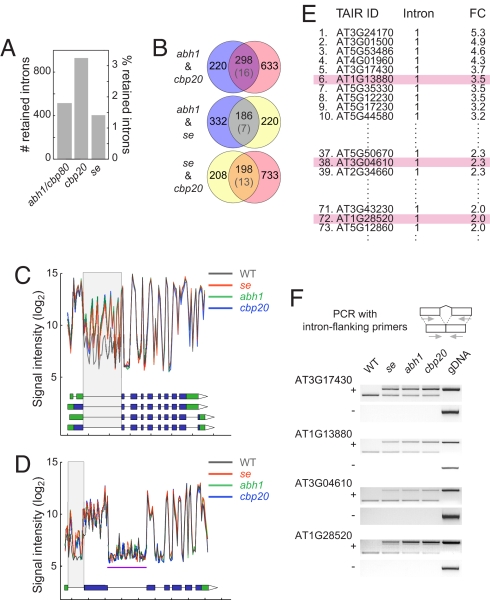
We identified 518 and 931 introns that gave substantially higher hybridization signals in abh1/cbp80 and cbp20 mutants, respectively, than in wild type (Table S2). Of these, a large fraction, 298, was identified in both mutants (Fig. 2A), with the overlap being much more than expected by chance (P < 10−10, χ2 test with Yate's continuity correction). The increased hybridization signals for introns are most easily interpreted as reflecting intron retention, consistent with the reduced splicing efficiency caused by loss of human or yeast CBC activity (20, 21).
Fig. 2.
Splicing defects in abh1/cbp80-285, cbp20 and se-1 mutants. (A) Number of retained introns in mutants. (B) Overlap of retained introns among the three mutants examined. Parentheses give overlap expected by chance. (C) As one example for an improperly spliced mRNA detected by tiling arrays, hybridization signals of probes representing the AT3G01500 gene are shown for wild-type, abh1/cbp80, cbp20 and se plants. Values are averaged from three biological replicates each. Gray box highlights retained intron. Annotated splice forms (TAIR 7) are shown below, with blue indicating coding sequence. Tick marks indicate 500 bases each. (D) As a second example, tiling array signals for AT3G04670 are shown. Note second intron (purple underline) that is unchanged in the mutants. (E) Excerpt from table of genes with introns retained in abh1/cbp80, cbp20, and se-1 mutants (see Table S2). The second column (“Intron”) indicates intron position from 5′ end. As discussed in the text, first introns are overrepresented. FC, fold change. Details for highlighted genes are shown in F. (F) Validation of intron retention in selected genes by conventional RT-PCR analysis. gDNA, genomic DNA control. “+” and “−” indicates reactions with and without reverse transcriptase. The upper band corresponds to unspliced form (as in gDNA), the lower band to spliced form.
We found 406 introns with elevated hybridization signals in se-1 mutants, which included 244 introns also affected in abh1/cbp80 or cbp20 mutants (Fig. 2 B–D). Again, this overlap is significantly higher than expected by chance (P < 10−10). In total, 140 introns were affected in all three mutants. These results imply that the CBC and SE have overlapping functions not only in pri-miRNA processing but also in pre-mRNA splicing. We generally detected only a single intron from each gene as being retained in the mutants (Fig. 2 C and D). In addition, first introns seemed to be most sensitive to loss of CBC and SE activity (Fig. S3), possibly suggesting that efficient splicing of introns close to the m7G cap have a stronger requirement for CBC and SE function than introns more downstream.
To confirm that a subset of introns is inefficiently spliced in se-1, abh1/cbp80, and cbp20 mutants, we analyzed several mRNAs in more detail, including ATG17430, which encodes a potential phosphate translocator; AT1G13880, which encodes an ELM2 domain-containing protein; and AT3G04610, which encodes FLK, a likely RNA-binding protein known to affect flowering time (40, 41). The mean changes in the hybridization signal for the affected introns ranged from 2- to >5-fold (Fig. 2E). RT-PCR analysis with intron-flanking primers revealed that the respective introns are quantitatively removed by splicing in wild type (Fig. 2F). In contrast, in se-1, abh1/cbp80, and cbp20 mutants, we easily detected RT-PCR products corresponding in size to unspliced transcripts (Fig. 2E). Because se-1 is a weak allele (7, 8, 17), we repeated the RT-PCR analysis with the se-3 allele, which carries a T-DNA insertion in the first exon and has much stronger developmental defects than se-1 (7). The results were similar as for the se-1 allele (Fig. S4). Taken together, these results demonstrate that SE and CBC have overlapping functions in pre-mRNA splicing.
No General Requirement of miRNAs or the NMD Pathway for Pre-mRNA Splicing.
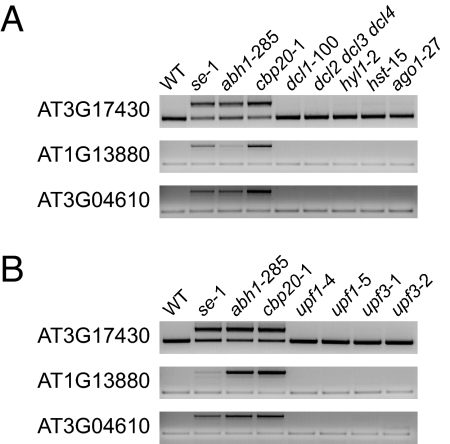
We have demonstrated that the CBC components ABH1/CBP80 and CBP20 and the zinc finger protein SE are required for both pri-miRNA processing and pre-mRNA splicing. Because SE had not previously been linked to mRNA processing, we considered the possibility that the splicing defects in se mutants are indirect effects, caused by lack of an miRNA that targets a gene involved in splicing. To test this hypothesis, we analyzed splicing patterns of several mRNAs that feature retained introns in se, abh1/cbp80, and cbp20 mutants in strains with mutations in other genes required for miRNA function. Mutants impaired in miRNA processing, such as dcl1 (2) or hyl1 (5), did not show any splicing defects for several mRNAs affected in abh1/cbp80, cbp20, or se mutants (Fig. 3A). Similarly, splicing appeared unaffected by loss of HASTY (HST), which is required for normal miRNA accumulation, potentially through effects on miRNA export from the nucleus (42) or ARGONAUTE1 (AGO1), the protein that slices miRNA target mRNAs (43, 44) (Fig. 3A). These observations suggest that the splicing defects observed in plants lacking the CBC or SE are not an indirect effect of impaired miRNA processing. Taken together, our results indicate that the CBC and SE have a function in mRNA splicing that is distinct from that of other miRNA processing factors.
Fig. 3.
Analysis of splicing in other miRNA-processing mutants and in NMD mutants. (A) Comparative RT-PCR analysis of mRNAs with introns retained in abh1/cbp80, cbp20 and se mutants in other miRNA processing mutants and related genotypes (dcl1, dcl2 dcl3 dcl4, hyl1, hst and ago1). (B) Comparative RT-PCR analysis of NMD mutants (upf1 and upf3).
mRNAs with premature stop codons, because of either DNA mutations or missplicing, are subject to nonsense-mediated mRNA decay (NMD) (45). The human CBC has been shown to be important for NMD through interaction of CBP80 with UPF1, an RNA helicase that associates with two components of the exon–junction complex, UPF2 and UPF3 (28, 45). Homologs of UPF1 and UPF3 are essential for NMD in A. thaliana as well (46–50). Because mRNAs with unspliced introns in cbp and se mutants also feature premature stop codons, we considered the possibility that the intron retention observed in these mutants might be an indirect consequence of NMD failure. To test this hypothesis, we performed RT-PCR analysis on RNA extracted from upf1 and upf3 mutants. In contrast to cbp and se mutants, we did not detect unspliced mRNAs in plants carrying two different mutant alleles each of upf1 and upf3 (Fig. 3B). We conclude that the CBC and SE in A. thaliana primarily promote splicing rather than degrade misspliced mRNAs.
Intron Dependence of Pri-miRNA Processing by the CBC and SE.
Several A. thaliana miRNA loci have been shown to produce a variety of pri-miRNA transcripts, some of them containing introns (11, 51–54). It seems unlikely, however, that only intron-containing pri-miRNA transcripts are affected, because most of the pri-miRNA transcripts we examined in detail (Fig. 1E) are not known to be spliced. We nevertheless wanted to determine whether the presence of an intron had a particularly strong effect on precursor processing in plants lacking the CBC or SE, because we had noticed before that miRNAs differ in their requirement for CBC and SE function (Fig. 1 B and D). In such a scenario, the observed changes in miRNA levels in cbp and se mutants would be at least partially an indirect consequence of pri-miRNA splicing defects.
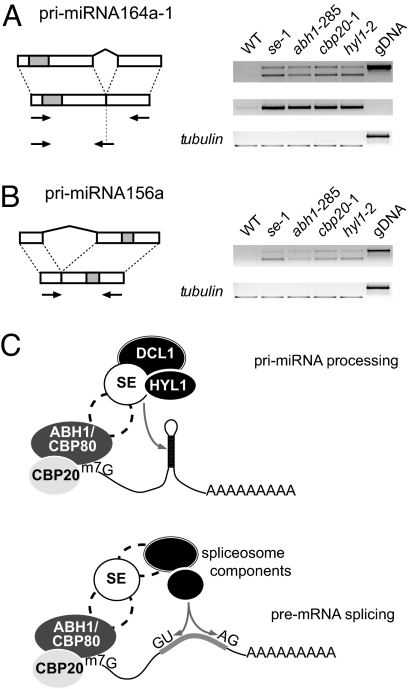
We examined this hypothesis in more detail using as a model miR164, the levels of which are strongly decreased in abh1/cbp80, cbp20 and se mutants (Fig. 1 B and D). One of the loci from which miR164 is produced is MIR164a, which gives rise to several pri-miRNAs, including pri-miRNA164a-1, from which an intron is removed by splicing (53). RT-PCR analysis revealed that cbp and se-1 mutants accumulate both the spliced and unspliced version of pri-miRNA164a-1, and that their changes in levels of both pri-miRNA164a-1 isoforms are very similar to those seen in the pri-miRNA processing mutant hyl1. Use of an oligonucleotide primer that spanned the exon–exon junction confirmed high levels of the correctly spliced form of pri-miRNA164a-1 in all four mutants (Fig. 4A). Similar results were obtained for another spliced pri-miRNA, pri-miRNA156a (Fig. 4B) (52). Thus, compared with other miRNA processing factors, there is no evidence for a particular requirement of the CBC and SE in processing of spliced pri-miRNAs and hence no evidence that the CBC and SE affect miRNA accumulation indirectly because of their splicing defects. We therefore conclude that the effects on pre-mRNA splicing and miRNA processing likely reflect independent roles of the CBC and SE in these two processes.
Fig. 4.
Accumulation of intron-containing pri-miRNAs. (A) Diagram of the pri-miRNA164a-1 transcript (53), with miRNA foldback indicated in gray, with intron-flanking and exon–exon-junction spanning oligonucleotide primers shown as arrows. RT-PCR results are shown on the right. (B) Diagram of the pri-miRNA156a transcript (52) and RT-PCR analysis. (C) Illustration comparing potential roles of CBC and SE in pri-miRNA processing and pre-mRNA splicing. SE might be part of a larger cap-binding complex (indicated by an additional component with a dashed outline) or might interact only transiently with the CPC.
Conclusions
Our results imply there are at least two different functions of the CBC and SE, in pre-mRNA splicing and pri-miRNA processing, and that the CBC and SE cooperate in both. The splicing defects set cbp and se mutants apart from mutants impaired in miRNA biogenesis or NMD. Pre-mRNA splicing, however, is not abolished in abh1/cbp80 or cbp20 mutants (Fig. 2F), consistent with these genes not being essential for viability. In addition, we generally detected reduced splicing efficiency only for individual introns of a given transcript, indicating that certain introns are particularly sensitive to loss of CBC or SE function (Fig. 2 B and C).
In other organisms, the CBC has been shown to affect splicing by facilitating the cotranscriptional assembly of the spliceosome (55). One interpretation of the differential effects of the CBC on splicing might be that the affected introns preferentially undergo cotranscriptional splicing, whereas other introns might be processed posttranscriptionally and therefore might be less sensitive to loss of the CBC. Alternatively, the CBC might act redundantly with other cap-binding proteins, such as eukaryotic translation initiation factor 4G (eIF4G), which is structurally related to CBC (56). Interestingly, the subnuclear localization pattern of SE partially overlaps with that of SR45, a plant-specific serine-arginine-rich (SR) protein required for splicing (12, 57).
We cannot entirely disregard the possibility that the CBC and SE indirectly affect miRNA processing through splicing of transcripts encoding miRNA processing factors. However, the tiling array data did not indicate that introns of the mRNAs encoding the two known main players, DCL1 and HYL1, are affected (Fig. S5). Furthermore, it has been previously established that SE directly interacts with DCL1 and HYL1. Using guilt by association as an argument, we suggest that, like SE, the CBC also has a rather direct role in miRNA processing.
Our analysis of intron-containing pri-miRNAs implies that the miRNA-processing function of CBC and SE does not rely on their role in pre-mRNA splicing. But what is the missing link between pre-mRNA splicing and pri-miRNA processing that requires both CBC and SE? Perhaps CBC facilitates the loading of the miRNA-processing machinery onto pri-miRNA, in analogy with its role in recruiting the splicing commitment complex onto pre-mRNAs. SE might then be a common mediator for interactions between the CBC and different processing complexes (Fig. 4C). Because standard approaches have not yet revealed any direct in vitro interaction between CBP80 or CBP20 and SE (S.L. and D.W., unpublished data), one direction for future work is the analysis of larger in vivo complexes in which the CBC and SE participate.
Materials and Methods
Plant Material and Growth Conditions.
All mutants were in the Columbia-0 (Col-0) background. abh1-285 (SALK_024285) and abh1-753 (SALK_016753) are from the SALK and dcl1-100 from the GABI-KAT T-DNA collections (58, 59). The other mutants have been described (6, 7, 17, 34, 46, 48, 60). Plants were grown in long days (16-h light/8-h dark) or continuous light at 23°C on soil or solid half-strength MS medium supplemented with 1% sucrose.
Tiling Array Analyses.
Targets (hybridization probes) for tiling array hybridization were generated from 1 μg of total RNA using the MessageAmp II-Biotin Enhanced Kit (Ambion) using unmodified NTPs instead of biotinylated NTPs, the GeneChip WT Double-Stranded cDNA Synthesis Kit, and the GeneChip WT Double-Stranded DNA Terminal Labeling Kit (Affymetrix). Targets were hybridized to Affymetrix Arabidopsis Tiling 1.0R arrays in triplicate. For washing on the Affymetrix Fluidics Station 450, the Affymetrix protocol FS450_0001 was used; for scanning, a GeneChip Scanner 3000 7G was used.
Raw data were quantile-normalized (61), and probes likely to be prone to cross-hybridization properties were removed from further analysis based on described parameters (62). For analysis of pri-miRNA expression, tiling array probes were mapped to the genome coordinates from 162 annotated miRNA foldbacks, including 200 additional base-pairs up- and downstream of the foldback (63). We applied the SAM algorithm to median expression levels from wild-type and mutants across the three replicates (64).
For analysis of intron retention, probes were mapped to known constitutive introns (TAIR7 annotation). Only genes with median exon intensities above the lower quartile in all three biological replicates and across all four genotypes were considered. Average fold changes were calculated between all mutants compared with wild-type and across three replicates. Subsequent analysis focused on introns showing a change of log2 ≥0.7 (≈1.6-fold or greater). Table S2 lists all introns (including corresponding TAIR-IDs and fold changes) that satisfy this criterion in se-1 and at least one of the two cbp mutants.
RNA and RT-PCR Analyses.
Please see SI Text. Oligonucleotide primers are listed in Table S3.
Supplementary Material
Acknowledgments.
We are grateful to E. Izaurralde, S. Henz, and the miRNA group of the Weigel laboratory for suggestions and critical reading of the manuscript. We thank the European Arabidopsis Stock Center (Nottingham, U.K.), J. Carrington (Oregon State University, Corvallis, OR), B. Davies (University of Leeds, Leeds, U.K.), I. Papp (Agricultural Biotechnology Center, Gödöllö, Hungary), S. Poethig (University of Pennsylvania, Philadelphia), M. Tsiantis (University of Oxford, Oxford, U.K.), and H. Vaucheret (INRA, Versailles, France) for mutant seeds. Our work on small RNAs is supported by European Community Grant FP6 IP SIROCCO (contract LSHG-CT-2006-037900). The development of tools for tiling array analysis is supported by European Community Grant FP6 IP AGRON-OMICS (contract LSHG-CT-2006-037704).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information–Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE11043).
See Commentary on page 8489.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802493105/DCSupplemental.
References
- 1.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- 2.Park W, et al. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finnegan EJ, Margis R, Waterhouse PM. Posttranscriptional gene silencing is not compromised in the Arabidopsis CARPEL FACTORY (DICER-LIKE1) mutant, a homolog of Dicer-1 from Drosophila. Curr Biol. 2003;13:236–240. doi: 10.1016/s0960-9822(03)00010-1. [DOI] [PubMed] [Google Scholar]
- 4.Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA. 2004;101:12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han MH, Goud S, Song L, Fedoroff N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc Natl Acad Sci USA. 2004;101:1093–1098. doi: 10.1073/pnas.0307969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vázquez F, Gasciolli V, Crété P, Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol. 2004;14:346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 7.Grigg SP, Canales C, Hay A, Tsiantis M. SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature. 2005;437:1022–1026. doi: 10.1038/nature04052. [DOI] [PubMed] [Google Scholar]
- 8.Lobbes D, et al. SERRATE: A new player on the plant microRNA scene. EMBO Rep. 2006;7:1052–1058. doi: 10.1038/sj.embor.7400806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang L, et al. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 2006;47:841–850. doi: 10.1111/j.1365-313X.2006.02835.x. [DOI] [PubMed] [Google Scholar]
- 10.Kurihara Y, Takashi Y, Watanabe Y. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA. 2006;12:206–212. doi: 10.1261/rna.2146906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song L, Han MH, Lesicka J, Fedoroff N. Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc Natl Acad Sci USA. 2007;104:5437–5442. doi: 10.1073/pnas.0701061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang Y, Spector DL. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol. 2007;17:818–823. doi: 10.1016/j.cub.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujioka Y, Utsumi M, Ohba Y, Watanabe Y. Location of a possible miRNA processing site in SmD3/SmB nuclear bodies in Arabidopsis. Plant Cell Physiol. 2007;48:1243–1253. doi: 10.1093/pcp/pcm099. [DOI] [PubMed] [Google Scholar]
- 14.Schwartz BW, Yeung EC, Meinke DW. Disruption of morphogenesis and transformation of the suspensor in abnormal suspensor mutants of Arabidopsis. Development. 1994;120:3235–3245. doi: 10.1242/dev.120.11.3235. [DOI] [PubMed] [Google Scholar]
- 15.Schauer SE, Jacobsen SE, Meinke DW, Ray A. DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plants Sci. 2002;7:487–491. doi: 10.1016/s1360-1385(02)02355-5. [DOI] [PubMed] [Google Scholar]
- 16.Clarke JH, et al. The SERRATE locus controls the formation of the early juvenile leaves and phase length in Arabidopsis. Plant J. 1999;20:493–501. doi: 10.1046/j.1365-313x.1999.00623.x. [DOI] [PubMed] [Google Scholar]
- 17.Prigge MJ, Wagner DR. The Arabidopsis SERRATE gene encodes a zinc-finger protein required for normal shoot development. Plant Cell. 2001;13:1263–1279. doi: 10.1105/tpc.13.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hugouvieux V, Kwak JM, Schroeder JI. An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell. 2001;106:477–487. doi: 10.1016/s0092-8674(01)00460-3. [DOI] [PubMed] [Google Scholar]
- 19.Bezerra IC, Michaels SD, Schomburg FM, Amasino RM. Lesions in the mRNA cap-binding gene ABA HYPERSENSITIVE 1 suppress FRIGIDA-mediated delayed flowering in Arabidopsis. Plant J. 2004;40:112–119. doi: 10.1111/j.1365-313X.2004.02194.x. [DOI] [PubMed] [Google Scholar]
- 20.Izaurralde E, et al. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 21.Lewis JD, Görlich D, Mattaj IW. A yeast cap binding protein complex (yCBC) acts at an early step in pre-mRNA splicing. Nucleic Acids Res. 1996;24:3332–3336. doi: 10.1093/nar/24.17.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis JD, Izaurralde E. The role of the cap structure in RNA processing and nuclear export. Eur J Biochem. 1997;247:461–469. doi: 10.1111/j.1432-1033.1997.00461.x. [DOI] [PubMed] [Google Scholar]
- 23.Izaurralde E, et al. A cap-binding protein complex mediating U snRNA export. Nature. 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- 24.Cheng H, et al. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 25.Balatsos NA, et al. Inhibition of mRNA deadenylation by the nuclear cap binding complex (CBC) J Biol Chem. 2006;281:4517–4522. doi: 10.1074/jbc.M508590200. [DOI] [PubMed] [Google Scholar]
- 26.Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 27.Chiu SY, Lejeune F, Ranganathan AC, Maquat LE. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev. 2004;18:745–754. doi: 10.1101/gad.1170204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hosoda N, Kim YK, Lejeune F, Maquat LE. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat Struct Mol Biol. 2005;12:893–901. doi: 10.1038/nsmb995. [DOI] [PubMed] [Google Scholar]
- 29.Colot HV, Stutz F, Rosbash M. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev. 1996;10:1699–1708. doi: 10.1101/gad.10.13.1699. [DOI] [PubMed] [Google Scholar]
- 30.Visa N, et al. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J Cell Biol. 1996;133:5–14. doi: 10.1083/jcb.133.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fortes P, et al. Luc7p, a novel yeast U1 snRNP protein with a role in 5′ splice site recognition. Genes Dev. 1999;13:2425–2438. doi: 10.1101/gad.13.18.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fortes P, et al. Genetic and physical interactions involving the yeast nuclear cap-binding complex. Mol Cell Biol. 1999;19:6543–6553. doi: 10.1128/mcb.19.10.6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hugouvieux V, et al. Localization, ion channel regulation, and genetic interactions during abscisic acid signaling of the nuclear mRNA cap-binding protein, ABH1. Plant Physiol. 2002;130:1276–1287. doi: 10.1104/pp.009480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papp I, et al. A mutation in the Cap Binding Protein 20 gene confers drought tolerance to Arabidopsis. Plant Mol Biol. 2004;55:679–686. doi: 10.1007/s11103-004-1680-2. [DOI] [PubMed] [Google Scholar]
- 35.Quesada V, Dean C, Simpson GG. Regulated RNA processing in the control of Arabidopsis flowering. Int J Dev Biol. 2005;49:773–780. doi: 10.1387/ijdb.051995vq. [DOI] [PubMed] [Google Scholar]
- 36.Caicedo AL, et al. Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc Natl Acad Sci USA. 2004;101:15670–15675. doi: 10.1073/pnas.0406232101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuhn JM, Breton G, Schroeder JI. mRNA metabolism of flowering-time regulators in wild-type Arabidopsis revealed by a nuclear cap binding protein mutant, abh1. Plant J. 2007;50:1049–1062. doi: 10.1111/j.1365-313X.2007.03110.x. [DOI] [PubMed] [Google Scholar]
- 38.Schmid M, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 40.Mockler TC, et al. Regulation of flowering time in Arabidopsis by K homology domain proteins. Proc Natl Acad Sci USA. 2004;101:12759–12764. doi: 10.1073/pnas.0404552101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim MH, et al. A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell. 2004;16:731–740. doi: 10.1105/tpc.019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park MY, et al. Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci USA. 2005;102:3691–3696. doi: 10.1073/pnas.0405570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qi Y, Denli AM, Hannon GJ. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell. 2005;19:421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 45.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 46.Hori K, Watanabe Y. UPF3 suppresses aberrant spliced mRNA in Arabidopsis. Plant J. 2005;43:530–540. doi: 10.1111/j.1365-313X.2005.02473.x. [DOI] [PubMed] [Google Scholar]
- 47.Kertesz S, et al. Both introns and long 3′-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res. 2006;34:6147–6157. doi: 10.1093/nar/gkl737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arciga-Reyes L, Wootton L, Kieffer M, Davies B. UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 2006;47:480–489. doi: 10.1111/j.1365-313X.2006.02802.x. [DOI] [PubMed] [Google Scholar]
- 49.Yoine M, Nishii T, Nakamura K. Arabidopsis UPF1 RNA helicase for nonsense-mediated mRNA decay is involved in seed size control and is essential for growth. Plant Cell Physiol. 2006;47:572–580. doi: 10.1093/pcp/pcj035. [DOI] [PubMed] [Google Scholar]
- 50.Yoine M, et al. The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis. Plant J. 2006;47:49–62. doi: 10.1111/j.1365-313X.2006.02771.x. [DOI] [PubMed] [Google Scholar]
- 51.Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie Z, et al. Expression of Arabidopsis MIRNA genes. Plant Physiol. 2005;138:2145–2154. doi: 10.1104/pp.105.062943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nikovics K, et al. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell. 2006;18:2929–2945. doi: 10.1105/tpc.106.045617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Warthmann N, Das S, Lanz C, Weigel D. Comparative analysis of the MIR319a microRNA locus in Arabidopsis and related Brassicaceae. Mol Biol Evol. 2008;25:892–902. doi: 10.1093/molbev/msn029. [DOI] [PubMed] [Google Scholar]
- 55.Gornemann J, Kotovic KM, Hujer K, Neugebauer KM. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 56.Marintchev A, Wagner G. eIF4G and CBP80 share a common origin and similar domain organization: implications for the structure and function of eIF4G. Biochemistry. 2005;44:12265–12272. doi: 10.1021/bi051271v. [DOI] [PubMed] [Google Scholar]
- 57.Ali GS, et al. Regulation of plant developmental processes by a novel splicing factor. PLoS ONE. 2007;2:e471. doi: 10.1371/journal.pone.0000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 59.Rosso MG, et al. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol. 2003;53:247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- 60.Morel JB, et al. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 62.Clark RM, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- 63.Backman TW, et al. Update of ASRP: The Arabidopsis small RNA project database. Nucleic Acids Res. 2008;36:D982–D985. doi: 10.1093/nar/gkm997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ghosh S, et al. Differential analysis for high density tiling microarray data. BMC Bioinformatics. 2007;8:359. doi: 10.1186/1471-2105-8-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.