Abstract
Tobacco etch virus (TEV) protease recognizes a 7-aa consensus sequence, Glu-Xaa-Xaa-Tyr-Xaa-Gln-Ser, where Xaa can be almost any amino acyl residue. Cleavage occurs between the conserved Gln and Ser residues. Because of its distinct specificity, TEV protease can be expressed in the cytoplasm without interfering with viability. Polypeptides that are not natural substrates of TEV protease are proteolyzed if they carry the appropriate cleavage site. Thus, this protease can be used to study target proteins in their natural environment in vivo, as well as in vitro. We describe two Tn5-based mini-transposons that insert TEV protease cleavage sites at random into target proteins. TnTIN introduces TEV cleavage sites into cytoplasmic proteins. TnTAP facilitates the same operation for proteins localized to the bacterial cell envelope. By using two different target proteins, SecA and TolC, we show that such modified proteins can be cleaved in vivo and in vitro by TEV protease. Possible applications of the site-specific proteolysis approach are topological studies of soluble as well as of inner and outer membrane proteins, protein inactivation, insertion mutagenesis experiments, and protein tagging.
Proteases are important tools in molecular biology. Until recently, however, they have been primarily applied to in vitro studies. To extend the use of proteases, we showed in a previous paper that the site-specific tobacco etch virus (TEV) protease can be used to selectively cleave an essential cytoplasmic protein in vivo (1).
To facilitate the generation of TEV protease recognition sites into proteins, we have constructed two mini-transposons that can insert into a gene to generate “sandwich” fusions of a short DNA fragment encoding the TEV protease cleavage site and the target gene that remains otherwise intact. These mini-transposons are derivatives of Tn5 that generate stable inserts, because Tn5 transposase is expressed in trans. To detect inserts in which the ORF of target gene and of TEV protease recognition sequence match, the transposons carry the bacterial reporter genes uidA or signal sequenceless phoA, both lacking a promoter and translation initiation signals. uidA encodes cytoplasmic β-glucuronidase (2). phoA encodes periplasmic alkaline phosphatase, which must be exported to the periplasm to be active (3). Signal sequenceless phoA is the most widely used tool for generating fusions to cytoplasmic membrane proteins for studying membrane protein topology (4). The presence of uidA and phoA fusions can be easily monitored on agar indicator plates containing specific dyes, i.e. 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-GLUC) (5) and 5-bromo-4-chloro-3-indolyl-phosphate, respectively. A related strategy was applied recently by Manoil and Bailey (6) in the construction of a set of transposons that insert 31 codons into target genes. Here, fusions to lacZ and phoA are generated to detect insertions into the reading frame of the target gene. These transposons have been shown to be useful for structure-function studies of LacY, LacI, and MalK (6–8).
TnTIN was used to introduce protease recognition sites into SecA, an essential cytoplasmic protein involved in secretion (9). One insert, located after amino acid 830, left SecA function intact. On coexpression of TEV protease, cleavage of this SecA derivative occurred in vivo. In addition, the purified SecA derivative was cleaved by TEV protease in vitro.
TnTAP was used to insert protease sites into the outer membrane protein TolC. TolC is involved in the uptake of antibiotics and colicins and in the export of the virulence factor hemolysin (10–12). TolC derivatives carrying TEV site insertions after amino acids 79, 157, and 306 were cleaved after adding TEV protease to whole cells.
MATERIALS AND METHODS
Bacteria and Plasmids.
Escherichia coli strains used are: MM52 [F− ΔlacU169 araD136 rbsR relA rpsL thi secA(Ts)] (13); ME7000 (rpsL Δ(add-uid-man) recA::cam); CC118 (araD139 Δ(ara, leu)7697 ΔlacX74 phoAΔ20 galE galK thi rpsE rpoB argEam recA1) (14); and MM118 (CC118 tolC recA::cam pcnB80 zid::Tn10) (15).
pDHB60 is a high copy number pBR322 derivative. It was derived from pDHB32 (16) by replacement of the malFG′ insert with the polylinker: GAATTCGAGCTCGCCCGGGGATCCTCTAGAGTCGACCTGCAGCCCAAGCTT. pMM70 encodes the wild-type tolC gene under its own promoter from pBR322. pMF8 expresses wild-type secA under its own promoter from pBR322 (17).
Construction of TnTIN.
Oligonucleotides containing a BamHI site at the 5′ end, the coding sequence for the TEV protease recognition sequence, internal NotI, NcoI, EcoRI, SalI, and NotI restriction sites, and a HindIII site at the 3′ end were annealed and filled in by using Klenow enzyme. Oligonucleotides used were: CGGATCCTGACTCTTATACACAAGTTTGAAAACCTGTACTTCCAGTCAGCGGCC- GCCATGGAATTGAATTCGTC and CCCAAGCTTCTGACTCTTATACACAAGTATTGCGGCCGCGTCGAC- GAATTCAATTCCATGGCG. Subsequently, the double-stranded DNA was cut with BamHI and HindIII and then ligated into pDHB60 cut with BamHI and HindIII to yield pPB201. The sequence of the insert was verified by nucleotide sequencing. The E. coli uidA gene was moved as a 1.9-kb NcoI–EcoRI fragment from pGUS N358−>S (CLONTECH) into pPB201 that was cut with NcoI and EcoRI, yielding pPB202. Finally, the neo gene conferring kanamycin resistance was cloned as a 1.2-kb SalI fragment from pUC4K (from Pharmacia) into pPB202 cut with SalI, yielding pPB203.
Transposition of TnTIN into a pACYC184 Derivative Containing Tn5 Transposase.
To clone Tn5 transposase into pACYC184 (18), pRZ7016MA.EK.LP (19) was cut with EcoRI and NcoI. The transposase-containing fragment was cloned into pACYC184 cut with EcoRI and NcoI, yielding pPB101. To allow transposition of TnTIN into pPB101, pPB101 and pPB203 were transformed into strain ME7000. During growth overnight, TnTIN transposed into pPB101. Plasmid DNA was prepared and digested with DraIII and NdeI. These restriction enzymes cleave pPB203, but not pPB101 or TnTIN. This restriction digest was transformed into strain ME7000. Selection for Tetr (from pPB101) and Kanr (from TnTIN) yielded strains containing plasmids where TnTIN transposed into pPB101. This event was verified by restriction mapping. One candidate was chosen and termed pPB301.
Construction of TnTAP.
To construct TnTAP, a signal sequenceless phoA gene lacking its stop codon was cloned from pSWFII (20) as a 1.2-kb BamHI–BstEII-digested fragment filled in with Klenow, into pPB203, which was cut with NcoI and EcoRI and then filled with Klenow, yielding pRL1. Because this phoA fragment contained an extra set of 19 bp outside end (OE) of Tn5, the extra OE had to be removed. For this purpose, a fragment lacking the extra 19-bp inverted repeat was generated via PCR by using the following oligonucleotide: TTCGGTACCTTGCCCTGTTCTGGAAAACCG. This primer starts just downstream of the extra 19-bp inverted repeat of the phoA fragment and has a KpnI restriction site at its 5′ end. The second primer, CATCCTGCAACTCTGCGGTAGAAACG, is located downstream of the BssHII site in phoA. The 458-bp PCR product was cut with KpnI and BssHII and ligated into pRL1 cut with KpnI and BssHII, yielding pMM100.
Subsequently, TnTAP was transposed into pPB101 by following the same procedure as given above for TnTIN, yielding pMM1.
Transposition of TnTIN into secA and TnTAP into tolC.
To isolate in-frame insertions of TnTIN into secA, pMF8 containing the wild-type secA gene first was transformed into strain ME7000 carrying pPB301. During growth overnight, TnTIN transposed into pMF8. Plasmids were purified and digested with BstEII and XbaI, which cleave pPB301, but not pMF8 or TnTIN. The restriction digest was transformed into strain ME7000. Selection for Ampr (from pMF8) and Kanr (from TnTIN) yielded strains containing plasmids where TnTIN transposed into pMF8. These strains were tested for insertions of TnTIN into the secA gene, which resulted in in-frame fusions of uidA to secA by plating on agar plates containing 40 μg/ml X-GLUC. Blue colonies (UidA+) were purified, and the expression of secA-tev-uidA fusions was tested by Western blotting that used polyclonal antibodies against SecA.
To isolate in-frame fusions of TnTAP to tolC, pMM70 containing the wild-type tolC gene first was transformed into strain MM118 carrying pMM1. After growth overnight, plasmid DNA was isolated and digested with NheI to destroy pMM1, but not pMM70 or TnTAP. The restriction digest was transformed into strain MM118. Selection for Ampr (from pMM70) and Kanr (from TnTAP) yielded strains containing plasmids where TnTAP transposed into pMM70. These strains were tested for the insertion of TnTAP into the tolC gene, which resulted in in-frame fusions of phoA to tolC by plating on agar plates containing 40 μg/ml of 5-bromo-4-chloro-3-indolyl-phosphate. Blue colonies (PhoA+) were purified, and the expression of tolC-tev-phoA fusions was tested by Western blotting that used polyclonal antibodies against TolC.
Deletion of uidA (or phoA) and neo from TnTIN (or TnTAP) Inserts in secA (or tolC).
Plasmids containing TnTIN (TnTAP) inserts in secA (tolC) were cut with NotI and religated, deleting uidA (phoA) and neo. The remaining insert of 72 bp encodes LTLIHKFENLYFQSAAAILVYKSQ. TEV protease recognition sequence is ENLYFQS. The resulting plasmids were verified by restriction analysis. Expression of SecA (TolC) proteins containing the desired insert was detected on Western blots, because these derivatives migrated at slightly higher molecular weight on 7% (10%) SDS/PAGE.
Sequencing of Fusion Joints Generated by TnTIN and TnTAP.
Fusion joints were sequenced by using primers TTCACGGGTTGGGGTTTCTACAG and GCAGTAATATCGCCCTGAGCAGC reading out of the reporter genes uidA and phoA into the target gene, respectively.
Expression of TEV Protease from a pACYC184 Derivative.
We wanted to express TEV protease under tac promoter control as a glutathione S-transferase (GST) hybrid from a pACYC184 derivative that is compatible with the pBR322-derived secA plasmids. For this construct, pGEX2T-27K (21) was digested with ApaI and BspHI filled in with Klenow. This 3-kb fragment, containing Ptac and the GST-TEV protease fusion was ligated into a derivative of pACYC184 (that contained the lacIq gene inserted into the EcoRI site) cut with XmnI and ApaI to yield pMM13.
Proteolysis of SecA Containing TEV Protease Cleavage Site in Vivo and in Vitro.
pME410 was transformed into strain ME7000 containing pMM13. Cells were grown with and without 100 μM isopropyl β-d-thiogalactoside used to induce expression of TEV protease. Cleavage of SecA was detected by Western blotting.
SecA410 containing a TEV protease cleavage site after amino acid 830 was purified by a method to be described elsewhere. One microgram of purified SecA in proteolysis buffer (50 mM Tris⋅HCl, pH 8.0/0.5 mM EDTA pH 8.0) was incubated in the presence of 5 mM DTT (final concentration) and 1 μl TEV protease (1 mg/ml stock solution from GIBCO/BRL). After 3 hr at 30°C, proteolysis was stopped by addition of 100 mM iodoacetamide (final concentration). Proteolysis of SecA was detected on SDS/PAGE followed by staining with Coomassie blue (22).
Proteolysis of Whole Cells.
Cells expressing TolC containing TEV protease sites were grown in Luria–Bertani medium (23) to an OD600 of 0.3. Cells were washed once in proteolysis buffer (see above). For proteolysis, 5 mM DTT and 2.5 μl TEV protease (70 μg/ml final concentration) was added to 42.5 μl cells. After 4 hr at 30°C, 100 mM iodoacetamide was added to inactivate TEV protease. After precipitation with trichloroacetic acid, samples were subjected to SDS/PAGE and Western blotting by using polyclonal antibodies against TolC (24, 25).
RESULTS
Construction of TnTIN and TnTAP.
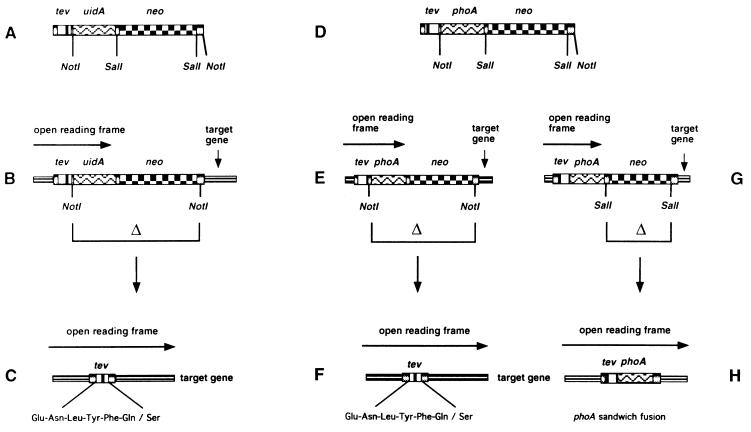
We have constructed a set of mini-transposons that introduce TEV protease cleavage sites into target proteins (Fig. 1). The basic architecture of TnTIN is: the 19-bp OE of Tn5 required for transposition, a 7-codon TEV protease cleavage site, uidA, neo, and another 19-bp OE of Tn5. TnTAP is basically the same except that uidA was replaced by signal sequenceless phoA. The Tn5 transposase, which acts on the flanking 19-bp OE sequences, is supplied in trans, generating stable insertions. When inserted in the correct orientation and reading frame, the transposons form translational fusions with the target gene. This protein fusion includes a 5′ portion of the target gene, TEV protease cleavage site, and either uidA or phoA. These translational fusions can be detected on indicator plates containing specific dyes, i.e., X-GLUC for uidA fusions and 5-bromo-4-chloro-3-indolyl-phosphate for phoA fusions. TnTIN is used for cytoplasmically localized target proteins, TnTAP for cell envelope proteins. The transposons carry the neo gene, conferring to kanamycin resistance, which can be used to select for cells carrying transposon insertions. Both transposons carry NotI restriction sites after the codons for TEV protease site and after neo. NotI deletions remove uidA or phoA and neo, leaving a 72-bp insert in an otherwise intact target gene. We chose the 8-bp cutter NotI, because this restriction site occurs very rarely in target genes. The insertion generated by both transposons is identical: LTLIHKFENLYFQSAAAILVYKSQ. The TEV protease recognition sequence is ENLYFQS.
Figure 1.
Structure and use of TnTIN and TnTAP. tev represents TEV protease cleavage site, uidA encodes β-glucuronidase, phoA encodes signal sequenceless alkaline phosphatase, and neo confers to kanamycin resistance. (A) Architecture of TnTIN. (B) TnTIN after insertion into a target gene. When inserted in the correct orientation and reading frame, the reading frame of the target gene extends into the tev site and uidA, resulting in a hybrid gene that encodes a hybrid protein composed of an N-terminal part of the target protein, TEV protease cleavage site, and β-glucuronidase. (C) Digestion by NotI removes the uidA and neo genes, generating a sandwich fusion of the 72-bp TEV protease cleavage site within the target gene. (D) Architecture of TnTAP. (E and G) TnTAP after insertion into a target gene. When inserted in the correct orientation and reading frame, the reading frame of the target gene extends into the tev site and phoA, resulting in a hybrid gene that encodes a hybrid protein composed of an N-terminal part of the target protein, TEV protease cleavage site, and signal sequenceless alkaline phosphatase. (F) Digestion by NotI removes the uidA and neo genes, generating a sandwich fusion of the 72-bp TEV protease cleavage site within the target gene. (H) Digestion by SalI removes the neo gene, generating a sandwich fusion between the target gene and phoA. The TEV protease site is present at the N terminus of signal sequenceless alkaline phosphatase.
An additional feature of TnTAP is that after a SalI deletion, the neo gene is removed, leaving a sandwich fusion of phoA and the target gene. This fusion occurs because the phoA gene lacks a stop codon so that the reading frame at the 3′ end of phoA remains open and extends into the 3′ end of the target gene. Thus, TnTAP is the first transposon that generates phoA sandwich fusions at random. To date, phoA sandwich fusions could be constructed only by using a cloning vector described earlier (20). The use of TnTAP for construction of phoA sandwich fusions will be described separately.
TnTIN Insertions into the secA Gene.
To test the function of TnTIN, we obtained derivatives of secA plasmid pMF8 into which the transposon had inserted. Initially, pMF8 and pPB301 were transformed into strain ME7000 (Δuid recA). Twenty-four Kanr colonies, which were blue on agar plates containing X-GLUC, were analyzed. Blue color on X-GLUC plates indicated the presence of a translational fusion of uidA to an ORF of pMF8. Western blots of these colonies with antibodies against SecA indicated that nine candidates had insertions within the secA gene (data not shown). Nucleotide sequencing indicated that we obtained fusions after codons 351 and 830 of SecA, respectively. Fusions to codon 830 expressed a large secA-TEV site-uidA fusion migrating at about 150 kDa on SDS/PAGE (Fig. 2). One of these candidates, termed secATnTIN410, was chosen for further characterization.
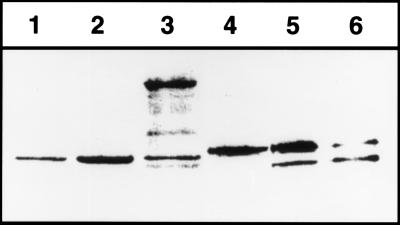
Figure 2.
Proteolysis of SecA410 in vivo. To monitor cleavage of SecA410 by TEV protease, a Western blot was prepared by using cells of ME7000 expressing either wild-type SecA (lane 1), wild-type SecA expressed from pMF8 (lane 2), secATnTIN410 (lane 3), SecA410 resulting from a cutback of TnTIN with NotI (lane 4), or SecA410 plus TEV protease expressed from pMM13 (lanes 5 and 6) after overnight growth in Luria–Bertani. The growth medium was without (lane 1–5) or supplemented with 100 μM isopropyl β-d-thiogalactoside (lane 6) to induce TEV protease. To detect SecA, polyclonal antibodies against SecA were used.
Characterization of secA-TEV Site-uidA Fusions.
Plasmids containing secA-TEV site-uidA fusions to codon 830 were digested with NotI and religated, yielding pME410. Deletion of uidA and neo generated a 24-codon insertion containing the TEV protease cleavage site. The presence of the insertion was detectable on SDS/PAGE because this SecA derivative, termed SecA410, migrated at slightly larger molecular weight than wild-type SecA. To test if TEV protease could cleave this SecA construct, pMM13 expressing TEV protease under ptac control was transformed into strain ME7000 containing pME410, expressing the secA-TEV cleavage site-secA sandwich fusion. Coexpression of TEV protease led to cleavage of SecA, indicating that the TEV protease recognition sequence was surface-exposed in the functional SecA dimer (see below) (Fig. 2). This finding agrees with the model that the C-terminal 70-aa residues of SecA are involved in lipid binding as well as SecB binding and therefore should be surface accessible (26). The efficiency of cleavage was 12 ± 15% when expression of TEV protease from the tac promoter was uninduced and 80 ± 10% when expression of TEV protease was induced by addition of isopropyl β-d-thiogalactoside to the growth medium (Fig. 2). Because in pMF8 expression of SecA is under the control of the wild-type promoter, incomplete cleavage of SecA might be because of its autoregulation: SecA expression is derepressed when protein secretion is not fully functional (9). This explanation is supported by the finding that the purified SecA410 was cleaved more efficiently by TEV protease in vitro (Fig. 3).
Figure 3.
Proteolysis of SecA410 in vitro. Cleavage of SecA by TEV protease in vitro was monitored by 7% SDS/PAGE. Lane 1, 1 μg of purified SecA410. Lane 2, 1 μg of purified SecA410 after incubation with 1 μg of TEV protease for 3 hr at 30°C.
To initially test to see if the insertion of the TEV protease site before and after cleavage by TEV protease would interfere with SecA function, we transformed pME410 and pMM13 into strain MM52 (secATs). MM52 does not grow at 42°C because of its conditional SecA defect. MM52 expressing the plasmid-encoded SecA410 derivative complemented the secA(Ts) allele and grew well at 42°C. Thus, SecA function was not compromised significantly by the insertion containing the TEV protease cleavage site. However, when TEV protease was coexpressed, the secATs mutation was no longer complemented as was indicated by the inability of the strain to grow on Luria–Bertani agar plates at 42°C.
Proteolysis of Purified SecA by TEV Protease in Vitro.
Because proteolysis of SecA occurred in vivo, we wanted to examine if similar results could be obtained when using purified SecA and TEV protease. SecA containing the TEV protease site after amino acid 830 was purified. Addition of purified TEV protease resulted in efficient cleavage of SecA (Fig. 3).
TnTAP Insertions into the tolC Gene.
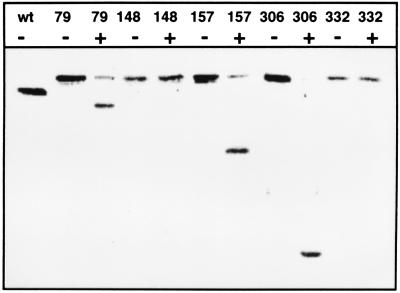
To test the function of TnTAP, we used the outer membrane protein TolC as a target. As with TnTIN insertions into secA, we obtained derivatives of tolC plasmid pMM70 into which the transposon had inserted. After analyzing 180 Kanr colonies, which were blue on agar plates containing 5-bromo-4-chloro-3-indolyl-phosphate, we isolated 115 fusions in which the TEV protease cleavage site was in the tolC reading frame. Sequencing of the fusion joints indicated that TnTAP inserted at 50 different positions in tolC. Five candidates were chosen for further characterization. Deletion of phoA and neo yielded tolC-TEV site-tolC sandwich fusions. Adding TEV protease to whole cells led to cleavage of TolC derivatives containing TEV protease sites after amino acids 79, 157, and 306. No cleavage occurred when the protease sites were inserted after amino acids 148 and 332 (Fig. 4). The TolC derivatives were expressed in pcnB mutant strain MM118. pcnB decreases the plasmid copy number of ColE1 ori plasmids (15), thus reducing the problem of overexpression. To obtain evidence for the functionality of the constructed TolC derivatives, we tested growth on MacConkey plates. TolC is known to confer to resistance against hydrophobic agents such as bile salts (27). Growth on MacConkey plates containing 0.15% bile salts is a convenient monitor of TolC function. Cells expressing TolC derivatives containing TEV protease sites after amino acid 79 and 306 grew as well as the wild type. Inserts after amino acid 157 and 332 conferred to slightly reduced growth, whereas the insert after amino acid 148 was apparently nonfunctional because no growth on MacConkey plates could be detected. In addition, we wanted to test to see if the expression of these TolC derivatives had a negative effect on the integrity of the outer membrane. Thus, we investigated hypersensitivity against the antibiotics rifampicin and chloramphenicol on Luria–Bertani agar plates containing 1, 5, and 50 μg/ml rifampicin and 0.1 and 1 μg/ml chloramphenicol. Because growth of cells expressing the TolC derivatives was essentially the same as that of wild-type cells, we concluded that the outer membrane was not hyperpermeabilized by the TolC constructs.
Figure 4.
Proteolysis of TolC derivatives on the extracellular surface of whole cells. Cleavage of TolC carrying TEV protease sites was monitored by Western blotting. TEV protease was added to intact cells for 3 hr at 30°C. After inactivation of TEV protease, cells were suspended in sample buffer and subjected to SDS/PAGE. wt, wild-type TolC. Numbers indicate the amino acid of TolC after which a TEV protease site was inserted. −, no TEV protease added to whole cells. +, TEV protease (70 μg/ml final concentration) added to whole cells.
DISCUSSION
We describe two mini-transposons, TnTIN and TnTAP, that fuse a short insert of 24 codons into target genes. The insert encodes the cleavage site of TEV protease that can proteolyze target proteins in vivo, in vitro, or on the extracellular side of whole cells when the recognition sequence is surface accessible. We showed that this site-specific proteolysis approach can be applied to soluble cytoplasmic as well as to integral outer membrane proteins. Because only surface-exposed TEV protease cleavage sites are proteolyzed, this approach can be used in topological studies. The insertion and cleavage of TEV protease sites may represent a novel tool for studying the topology of outer membrane proteins. Experiments currently are being carried out to test this idea.
The 24-codon insertions can be used to detect regions in proteins tolerating additional sequence, which may provide helpful information for protein engineering and structure-function studies. Sites that can accommodate the 24-aa insertion without loss of activity can be used to inactivate essential proteins in vivo by site-specific proteolysis via coexpressed TEV protease. This approach may serve as an alternative or addition to existing methods, e.g., conditional lethal or suppressible nonsense mutations and dilution experiments. It remains to be seen, however, what fraction of all possible inserts in permissive sites will yield proteins that are effectively inactivated, either as a primary or secondary consequence of cleavage. Examples for secondary consequences of cleavage are dissociation of proteolytic fragments and/or further degradation of these fragments by cellular proteases. For inactivation of essential proteins, the experimental system can be optimized such that TEV protease is under control of the tight arabinose promoter (28) to minimize cleavage of the target protein when expression of TEV protease is not induced. When, as in the case of SecA, the expression of the target protein is autoregulated by using a foreign promoter, e.g., the lac promoter, will exclude increased expression after inactivation of the target protein.
The transposons provide a technically simple and rapid alternative to inserting protease cleavage sites via linker or oligonucleotide mutagenesis. Another advantage is that transposition occurs rather randomly, as was detected when sequencing 115 insertions into the tolC gene. Random insertions may be desired for searching permissive sites in target proteins for which detailed structural information is not available.
Because TEV protease can be actively expressed in yeast cells without affecting viability (29), site-specific proteolysis may be applied in eukaryotes and cell cultures as well. Also, because the use of Tn5 is not restricted to E. coli, and uidA is commonly used as a reporter gene in plant molecular biology, we anticipate general applicability.
Acknowledgments
We thank Jon Beckwith for challenging us, many years ago, to devise an in vivo selection for E. coli mutants defective in membrane protein insertion. Our efforts have directly resulted in this method. We thank J. Barondess for discussions, R. Benz and I. Gentschev for antibodies to TolC and bacterial strains, Kelly Hughes for reading the manuscript, D. Oliver for a secA plasmid, K.-L. Schimz for antibodies to SecA, W. Reznikoff for transposase plasmids, Kate Wilson for strains and discussions, and C. Manoil and B. Traxler for sharing unpublished results. We are grateful to W. Boos in whose laboratory part of this work was carried out and to D. Pette and SFB156 for support. This work was supported by a grant from the Deutsche Forschungsgemeinschaft. D.B. was supported by National Institutes of Health Grants GM54160 and GM41883, and M.M. was supported by a fellowship from the Studienstiftung des deutschen Volkes.
ABBREVIATIONS
- TEV
tobacco etch virus
- X-GLU
5-bromo-4-chloro-3-indolyl-β-d-glucuronide
- OE
outside end
References
- 1.Mondigler M, Ehrmann M. J Bacteriol. 1996;178:2986–2988. doi: 10.1128/jb.178.10.2986-2988.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farrell L B, Beachy R N. Plant Mol Biol. 1990;15:821–825. doi: 10.1007/BF00039422. [DOI] [PubMed] [Google Scholar]
- 3.Chang C N, Kuang Q-J, Chen E Y. Gene. 1986;44:121–125. doi: 10.1016/0378-1119(86)90050-8. [DOI] [PubMed] [Google Scholar]
- 4.Traxler B, Boyd D, Beckwith J. J Membrane Biol. 1993;132:1–11. doi: 10.1007/BF00233047. [DOI] [PubMed] [Google Scholar]
- 5.Wilson K J, Sessitsch A, Corbo J C, Giller K E, Akkermans A D, Jefferson R A. Microbiology. 1995;141:1691–1705. doi: 10.1099/13500872-141-7-1691. [DOI] [PubMed] [Google Scholar]
- 6.Manoil C, Bailey J. J Mol Biol. 1997;267:250–263. doi: 10.1006/jmbi.1996.0881. [DOI] [PubMed] [Google Scholar]
- 7.Nelson B D, Manoil C, Traxler B. J Bacteriol. 1997;179:3721–3728. doi: 10.1128/jb.179.11.3721-3728.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lippincott J, Traxler B. J Bacteriol. 1997;179:1337–1343. doi: 10.1128/jb.179.4.1337-1343.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliver D B. Mol Microbiol. 1993;7:159–165. doi: 10.1111/j.1365-2958.1993.tb01107.x. [DOI] [PubMed] [Google Scholar]
- 10.Wandersman C, Delepelaire P. Proc Natl Acad Sci USA. 1990;87:4776–4780. doi: 10.1073/pnas.87.12.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benz, R., Maier, E. & Gentschev, I. (1993) Int. J. Med. Microbiol. Virol. Parasitol. Infect. Dis. 187–196.
- 12.Koronakis V, Li J, Koronakis E, Stauffer K. Mol Microbiol. 1997;273:617–626. doi: 10.1046/j.1365-2958.1997.d01-1880.x. [DOI] [PubMed] [Google Scholar]
- 13.Oliver D B, Beckwith J. Cell. 1981;25:765–772. doi: 10.1016/0092-8674(81)90184-7. [DOI] [PubMed] [Google Scholar]
- 14.Manoil C, Beckwith J. Proc Natl Acad Sci USA. 1985;82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopilato J, Bortner S, Beckwith J. Mol Gen Genet. 1986;205:285–290. doi: 10.1007/BF00430440. [DOI] [PubMed] [Google Scholar]
- 16.Boyd D, Manoil C, Beckwith J. Proc Natl Acad Sci USA. 1987;84:8525–8529. doi: 10.1073/pnas.84.23.8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt M G, Oliver D B. J Bacteriol. 1989;171:643–649. doi: 10.1128/jb.171.2.643-649.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang A C Y, Cohen S N. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiegand T W, Reznikoff W S. J Mol Biol. 1994;235:486–495. doi: 10.1006/jmbi.1994.1008. [DOI] [PubMed] [Google Scholar]
- 20.Ehrmann M, Boyd D, Beckwith J. Proc Natl Acad Sci USA. 1990;87:7574–7578. doi: 10.1073/pnas.87.19.7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parks T D, Leuther K K, Howard E D, Johnston S A, Dougherty W G. Anal Biochem. 1994;216:413–417. doi: 10.1006/abio.1994.1060. [DOI] [PubMed] [Google Scholar]
- 22.Neuhoff V, Arold N, Taube D, Ehrhardt W. Electrophoresis. 1988;9:255–262. doi: 10.1002/elps.1150090603. [DOI] [PubMed] [Google Scholar]
- 23.Miller J. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 24.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 26.Breukink E, Nouwen N, van Raalte A, Mizushima S, Tommassen J, de Kruijff B. J Biol Chem. 1995;270:7902–7907. doi: 10.1074/jbc.270.14.7902. [DOI] [PubMed] [Google Scholar]
- 27.Fralick J A. J Bacteriol. 1996;178:5803–5805. doi: 10.1128/jb.178.19.5803-5805.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guzman L-M, Belin D, Carson M, Beckwith J. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith T A, Kohorn B D. Proc Natl Acad Sci USA. 1991;88:5159–5162. doi: 10.1073/pnas.88.12.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]