Abstract
More than 50% of transitional cell carcinomas of the bladder show loss of heterozygosity of a region spanning the TSC1 locus at 9q34 and mutations of TSC1 have been identified in 14.5% of tumours. These comprise nonsense mutations, splicing mutations, small deletions and missense mutations. Missense mutations are only rarely found in the germline in TSC disease. Therefore, we have examined six somatic missense mutations found in bladder cancer to determine whether these result in loss of function. We describe loss of function via distinct mechanisms. Five mutations caused mutually exclusive defects at mRNA and protein levels. Of these, two mutations caused pre-mRNA splicing errors that were predicted to result in premature protein truncation and three resulted in markedly reduced stability of exogenous TSC1 protein. Primary tumours with aberrant TSC1 pre-mRNA splicing were confirmed as negative for TSC1 expression by immunohistochemistry. Expression was also significantly reduced in a tumour with a TSC1 missense mutation resulting in diminished protein half-life. A single TSC1 missense mutation identified in a tumour with retained heterozygosity of the TSC1 region on chromosome 9 caused an apparently TSC2- and mTOR-independent localization defect of the mutant protein. We conclude that although TSC1 missense mutations do not play a major role in causation of TSC disease, they represent a significant proportion of somatic loss of function mutations in bladder cancer.
INTRODUCTION
Tuberous Sclerosis Complex (TSC) is an autosomal dominant tumour suppressor gene syndrome with an incidence of 1 in 6000–10 000 births. TSC is characterized by the development of benign growths, called hamartomas, in the kidneys, heart, brain and skin, and patients present clinically with a variety of developmental disorders (1). TSC is caused by mutations affecting either of the tumour suppressor genes TSC1 or TSC2. TSC1 on chromosome 9q34 encodes hamartin (2) and TSC2 on chromosome 16p13.3 encodes tuberin (3). Approximately half of large TSC families show linkage to 9q34 and half to 16p13.3 (4–6). Tumour development in TSC patients is thought to occur as the result of a somatic ‘second-hit’ in either TSC1 or TSC2, according to Knudson’s tumour suppressor model (7). Loss of heterozygosity (LOH) of TSC1 or TSC2 has been reported in some TSC hamartomas, such as renal angiomyolipomas. However, loss of the wild-type allele in brain lesions is rare, suggesting the possibility of tissue-specific haploinsufficiency of TSC genes (8–10).
Co-localization and co-immunoprecipitation of TSC1 and TSC2 in mammalian cells (11,12) and direct binding in yeast two-hybrid assays provide a tentative explanation for the similar disease phenotype in TSC patients with mutations in either TSC1 or TSC2 genes (2,13). Functionally, the TSC1/TSC2 complex is positioned at the centre of multiple growth signalling pathways and is a key integrator of signals controlling protein translation and cell growth (14). Activation of the TSC1/TSC2 complex in growth-limiting conditions attenuates signalling through mTOR via specific GTPase activating protein (GAP) activity of TSC2 towards RHEB (15,16).
While epithelial malignancy is not a common feature of TSC, studies in this laboratory and others have implicated loss of function of TSC1 in bladder tumorigenesis (17–19). Loss of heterozygosity (LOH) for markers on chromosome 9 is observed in more than 50% of bladder tumours of all grades and stages (20) and sub-chromosomal LOH analyses have identified the TSC1 locus at 9q34 as a common critical region of deletion between markers D9S149 and D9S66 (19,21–23). To date, we have screened 154 bladder tumours by fluorescent single strand conformation polymorphism (F-SSCP) analysis and direct sequencing, and found an overall mutation frequency of 14.5%. The mutation spectrum comprises nonsense (35%), missense (26%), frameshift (26%), in-frame deletions (3%) and splicing (10%) mutations (24) (Platt et al., in preparation). In all cases but one, TSC1 missense mutations were tumour-specific somatic events. TSC1 is the only gene on 9q that has been found to be mutated in bladder tumours, and may therefore be the critical gene on this chromosome arm implicated in >50% of all bladder tumours.
Missense mutations of TSC1 have not routinely been confirmed as functionally inactivating in TSC disease, though two recent reports provide evidence that in a few cases these are likely to be disease-causing (25,26). Here we sought to determine whether the TSC1 missense mutations identified in bladder tumours constituted inactivating mutations. We anticipated that discrete amino acid changes of mutant proteins might allow the identification of functionally important residues. Wild-type and mutant TSC1 constructs were retrovirally delivered into TSC1-null bladder tumour cell lines and functionally characterized. All somatic TSC1 missense mutations perturbed TSC1 function by causing aberrant splicing, protein instability or protein mislocalization. Defects were confirmed in primary tumours by RT–PCR analysis of mutant transcripts and immunohistochemical analysis.
RESULTS
Missense mutations of TSC1 identified in bladder tumours
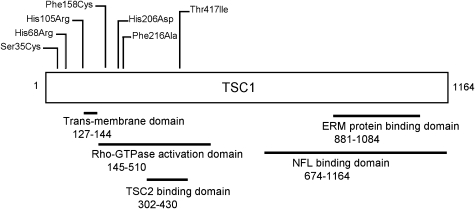
Previously, we identified 8 mutations including 2 missense mutations in a series of 62 bladder tumours (24). Screening of an additional 92 tumours (Platt et al., in preparation) identified an additional 15 mutations of which 4 were missense mutations. In total, therefore, 6 missense mutations have been identified in 154 tumours, representing 26% of all mutations found (Table 1). Mutations were determined as tumour-specific by genotyping of paired tumour and blood samples. 1250C>T (Thr417Ile) was previously described in the germline in TSC disease in Japanese patients, but was not confirmed as causative of TSC (27,28). 1250C>T was also identified in our laboratory in the patient’s constitutional DNA, and the tumour sample retained heterozygosity for microsatellite markers at the TSC1 gene locus (24). Threonine 417 was previously identified as a site of CDK1-dependent phosphorylation (29). To determine the biological significance of this variant, 1250C>T (Thr417Ile) was characterized here alongside tumour-specific missense mutations. Five of six other tumours with TSC1 missense mutations showed LOH for markers at the TSC1 locus (Table 1). There was no relationship between mutation and tumour stage or grade. Missense mutations were predominantly N-terminal but did not localize to a common functional domain (Fig. 1).
Table 1.
TSC1 missense variants identified in bladder tumours
| Mutation | Exon | Amino-acid change | BLOSUM62 score | Tumour stage/grade | Tumour 9q LOH status |
|---|---|---|---|---|---|
| 104C>G | 3 | Ser35Cys | −1 | T1 G3 | Loss |
| 203A>G | 4 | His68Arg | 0 | T2 G2 | Loss |
| 314A>G | 5 | His105Arg | 0 | Ta G2 | Loss |
| 473T>G | 6 | Phe158Cys | −2 | Ta G1 | Loss |
| 616C>G | 7 | His206Asp | −1 | Ta G1 | Loss |
| 648T>A | 7 | Phe216Ala | −2 | Ta G2 | Retention |
| 1250C>Ta | 12 | Thr417lle | −1 | T2 G3 | Retention |
aThis variant was also found in the patient’s normal DNA.
Figure 1.
Positions of amino acid substitutions in relation to described functional domains of hamartin.
To assess possible functional implications of amino acid substitutions, conservation of missense mutant residues was determined in TSC1 orthologs. His68, Phe158 and Phe216 were conserved among Rattus, Mus, Drosophila, Fugu and Gallus orthologs. His105, His206 and Thr417 differed only in Drosophila, which shares 31% identity with human TSC1. Ser35 differed in Drosophila, Fugu and Gallus orthologs. According to the BLOSUM62 scoring matrix (30), His68Arg and His105Arg substitutions are considered conservative. Ser35Cys, His206Asp and Thr417Ile are less conservative and Phe158Cys and Phe216Asp are least conservative (Table 1).
Re-expression of missense mutant TSC1 proteins in TSC1-null urothelial cells
We anticipated that missense mutations might lead to loss of function at the amino acid level. Thus, our initial approach was to express wild-type and missense mutant TSC1 cDNAs in TSC1-null bladder cell lines and investigate function of the mutant proteins. Missense mutant proteins were C-terminally FLAG- or GFP-tagged, expressed in 97-1 and HCV29 cells and characterized for TSC2 binding activity, and mTOR suppressive activity in nutrient-starved conditions.
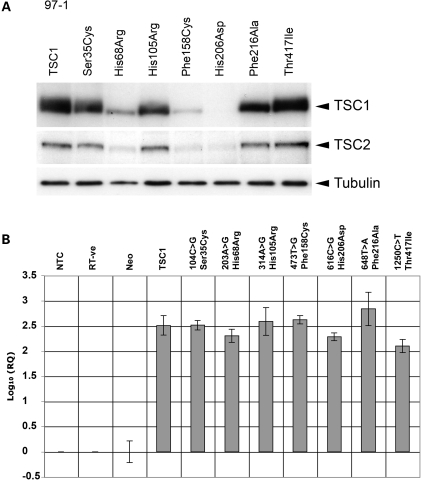
Expression of mutant proteins was investigated by immunoblotting following retroviral transduction and selection of neomycin resistant mass cell populations. Expression levels of mutant proteins were markedly different; Ser35Cys, His105Arg, Phe216Asp and Thr417Ile were expressed at high levels similar to wild-type protein, whereas His68Arg and Phe158Cys were much reduced and His206Asp was not detected (Fig. 2A). Repeated independent infections of these two cell lines and of telomerase-immortalized normal human urothelial cells (TERT-NHUC) and assessment of mass populations of cells following selection resulted in entirely reproducible expression levels of all mutant proteins (data not shown). The transcription of missense RNA was confirmed by real time RT–PCR and uniform levels of wild-type and missense transcripts were detected (Fig. 2B). The consistently reduced or absent protein expression of His68Arg and Phe158Cys and His206Asp missense mutant forms despite the presence of RNA expression suggested possible effects of these missense changes on protein stability.
Figure 2.
(A) Immunoblot showing levels of wild-type and mutant TSC1-FLAG proteins and endogenous TSC2 in 97-1 cell lines. (B) Measurement of TSC1 RNA levels by real time RT–PCR analysis of wild-type and mutant TSC1-FLAG mRNA transcript levels in 97-1 cell lines. RT−ve is reverse transcriptase-negative control, NTC is no template control. TSC1 expression is standardized to SDHA and normalized to the 97-1 Neo cell line.
Mutant TSC1 proteins retain interaction with TSC2
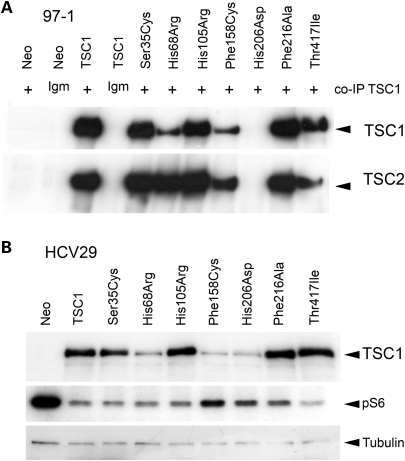
The interaction between TSC1 and TSC2 appears to be important in maintaining the stability of each of the proteins. TSC1/TSC2 binding specifically augments TSC2 expression by limiting its ubiquitination (31,32). TSC2 GAP activity towards RHEB defines TSC1/2 mediated control of mTOR signalling, and phosphorylation of downstream effectors of mTOR is constitutive and refractory to amino acid withdrawal in cells lacking TSC1 or TSC2 (33–35). Where expressed, missense mutant TSC1 proteins stabilized TSC2 levels in 97-1 cells (Fig. 2A) and co-immunoprecipitated with it (Fig. 3A). However, our data cannot exclude minor effects on interaction or TSC2 stability.
Figure 3.
(A) Immunoblot showing TSC1 and TSC2 in 97-1 Neo control and FLAG-tagged wild-type and mutant TSC1 cell line lysates immunoprecipitated with anti-TSC1 and non-specific mouse IgM antibodies. (B) Immunoblot showing expression levels of GFP-tagged wild-type and missense mutant TSC1 protein in HCV29 cell lines. Also shown are levels of S6 phosphorylation in transiently amino acid starved cells.
While His206Asp-FLAG was undetected in transduced 97-1 cells, we were able to achieve low-level expression of His206Asp-GFP in HCV29 cells, suggesting some stabilizing effect of the GFP tag. TSC2 co-immunoprecipitation and S6 phosphorylation assays were not accurately quantitative, but expression of all missense mutant proteins in TSC1-null cells reduced S6 phosphorylation in amino acid starved conditions, relative to vector alone (Fig. 3B), indicating that none of these mutant forms of TSC1 abolish interaction with TSC2.
Aberrant RNA splicing caused by TSC1 missense mutations
High-level expression and TSC2 binding activity of TSC1 Ser35Cys, His105Arg and Phe216Ala proteins suggested no functional effect of amino acid substitutions caused by 104C>G, 314A>G and 648T>A missense mutations. However, the expression of proteins from exogenous missense cDNAs did not allow the assessment of possible effects of mutations at the pre-mRNA level. We speculated that TSC1 missense mutations may cause loss of function through introduction of splicing errors in mutant transcripts in vivo. Therefore, wild-type and missense mutant TSC1 pre-mRNA sequences were screened for effects of mutations on splice site definition using the neural network algorithm (http://www.fruitfly.org/seq_tools/splice.html).
Interestingly, significant differences were seen between splice site scores of wild-type and 104C>G and 314A>G TSC1 transcripts. The 104C>G mutation, positioned 3 bp upstream of the TSC1 exon 3/4 junction, resulted in a reduced exon 3/4 splice motif score. The 314A>G mutation resulted in the introduction of a high-scoring 5′ donor splice site, immediately upstream of the A/G transversion, by generation of a novel consensus splice motif. No differences in splice site scores were observed between wild-type and 648T>A or 1250C>T transcripts or between wild-type and mutant transcripts that generated low exogenous protein expression.
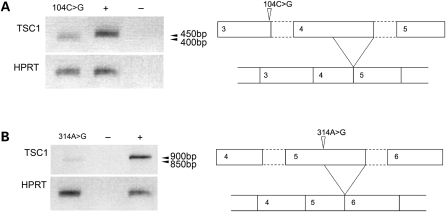
To examine splicing of missense mutant TSC1 transcripts, fragments spanning the mutation site and flanking intron/exon junctions were amplified from respective tumour cDNAs by RT–PCR and sequenced. An RT–PCR product reproducibly amplified from cDNA of the tumour containing 104C>G showed increased electrophoretic mobility relative to a control fragment amplified from TERT-NHUC cDNA (Fig. 4A). Sequencing of the tumour product revealed use of a cryptic 5′ donor site in exon 4, the adjacent downstream exon to the 104C>G mutant exon (Supplementary Material, Fig. S1). The reduced splice motif score at the exon 3/4 junction was shown to have no effect on exon 3/4 splicing. If translated, this 104C>G mutant transcript would generate 26 new amino acids from an alternate reading frame and truncate prematurely at residue 79.
Figure 4.
Semi quantitative RT–PCR analysis of TSC1 RNA in normal urothelial cells and TSC1 mutant tumour samples, showing increased electrophoretic mobility of RT–PCR products amplified from cDNA from tumours containing TSC1 104C>G (A) and 314A>G (B) missense mutations. Controls are amplification reactions using RT +ve (+) and RT−ve (−) cDNA from pooled TERT-NHUC. Lower panels show HPRT RT–PCR products. Also shown are schematic representations of aberrant splicing events associated with TSC1 104C>G (A) and TSC1 314A>G (B) mutations.
A faster migrating band was also reproducibly amplified from cDNA of the tumour containing 314A>G (Fig. 4B), compared to control TERT-NHUC cDNA. Sequence analysis of PCR products revealed a 50-nucleotide deletion in the transcript generated from the missense allele. The 314A>G mutation created a de novo 5′ donor splice site immediately upstream of 314A>G and resulted in the splicing of 50 nucleotides from the 3′ end of exon 5. The effect seen at the mRNA level was entirely consistent with the prediction made by in silico analysis. If translated, the 314A>G mutant transcript is expected to generate two new C-terminal amino acids and truncate prematurely at residue 107.
Exon skipping through introduction of a de novo 5′ splice site by point mutation is a relatively well-described mechanism of aberrant pre-mRNA splicing (36–38). No normal transcript was amplified from cDNA from tumours containing TSC1 104C>G or 314A>G mutations, suggesting that all splicing occurs via the novel sites. The abundance of missense transcripts appeared lower than normal transcript by semi-quantitative RT–PCR, when standardized to HPRT. However, by real time RT–PCR, TSC1 transcript abundance was relatively higher in tumours with TSC1 missense mutations causing transcript splicing or protein stability defects, than in uncultured or cultured TERT-NHUC, when standardized to SDHA (data not shown).
Missense mutant TSC1 proteins exhibit reduced protein stability
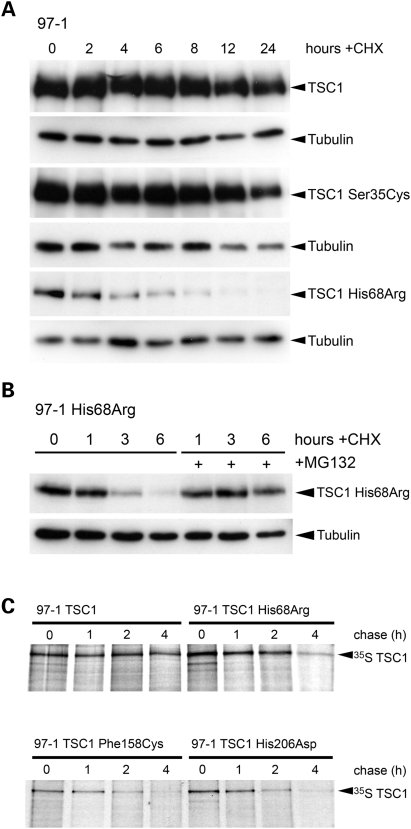
The demonstration of consistently low-level exogenous protein expression, despite uniform mRNA transcript abundance, suggested diminished stability of TSC1 His68Arg, Phe158Cys and His206Asp mutant proteins. By inhibiting protein synthesis with cycloheximide, we determined that His68Arg, Phe158Cys and His206Asp mutant proteins were turned-over relatively faster than wild-type TSC1, in a proteasome-dependent manner (Fig. 5 A and B and data not shown). Moreover, by 35S labelling, we were able to show that His68Arg, Phe158Cys and His206Asp proteins had markedly shorter half-lives than wild-type TSC1 (Fig. 5C).
Figure 5.
(A) Immunoblot showing turnover of wild-type and S35C and H68R missense TSC1 proteins in cycloheximide (CHX)-treated cells. Cells were cultured in full growth medium supplemented with 100 µg/ml CHX or DMSO vehicle alone, and lysed at time-points indicated. Tubulin is shown as a loading control. (B) Immunoblot showing stabilization of TSC1 H68R protein levels by proteasome inhibition. Cells were cultured in full growth medium and treated with 100 µg/ml CHX or DMSO vehicle alone or pre-treated with 40 µM MG-132. (C) Half-lives of wild-type TSC1 and His68Arg, Phe158Cys and His206Asp mutant proteins, as determined by 35S pulse chase analysis. Cells were pulsed with 250 µCi 35S in cysteine and methionine-free medium and chased in full growth medium supplemented with 200 nM cysteine and methionine. Cells were lysed at time-points as indicated and lysates immunoprecipitated with an anti-TSC1 antibody. Degradation of 35S-labelled TSC1 was determined of immunoprecipitated lysates by SDS–PAGE analysis and autoradiography.
Phe216Ala TSC1 protein shows altered localization
Previous studies have described a granular, cytoplasmic localization of endogenous TSC1 in vitro and in vivo and also of overexpressed TSC1 in COS-7 cells (39–41). Localization of monomeric TSC1 and of TSC1 complexed with TSC2 is likely influenced by culture conditions given that components of the Akt-mTOR signalling cascade are membrane localized when activated. In addition, TSC2 is reported to shuttle into the nucleus in a cell cycle and phosphorylation-dependent manner (42–44).
It was anticipated that the characterization of discrete missense amino acid changes may offer insight into potentially mTOR-independent or bladder-specific functions of TSC1. Our results indicated that TSC1 104C>G, 203A>G, 314A>G, 473T>G and 616C>G missense mutations cause loss of function by generic mechanisms of altered message or reduced protein stability. However, sequencing of RT–PCR products amplified from cDNA from the tumour containing the TSC1 648T>A mutation showed no altered splicing of the mutant transcript. Also, at the protein level, Phe216Ala was expressed at high levels similar to wild-type TSC1. The substitution of a phenylalanine residue for an alanine residue constitutes the loss of a high-molecular weight, hydrophobic benzyl group and is assigned a non-conservative –2 BLOSUM62 score. The lack of effect on splicing or protein stability raised the possibility that this mutation may cause a defect in TSC1 protein function per se.
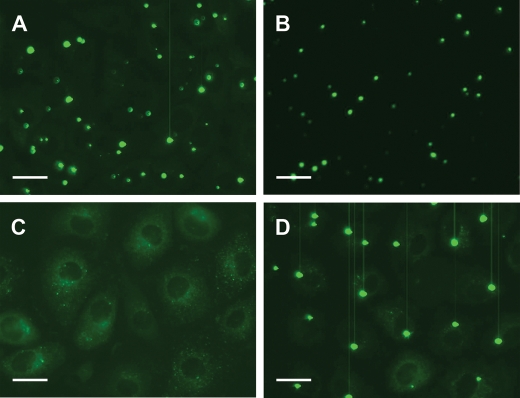
We determined the localization of GFP-tagged mutant proteins in nutrient replete and deficient conditions. Wild-type, Phe216Ala, Ser35Cys, His68Arg and Thr417Ile proteins were compared. In complete growth medium, wild-type TSC1 and Ser35Cys, His68Arg and Thr417Ile mutant proteins showed diffuse punctate cytoplasmic distribution with distinct cytoplasmic foci. In amino acid deficient medium, these proteins became localized almost exclusively to large cytoplasmic bodies. Intriguingly, the Phe216Ala substitution markedly altered the localization of TSC1; Phe216Ala was exclusively cytoplasmic and diffuse in full growth medium and did not redistribute to discrete bodies in amino acid deficient conditions (Fig. 6). However, it was shown to stabilize and to co-immunoprecipitate with endogenous TSC2, and to attenuate growth signalling through mTOR in starved conditions (Fig. 3B). These observations were reproduced in TSC1-transduced 97-1 and HCV29 cell lines and TERT-NHUC and suggest that localization of TSC1 to cytoplasmic foci is not a requirement for TSC2 binding or negative regulation of mTOR. Also, defective localization of Phe216Ala is likely TSC2- and mTOR-independent, possibly suggesting an independent function of TSC1 at cytoplasmic foci.
Figure 6.
Localization of wild-type TSC1 (A) and TSC1 Ser35Cys (B), Phe216Ala (C) and Thr417Ile (D) mutant proteins in amino acid-starved 97-1 cells. Cells were cultured in full growth medium on highly optically clear microscopy dishes to sub-confluence and amino acid starved for 24 h. TSC1-GFP was observed by UV microscopy of live cells. Scale bars show 100 µM.
Unlike the other five tumours with missense mutations, the tumour containing the TSC1 648T>A mutation did not show LOH of 9q, as confirmed by analysis of pure microdissected tumour cell populations (data not shown). Both normal and mutant alleles were detected by sequencing of tumour cDNA.
TSC1 protein expression in bladder tumour tissues
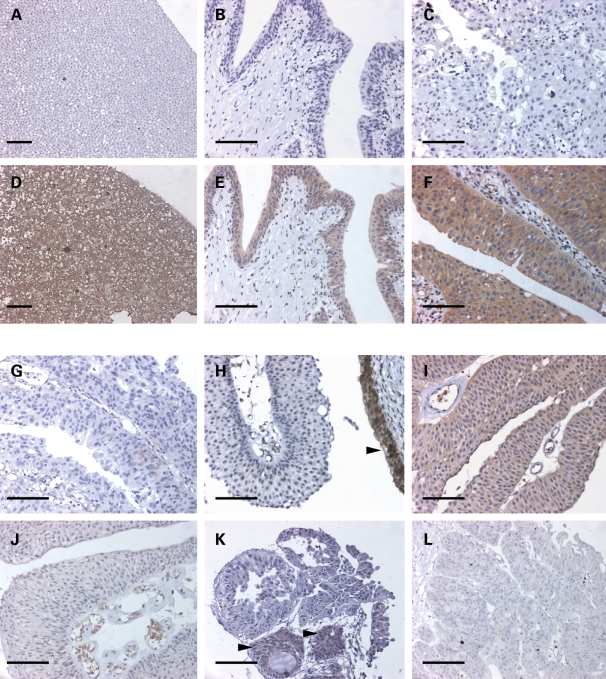
To examine the effects of pre-mRNA splicing defects and protein stability defects caused by missense mutations at the tumour level, TSC1 mutant and wild-type tumours were screened for expression of TSC1 protein by immunohistochemistry. Sensitivity and specificity of a rabbit monoclonal anti-TSC1 antibody was confirmed by staining of paraffin-embedded pellets of TSC1-null HCV29 cells and HCV29 in which wild-type TSC1 had been re-expressed (Fig. 7A and D). Normal ureter showed strong cytoplasmic TSC1 expression in the urothelium (Fig. 7E). Tumours with wild-type TSC1 showed strong cytoplasmic staining (Fig. 7F), and a tumour with a TSC1 73–77Δ 5 small deletion causing premature truncation at residue 27, showed no TSC1 expression (Fig. 7C). Both tumours with homozygous TSC1 missense mutations causing pre-mRNA splicing defects were negative for TSC1 expression (Fig. 7G and H). Figure 7H shows normal urothelium with strong TSC1 staining adjacent to immunonegative TSC1 314A>G mutant tumour cells. The 648T>A (Phe216Ala) mutant tumour showed moderate TSC1 expression consistent with normal stability of the Phe216Ala protein and retention of chromosome 9q heterozygosity in the tumour (Fig. 7I). Of the three missense mutations causing reduced protein stability (TSC1 203A>G, 473T>G and 616C>G), tumour material was available only for the 616C>G (His206Asp) mutant sample. The 616C>G mutation was identified from tumour material resected in 2003, and TSC1 expression was shown to be low in three tumour resections from this patient in successive years (Fig. 7J–L).
Figure 7.
TSC1 immunostaining of HCV29 Neo (A) and TSC1 (D) cell pellets, normal ureter (negative (B) and positive (E) antibody controls) and TSC1 73–77Δ 5 (C) and TSC1 wild-type (F) bladder tumours. TSC1 staining of TSC1 missense mutant bladder tumours; TSC1 104C>G (G), 314A>G (H), 648T>A (I) and 616C>G (J–L) mutant tumours. Arrow in (H) shows normal urothelium with strong TSC1 staining adjacent to immunonegative tumour cells. (J) to (L) show TSC1 staining in tumours resected from the same patient in 2001 (J), 2003 (K) and 2004 (L). Arrows in (K) show positive TSC1 immunoreactivity in von Brunn’s nests. Scale bars show 500 µM.
DISCUSSION
We have demonstrated that bladder tumour-derived TSC1 missense mutations result in loss of TSC1 function and that this occurs via distinct mechanisms. An overall TSC1 mutation frequency of ∼14.5% is found in bladder cancer (24) (Platt et al., in preparation) and missense mutations comprise 26% of mutations found to date. The identification of deleterious missense mutations in bladder tumours indicates a causative role of loss of TSC1 function via this mechanism in bladder tumorigenesis. The vast majority of TSC1 mutations in TSC disease are predicted to be protein truncating in nature, and no significant genotype/phenotype correlations have been observed (2,5,45–48). Non-chain terminating TSC1 mutations (missense or in-frame deletions) are rare in TSC disease (27,47,49,50) (www.LOVD.nl/TSC1). Although missense mutations have been reported previously, most have not been confirmed as disease-causing. Several have been revealed as rare polymorphisms or associated with other nonsense mutations in the same patient (46,50). Others have been disregarded as potentially disease-causing on the basis of conservative amino acid changes, or have not been characterized further (27,51). However, a recent publication by Jansen et al. (26) that identified three missense mutations (L916R, M224R and E412V) in affected individuals reported functional analyses on two of these. It was reported that E412V affected RNA splicing. Interestingly, in transfections of constructs of wild-type TSC1 and the M224R variant, the latter showed lower levels of protein expression that were unable to completely suppress S6 phosphorylation, results similar to those described here for missense variants with reduced protein half-life.
We have shown that two of six bladder tumour-derived missense mutations result in pre-mRNA splicing defects, three lead to reduced stability of mutant proteins and intriguingly, one mutation causes a TSC2- and mTOR-independent localization defect of the mutant protein. Wild-type TSC1 showed a granular cytoplasmic distribution in cells in full growth medium and became localized almost exclusively to large cytoplasmic bodies in amino acid starved cells. In contrast, TSC1 was cytoplasmic and diffuse in serum-starved cells. Preliminary results indicated that TSC1 bodies were dynamic, non-aggresomal structures (data not shown). Interestingly, the Phe216Ala substitution abolished the localization of TSC1 to these bodies. As expected from its position outside the recognized TSC2 binding domain, TSC1 Phe216Ala retained TSC2 binding activity and mTOR suppressive activity in amino acid starved conditions, suggesting that localization of TSC1 to cytoplasmic foci was not a requirement for TSC2 binding or for suppression of mTOR signalling. This may also suggest that TSC1 has a separable function at cytoplasmic foci that is independent of TSC2 and independent of negative regulation of mTOR by TSC1/TSC2. However, it is not clear whether the formation of intensely staining bodies in conditions of over-expression of TSC1 and/or TSC2 (39,52) is relevant to the normal physiological condition. Further investigation of the function of Phe216Ala is now required, ideally at levels of expression that are closer to normal and in both TSC1-null cells and those expressing endogenous wild-type TSC1.
Although the missense variant 1250C>T (T417I) was found in the patient’s germline and this patient showed no symptoms of TSC, we included this variant in our analyses as threonine 417 has previously been identified as a site of CDK1-dependent phosphorylation (29). We found no evidence for a functional defect and conclude that this represents a rare polymorphism. As this patient was Japanese, as were both TSC patients in whom this variant was reported previously (27,28), this rare variant may be confined to the Japanese population.
Evidence suggests that cis-acting mutations affecting splicing of some tumour suppressor genes can have causal roles in tumour initiation and progression (53). Predicted and confirmed effects of TSC-related and bladder tumour-derived TSC1 missense changes on pre-mRNA splicing reported here underscore the importance of using RNA-based techniques, together with conventional mutation detection methods, to effectively identify disease-causing mutations. One of the mutations that deleteriously affected splicing (104C>G) had an unexpected effect. The use of a cryptic 5′ donor site in an adjacent downstream exon is a non-conventional splicing event; disease-associated mutations at splice junctions typically result in skipping of the mutant exon (38,54,55). The 104C>G mutation was shown to reduce the exon 3 5′ splice site motif score and to disrupt an enhancer sequence spanning exon 3/4. 104C>G also disrupts a U1 snRNA-binding motif, spanning the 5′ splice site from position –3 to +8 (Supplementary Material, Fig. S1). In combination with a normally low-scoring exon 4 3′ acceptor site, these factors may result in reduced splicing efficiency of introns 3–4. Spliceosome assembly is directed by juxtaposed splicing elements, and were introns 4–5 to splice first, splicing machinery at the 3′ acceptor site of exon 4 may stimulate recruitment of U1 snRNA to possible binding sites in exon 4, and result in use of a cryptic 5′ donor site.
In bladder tumours as in TSC disease, nonsense, deletion and frameshift TSC1 mutations result in premature protein truncation and loss of protein function. We have now shown that missense mutations cause loss of function by aberrant splicing or reduced protein stability. The identification of tumour-specific TSC1 mutations in the context of chromosome 9 LOH argues strongly for a direct role of loss of TSC1 function in the aetiology of these tumours. These data are consistent with TSC1 acting as a tumour suppressor gene in bladder cancer in accordance with Knudsen’s two-hit hypothesis. Biallelic inactivation of TSC1 or TSC2 may not be necessarily required in some TSC-related tumours (9). The discrepancy between frequency of TSC1 mutation and frequency of LOH in the TSC1 gene region in bladder tumours suggests that haploinsufficiency of TSC1 may contribute to tumour development in some cases. LOH of chromosome 9 is a particularly frequent event in bladder cancer and to date, TSC1 is the only gene on 9q found to be mutated in bladder tumours. The contribution of partial loss of TSC1 to clonal expansion of tumour cells with 9q LOH is unknown, and haploinsufficiency remains a possibility in those bladder tumours with 9q LOH and no TSC1 mutation. LOH at the TSC1 locus may accompany another event driving loss of chromosome 9. Deletion of the CDKN2A locus at 9p21, which occurs in up to 50% of bladder tumours (56,57), or of an as yet unidentified chromosome 9 tumour suppressor gene, may constitute such a driving force. Loss of one copy of TSC1 may therefore be an advantageous gratuitous hit or ‘passenger event’ (58).
TSC is not a cancer prone syndrome and TSC lesions very rarely progress to malignancy. That TSC patients do not have an increased risk of developing bladder cancer, or other sporadic cancers, may be explained by the requirement of cumulative genetic insults and the typically late onset of malignant disease. The timing and order of initiating and subsequent genetic events in hamartoma and bladder tumour development is likely to be critical in determining malignant potential. Why TSC1 appears to be involved in bladder cancer and not other epithelial cancers is an unresolved question.
Frequent LOH of 9q is found in other tumour types, including ovarian carcinoma, gallbladder carcinoma, nasopharyngeal carcinoma and non-small cell lung cancer (59–65). However, reports of TSC1 and TSC2 mutation status in sporadic tumours other than bladder are very few. No mutations were observed in sporadic glial and glioneuronal tumours or renal cell carcinomas (66,67). Fifty-three and 39% of lung adenocarcinomas and precursor lesions, respectively, were found to have LOH of 9q (68). Subsequently, a screen of 47 lung adenocarcinomas identified three confirmed TSC1 mutations. However, LOH and mutations were not detected simultaneously (69).
In ovarian carcinoma, gallbladder carcinoma, nasopharyngeal carcinoma and lung adenocarcinoma, LOH or deletion is described in both TSC gene regions (63,68,70,71). If loss of TSC1/2 complex function is the pathogenic effect of loss of TSC1, it may be expected that bladder tumours would also show loss of TSC2 function. In an LOH screen of 16p, we have detected LOH for markers at the TSC2 locus in 16% of bladder tumours (Platt et al., in preparation). It will be important to screen for mutations in the retained copy of TSC2 in cases with LOH to determine whether there is also an involvement of TSC2 or whether there is an important independent role of TSC1 in urothelial cells.
MATERIALS AND METHODS
Plasmids
TSC1 (nucleotides 123–3757 with respect to Ensembl sequence ENST00000298552) was amplified from TERT-NHUC cDNA using the Advantage cDNA PCR kit (Clontech, Saint-Germain-en-Laye, France) and TSC1 FWD (GAAACTGAAGTACCAGTTGT) and REV (GCAAGTTAACACTGATTGACCATC) primers. TSC1 was cloned in-frame with a C-terminal FLAG or GFP tag in the pFB Neo retroviral vector, a derivative of pBABE (Stratagene, Amsterdam, The Netherlands) modified to contain the Neomycin resistance cassette. Missense mutant constructs were generated by site-directed mutagenesis using QuickChange (Stratagene) according to the manufacturer’s instructions. All constructs were fully sequenced using the BigDye® terminator cycle sequencing kit version 1.1 (Applied Biosystems, Warrington, UK).
Cell lines
Two urothelial cell lines with loss of function mutations in TSC1 (24) were used in this study. 97-1 is derived from a papillary myoinvasive bladder urothelial carcinoma of grade G1/2 and stage pT1/2 (72,73). 97-1 cells have a homozygous TSC1 2295C>T mutation causing premature protein truncation at amino acid residue 692. 97-1 were maintained in Ham’s F12 medium (Invitrogen, Paisley, UK) with 1% foetal calf serum (FCS), 1% insulin-transferrin-selenium (Sigma, Dorset, UK), 1% MEM non-essential amino acids (NEAA; Invitrogen) and 25 mm hydrocortisone (Sigma). HCV29 was established from non-malignant ureteric epithelium of a patient with bladder cancer (74) and contains a homozygous 384C>T mutation that is predicted to cause protein truncation at residue Q55. HCV29 were maintained in RPMI-1640 medium supplemented with 10% FCS and 1% L-glutamine. For starvation experiments, cells were washed once in PBS and cultured overnight in medium minus all amino acids (Cancer Research UK Media Production Services).
Retroviral transduction
Phoenix-A packaging cells (ATCC) were maintained in Dulbecco's modification of Eagle's medium (DMEM) supplemented with 10% FCS and 1% L-glutamine. Phoenix-A were transfected by incubation with 1 µg of retroviral plasmid DNA in 3 µl siPort XP-1™ (Ambion, Huntingdon, UK) transfection agent in 100 µl PBS for 24 h. Virus supernatant was harvested at 48 h post-transfection. Second and third harvests were taken subsequently at 24 h intervals. Virus supernatant was passed through a 0.45 µM filter (PALL, Portsmouth, UK) and was either snap frozen as 1.5 ml aliquots at –80°C or used immediately. Supernatants were diluted 1:1 with target cell line normal growth medium and supplemented with 8 µg/ml polybrene (Sigma). Target cells were infected at 50% confluence by overnight incubation in virus-containing medium, and were cultured for a further 24 h in full growth medium before beginning selection.
Primary and immortalized normal human urothelial cells
Primary normal human urothelial cells (NHUC) were isolated from samples of normal ureter collected during nephrectomy or bladder reconstruction surgery at St James’s University Hospital, Leeds as described (75). Immortalized NHUC (TERT-NHUC) were generated by transduction of primary cells with the human telomerase reverse transcriptase (TERT) gene (76). Both primary and immortalized cells were maintained in PromoCell keratinocyte growth medium 2 (PromoCell, Heidelberg, Germany) containing 0.09 mm CaCl2 and supplemented with 30 ng/ml cholera toxin.
Western blotting
Sub-confluent cells were washed once in ice-cold phosphate buffered saline (PBS) and lysed in RIPA buffer containing 0.1% protease and phosphatase inhibitor cocktails (Sigma). Cell debris was pelleted by centrifugation at 12 700 g for 10 min at 4°C. Protein was quantified using the BIO-RAD protein assay (Bio-Rad Laboratories, Hemel Hempstead, UK) and samples analysed by SDS–PAGE. Proteins were immobilized onto Hybond-C nitrocellulose (Amersham Biosicences, Little Chalfont, UK) by semi-dry electrophoretic transfer. Blots were blocked in 3% non-fat dried milk in Tris buffered saline (TBS) (0.05 M Tris; 138 mM NaCl; 2.7 mM KCl; pH 7.6) 0.1% [v/v] Tween 20 (Sigma) for 30 min at RT with orbital shaking. Primary antibodies were diluted in 5% BSA (Sigma) in TBS 0.1% [v/v] Tween 20 and blots probed for 1 h at RT or overnight at 4°C with orbital shaking. Blots were washed for 5 × 5 min in >4 ml/cm2 TBS 0.1% [v/v] Tween 20 with orbital shaking between primary and secondary antibody incubations and before developing. Secondary HRP-conjugated antibodies were diluted in 3% non-fat dried milk in TBS 0.1% [v/v] Tween 20 and blots probed for 1 h at RT or overnight at 4°C with shaking. Horseradish peroxidase (HRP)-labelled antigen was detected by chemiluminescence using ECL Plus western blotting detection reagent (Amersham). Primary antibodies used were anti-TSC1 (Invitrogen, Paisley, UK) at 1:1000, anti-TSC2 C-20 (Santa Cruz Biotechnology, CA, USA) at 1:500, anti-ribosomal phospho-S6 Ser235/6 (Cell Signaling Technology Inc., Danvers, MA, USA) at 1:3000 and anti-α tubulin (Serotec, Kidlington, UK) at 1:500.
RNA extraction
RNA was made using the RNeasy™ kit (Qiagen, Crawley, UK) according to the manufacturer’s instructions. DNase treated RNA was purified by RNeasy column purification and eluted in RNase free water. RNA was reverse transcribed into first-strand cDNA using SuperScript™ II reverse transcriptase and Oligo-dT primers (Invitrogen) according to the manufacturer’s instructions.
Real time RT–PCR
Quantitation of RNA transcripts was performed by SYBR Green assay (Applied Biosystems). In each reaction 12.5 µl SYBR Green reagent was combined with 1 µl cDNA and 750 nM each of TSC1 27 FWD (CAGTCAGGTTTCCCAAAAGC) and TSC1 84 REV (GAGTTCTTGAACAGGCAGCTG) in a volume of 20 µl. Cycle parameters were initialization at 50°C for 2 min, incubation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 60°C for 1 min and 95°C for 15 s. The SDHA (succinate dehydrogenase complex, subunit A, flavoprotein) gene transcript was amplified using primers SDHA FWD; TGGGAACAAGAGGGCATCTG and REV; CCACCACTGCATCAAATTCATG as an internal control for each sample. Test and control reactions were performed in triplicate and average Ct values normalized to expression levels of uncultured urothelium or low passage cultured NHUC. PCR reactions and analyses were done on the ABI PRISM 7700 Sequence Detector (Applied Biosystems).
RT–PCR amplification of wild-type and missense mutant TSC1 transcripts
TSC1 fragments spanning exons 2–5 and 2–9 were amplified from TSC1 104C>G and 314A>G mutant tumour cDNA, respectively, in a reaction comprising 1 µg cDNA template, 0.625U AmpliTaq Gold, 1 × Gold PCR Buffer, 2.5 mm MgCl2, 200 µM each of dATP, dCTP, dGTP and dTTP and 300 nM of each primer (TSC1 1F; GAAACTGAAGTACCAGTTGT and TSC1 1R; GCTTGAGAGAGCTTATGCTT or TSC1 2R; GCACACTCGATCACAACATC) in a 25 µl volume. Cycling conditions were 95°C for 5 min, 25 or 35 cycles of 95°C for 40 s, 56°C for 40 s and 72°C for 1 min followed by an extension step of 72°C for 10 min. Amplification of an approximately 900 bp HPRT (hypoxanthine phosphoribosyl transferase 1) gene fragment served as a positive control (reactions conditions were as previously except for use of primers HPRT FWD; GACACTGGCAAAACAATGCA and REV; CTTCGTGGGGTCCTTTTCACC).
Fluorescence imaging
Cells were seeded onto IbiTreat tissue culture-treated, highly optically clear plastic µ-dishes (Thistle Scientific, Glasgow, UK). Cells were imaged using an inverted Zeiss AxioPlan microscope with Zeiss oil-immersion objective lenses and an excitation/emission filterset optimized for GFP. Images were captured using Ludl shutters and a Hamamatsu ORCA II ER camera linked to a computer running Volocity (Improvision, Coventry, UK).
Immunohistochemistry
Paraffin wax-embedded bladder tumour biopsies were assessed for TSC1 expression by immunohistochemistry using the streptavidin/peroxidase method. Sections were dewaxed in xylene and rehydrated through graded ethanols then endogenous peroxidase activity blocked in 3% hydrogen peroxide. Antigen retrieval was done by pressure-cooking in 0.01 M citric acid buffer, pH 6.0. Slides were blocked in avidin/biotin solutions (Vector Laboratories) followed by normal goat serum (1:10), then incubated with TSC1 primary antibody (Epitomics; Insight Biotechnology, Wembley, UK) at 1:100 overnight. Slides were then incubated with goat anti-rabbit biotinylated secondary antibody (Dako Cytomation, Glostrup, Denmark) at 1:400 for 30 min. Antibody binding was visualized using 3,3′diamino-benzidine tetrahydrochloride (DAB, Vector Laboratories) according to the manufacturer’s instructions. Finally, sections were counterstained with haematoxylin, dehydrated, cleared and mounted.
Protein stability measurements
Turnover of exogenous TSC1 protein was investigated by inhibition of protein synthesis by cycloheximide (Sigma) treatment. Cells were grown to sub-confluence in full growth medium and treated with cycloheximde at 100 µg/ml or DMSO vehicle alone for up to 24 h. Proteasome dependence of TSC1 turnover was determined by co-treatment with 40 µM MG-132 (Biomol, Exeter, UK) or DMSO vehicle.
For 35S pulse chase labelling, 97-1 wild-type and mutant TSC1 cell lines were incubated for 45 min in 1 ml Ham’s F12 minus Cys/Met (Cancer Research UK Media Production Services) containing 1% dialysed FBS (Sigma) and supplements. Cells were labelled with 250 µCi/ml 35S Pro-mix (Amersham) for 45 min. Cells were washed with 2 ml warm PBS and chased in Ham’s F12 complete medium (Invitrogen), supplemented with 2 mm methionine (Sigma) and 2 mm cysteine (Sigma). Cell lysates were prepared in RIPA lysis buffer and clarified by centrifugation. Supernatants were incubated with 1.25 µg mouse anti-TSC1 IgG1 antibody (Invitrogen) for 1 h at 4°C with rotation. Immuno-complexes were precipitated with 50 µl protein-G sepharose incubated overnight at 4°C. Beads were washed in 1 ml ice-cold RIPA buffer and re-suspended in 80 µl 2 × SDS buffer. Samples were analysed by SDS–PAGE and autoradiography.
SUPPLEMENTARY MATERIAL
FUNDING
This work was funded by a Programme grant from Cancer Research UK (C6228/A5433).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Filomena Esteves for help with immunohistochemistry and Dr Claire Taylor (Cancer Research UK Mutation Detection Facility, Leeds) and Professor Ian Eperon (University of Leicester) for advice on RNA splicing.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Gomez M., Sampson J., Whittemore V. The Tuberous Sclerosis Complex. Oxford University Press; 1999. [Google Scholar]
- 2.van Slegtenhorst M., de Hoogt R., Hermans C., Nellist M., Janssen B., Verhoef S., Lindhout D., van den Ouweland A., Halley D., Young J., et al. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science. 1997;277:805–808. doi: 10.1126/science.277.5327.805. [DOI] [PubMed] [Google Scholar]
- 3.Consortium. Identification and characterization of the tuberous sclerosis gene on chromosome 16. The European Chromosome 16 Tuberous Sclerosis Consortium. Cell. 1993;75:1305–1315. doi: 10.1016/0092-8674(93)90618-z. [DOI] [PubMed] [Google Scholar]
- 4.Cheadle J.P., Reeve M.P., Sampson J.R., Kwiatkowski D.J. Molecular genetic advances in tuberous sclerosis. Hum. Genet. 2000;107:97–114. doi: 10.1007/s004390000348. [DOI] [PubMed] [Google Scholar]
- 5.Jones A.C., Daniells C.E., Snell R.G., Tachataki M., Idziaszczyk S.A., Krawczak M., Sampson J.R., Cheadle J.P. Molecular genetic and phenotypic analysis reveals differences between TSC1 and TSC2 associated familial and sporadic tuberous sclerosis. Hum. Mol. Genet. 1997;6:2155–2161. doi: 10.1093/hmg/6.12.2155. [DOI] [PubMed] [Google Scholar]
- 6.Povey S., Burley M.W., Attwood J., Benham F., Hunt D., Jeremiah S.J., Franklin D., Gillett G., Malas S., Robson E.B., et al. Two loci for tuberous sclerosis: one on 9q34 and one on 16p13. Ann. Hum. Genet. 1994;58:107–127. doi: 10.1111/j.1469-1809.1994.tb01881.x. [DOI] [PubMed] [Google Scholar]
- 7.Knudson A.G., Jr Mutation and cancer: statistical study of retinoblastoma. Proc. Natl Acad. Sci. USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henske E.P., Scheithauer B.W., Short M.P., Wollmann R., Nahmias J., Hornigold N., van Slegtenhorst M., Welsh C.T., Kwiatkowski D.J. Allelic loss is frequent in tuberous sclerosis kidney lesions but rare in brain lesions. Am. J. Hum. Genet. 1996;59:400–406. [PMC free article] [PubMed] [Google Scholar]
- 9.Niida Y., Stemmer-Rachamimov A.O., Logrip M., Tapon D., Perez R., Kwiatkowski D.J., Sims K., MacCollin M., Louis D.N., Ramesh V. Survey of somatic mutations in tuberous sclerosis complex (TSC) hamartomas suggests different genetic mechanisms for pathogenesis of TSC lesions. Am. J. Hum. Genet. 2001;69:493–503. doi: 10.1086/321972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramesh V. Aspects of tuberous sclerosis complex (TSC) protein function in the brain. Biochem. Soc. Trans. 2003;31:579–583. doi: 10.1042/bst0310579. [DOI] [PubMed] [Google Scholar]
- 11.Plank T.L., Yeung R.S., Henske E.P. Hamartin, the product of the tuberous sclerosis 1 (TSC1) gene, interacts with tuberin and appears to be localized to cytoplasmic vesicles. Cancer Res. 1998;58:4766–4770. [PubMed] [Google Scholar]
- 12.Johnson M.W., Emelin J.K., Park S.H., Vinters H.V. Co-localization of TSC1 and TSC2 gene products in tubers of patients with tuberous sclerosis. Brain Pathol. 1999;9:45–54. doi: 10.1111/j.1750-3639.1999.tb00209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hodges A.K., Li S., Maynard J., Parry L., Braverman R., Cheadle J.P., DeClue J.E., Sampson J.R. Pathological mutations in TSC1 and TSC2 disrupt the interaction between hamartin and tuberin. Hum. Mol. Genet. 2001;10:2899–2905. doi: 10.1093/hmg/10.25.2899. [DOI] [PubMed] [Google Scholar]
- 14.Kwiatkowski D.J., Manning B.D. Tuberous sclerosis: a GAP at the crossroads of multiple signaling pathways. Hum. Mol. Genet. 2005;14(Spec No. 2):R251–R258. doi: 10.1093/hmg/ddi260. [DOI] [PubMed] [Google Scholar]
- 15.Inoki K., Li Y., Xu T., Guan K.L. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes. Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tee A.R., Manning B.D., Roux P.P., Cantley L.C., Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 17.Adachi H., Igawa M., Shiina H., Urakami S., Shigeno K., Hino O. Human bladder tumors with 2-hit mutations of tumor suppressor gene TSC1 and decreased expression of p27. J. Urol. 2003;170:601–604. doi: 10.1097/01.ju.0000074621.74361.10. [DOI] [PubMed] [Google Scholar]
- 18.Edwards J., Duncan P., Going J.J., Watters A.D., Grigor K.M., Bartlett J.M. Identification of loci associated with putative recurrence genes in transitional cell carcinoma of the urinary bladder. J. Pathol. 2002;196:380–385. doi: 10.1002/path.1052. [DOI] [PubMed] [Google Scholar]
- 19.Hornigold N., Devlin J., Davies A.M., Aveyard J.S., Habuchi T., Knowles M.A. Mutation of the 9q34 gene TSC1 in sporadic bladder cancer. Oncogene. 1999;18:2657–2661. doi: 10.1038/sj.onc.1202854. [DOI] [PubMed] [Google Scholar]
- 20.Cairns P., Shaw M.E., Knowles M.A. Initiation of bladder cancer may involve deletion of a tumour-suppressor gene on chromosome 9. Oncogene. 1993;8:1083–1085. [PubMed] [Google Scholar]
- 21.Habuchi T., Devlin J., Elder P.A., Knowles M.A. Detailed deletion mapping of chromosome 9q in bladder cancer: evidence for two tumour suppressor loci. Oncogene. 1995;11:1671–1674. [PubMed] [Google Scholar]
- 22.Simoneau M., Aboulkassim T.O., LaRue H., Rousseau F., Fradet Y. Four tumor suppressor loci on chromosome 9q in bladder cancer: evidence for two novel candidate regions at 9q22.3 and 9q31. Oncogene. 1999;18:157–163. doi: 10.1038/sj.onc.1202277. [DOI] [PubMed] [Google Scholar]
- 23.van Tilborg A.A., Groenfeld L.E., van der Kwast T.H., Zwarthoff E.C. Evidence for two candidate tumour suppressor loci on chromosome 9q in transitional cell carcinoma (TCC) of the bladder but no homozygous deletions in bladder tumour cell lines. Br. J. Cancer. 1999;80:489–494. doi: 10.1038/sj.bjc.6690383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knowles M.A., Habuchi T., Kennedy W., Cuthbert-Heavens D. Mutation spectrum of the 9q34 tuberous sclerosis gene TSC1 in transitional cell carcinoma of the bladder. Cancer Res. 2003;63:7652–7656. [PubMed] [Google Scholar]
- 25.Au K.S., Williams A.T., Roach E.S., Batchelor L., Sparagana S.P., Delgado M.R., Wheless J.W., Baumgartner J.E., Roa B.B., Wilson C.M., et al. Genotype/phenotype correlation in 325 individuals referred for a diagnosis of tuberous sclerosis complex in the United States. Genet. Med. 2007;9:88–100. doi: 10.1097/gim.0b013e31803068c7. [DOI] [PubMed] [Google Scholar]
- 26.Jansen F.E., Braams O., Vincken K.L., Algra A., Anbeek P., Jennekens-Schinkel A., Halley D., Zonnenberg B.A., van den Ouweland A., van Huffelen A.C., et al. Overlapping neurologic and cognitive phenotypes in patients with TSC1 or TSC2 mutations. Neurology. 2007;70:908–915. doi: 10.1212/01.wnl.0000280578.99900.96. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H., Nanba E., Yamamoto T., Ninomiya H., Ohno K., Mizuguchi M., Takeshita K. Mutational analysis of TSC1 and TSC2 genes in Japanese patients with tuberous sclerosis complex. J. Hum. Genet. 1999;44:391–396. doi: 10.1007/s100380050185. [DOI] [PubMed] [Google Scholar]
- 28.Yamashita Y., Ono J., Okada S., Wataya-Kaneda M., Yoshikawa K., Nishizawa M., Hirayama Y., Kobayashi E., Seyama K., Hino O. Analysis of all exons of TSC1 and TSC2 genes for germline mutations in Japanese patients with tuberous sclerosis: report of 10 mutations. Am. J. Med. Genet. 2000;90:123–126. [PubMed] [Google Scholar]
- 29.Astrinidis A., Senapedis W., Henske E.P. Hamartin, the tuberous sclerosis complex 1 gene product, interacts with polo-like kinase 1 in a phosphorylation-dependent manner. Hum. Mol. Genet. 2006;15:287–297. doi: 10.1093/hmg/ddi444. [DOI] [PubMed] [Google Scholar]
- 30.Henikoff S., Henikoff J.G. Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benvenuto G., Li S., Brown S.J., Braverman R., Vass W.C., Cheadle J.P., Halley D.J., Sampson J.R., Wienecke R., DeClue J.E. The tuberous sclerosis-1 (TSC1) gene product hamartin suppresses cell growth and augments the expression of the TSC2 product tuberin by inhibiting its ubiquitination. Oncogene. 2000;19:6306–6316. doi: 10.1038/sj.onc.1204009. [DOI] [PubMed] [Google Scholar]
- 32.Chong-Kopera H., Inoki K., Li Y., Zhu T., Garcia-Gonzalo F.R., Rosa J.L., Guan K.L. TSC1 stabilizes TSC2 by inhibiting the interaction between TSC2 and the HERC1 ubiquitin ligase. J. Biol. Chem. 2006;281:8313–8316. doi: 10.1074/jbc.C500451200. [DOI] [PubMed] [Google Scholar]
- 33.Goncharova E.A., Goncharov D.A., Eszterhas A., Hunter D.S., Glassberg M.K., Yeung R.S., Walker C.L., Noonan D., Kwiatkowski D.J., Chou M.M., et al. Tuberin regulates p70 S6 kinase activation and ribosomal protein S6 phosphorylation. A role for the TSC2 tumor suppressor gene in pulmonary lymphangioleiomyomatosis (LAM) J. Biol. Chem. 2002;277:30958–30967. doi: 10.1074/jbc.M202678200. [DOI] [PubMed] [Google Scholar]
- 34.Kwiatkowski D.J., Zhang H., Bandura J.L., Heiberger K.M., Glogauer M., el-Hashemite N., Onda H. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum. Mol. Genet. 2002;11:525–534. doi: 10.1093/hmg/11.5.525. [DOI] [PubMed] [Google Scholar]
- 35.Onda H., Crino P.B., Zhang H., Murphey R.D., Rastelli L., Gould Rothberg B.E., Kwiatkowski D.J. Tsc2 null murine neuroepithelial cells are a model for human tuber giant cells, and show activation of an mTOR pathway. Mol. Cell. Neurosci. 2002;21:561–574. doi: 10.1006/mcne.2002.1184. [DOI] [PubMed] [Google Scholar]
- 36.Buratti E., Chivers M., Kralovicova J., Romano M., Baralle M., Krainer A.R., Vorechovsky I. Aberrant 5′ splice sites in human disease genes: mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res. 2007;35:4250–4263. doi: 10.1093/nar/gkm402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cartegni L., Krainer A.R. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 38.Nakai K., Sakamoto H. Construction of a novel database containing aberrant splicing mutations of mammalian genes. Gene. 1994;141:171–177. doi: 10.1016/0378-1119(94)90567-3. [DOI] [PubMed] [Google Scholar]
- 39.Nellist M., van Slegtenhorst M.A., Goedbloed M., van den Ouweland A.M., Halley D.J., van der Sluijs P. Characterization of the cytosolic tuberin-hamartin complex. Tuberin is a cytosolic chaperone for hamartin. J. Biol. Chem. 1999;274:35647–35652. doi: 10.1074/jbc.274.50.35647. [DOI] [PubMed] [Google Scholar]
- 40.Wei J., Li P., Chiriboga L., Mizuguchi M., Yee H., Miller D.C., Greco M.A. Tuberous sclerosis in a 19-week fetus: immunohistochemical and molecular study of hamartin and tuberin. Pediatr. Dev. Pathol. 2002;5:448–464. doi: 10.1007/s10024-001-0210-3. [DOI] [PubMed] [Google Scholar]
- 41.Yamamoto Y., Jones K.A., Mak B.C., Muehlenbachs A., Yeung R.S. Multicompartmental distribution of the tuberous sclerosis gene products, hamartin and tuberin. Arch. Biochem. Biophys. 2002;404:210–217. doi: 10.1016/s0003-9861(02)00300-4. [DOI] [PubMed] [Google Scholar]
- 42.Clements D., Mayer R.J., Johnson S.R. Subcellular distribution of the TSC2 gene product tuberin in human airway smooth muscle cells is driven by multiple localization sequences and is cell-cycle dependent. Am. J. Physiol. Lung Cell. Mol. Physiol. 2007;292:L258–L266. doi: 10.1152/ajplung.00354.2005. [DOI] [PubMed] [Google Scholar]
- 43.Rosner M., Freilinger A., Hengstschlager M. Akt regulates nuclear/cytoplasmic localization of tuberin. Oncogene. 2007;26:521–531. doi: 10.1038/sj.onc.1209812. [DOI] [PubMed] [Google Scholar]
- 44.York B., Lou D., Noonan D.J. Tuberin nuclear localization can be regulated by phosphorylation of its carboxyl terminus. Mol. Cancer Res. 2006;4:885–897. doi: 10.1158/1541-7786.MCR-06-0056. [DOI] [PubMed] [Google Scholar]
- 45.Ali J.B., Sepp T., Ward S., Green A.J., Yates J.R. Mutations in the TSC1 gene account for a minority of patients with tuberous sclerosis. J. Med. Genet. 1998;35:969–972. doi: 10.1136/jmg.35.12.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwiatkowska J., Jozwiak S., Hall F., Henske E.P., Haines J.L., McNamara P., Braiser J., Wigowska-Sowinska J., Kasprzyk-Obara J., Short M.P., et al. Comprehensive mutational analysis of the TSC1 gene: observations on frequency of mutation, associated features, and nonpenetrance. Ann. Hum. Genet. 1998;62:277–285. doi: 10.1046/j.1469-1809.1998.6240277.x. [DOI] [PubMed] [Google Scholar]
- 47.Niida Y., Lawrence-Smith N., Banwell A., Hammer E., Lewis J., Beauchamp R.L., Sims K., Ramesh V., Ozelius L. Analysis of both TSC1 and TSC2 for germline mutations in 126 unrelated patients with tuberous sclerosis. Hum. Mutat. 1999;14:412–422. doi: 10.1002/(SICI)1098-1004(199911)14:5<412::AID-HUMU7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 48.Young J., Povey S. The genetic basis of tuberous sclerosis. Mol. Med. Today. 1998;4:313–319. doi: 10.1016/s1357-4310(98)01245-3. [DOI] [PubMed] [Google Scholar]
- 49.Dabora S.L., Jozwiak S., Franz D.N., Roberts P.S., Nieto A., Chung J., Choy Y.S., Reeve M.P., Thiele E., Egelhoff J.C., et al. Mutational analysis in a cohort of 224 tuberous sclerosis patients indicates increased severity of TSC2, compared with TSC1, disease in multiple organs. Am. J. Hum. Genet. 2001;68:64–80. doi: 10.1086/316951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones A.C., Shyamsundar M.M., Thomas M.W., Maynard J., Idziaszczyk S., Tomkins S., Sampson J.R., Cheadle J.P. Comprehensive mutation analysis of TSC1 and TSC2-and phenotypic correlations in 150 families with tuberous sclerosis. Am. J. Hum. Genet. 1999;64:1305–1315. doi: 10.1086/302381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Slegtenhorst M., Verhoef S., Tempelaars A., Bakker L., Wang Q., Wessels M., Bakker R., Nellist M., Lindhout D., Halley D., et al. Mutational spectrum of the TSC1 gene in a cohort of 225 tuberous sclerosis complex patients: no evidence for genotype-phenotype correlation. J. Med. Genet. 1999;36:285–289. [PMC free article] [PubMed] [Google Scholar]
- 52.Nellist M., Verhaaf B., Goedbloed M.A., Reuser A.J., van den Ouweland A.M., Halley D.J. TSC2 missense mutations inhibit tuberin phosphorylation and prevent formation of the tuberin-hamartin complex. Hum. Mol. Genet. 2001;10:2889–2898. doi: 10.1093/hmg/10.25.2889. [DOI] [PubMed] [Google Scholar]
- 53.Srebrow A., Kornblihtt A.R. The connection between splicing and cancer. J. Cell Sci. 2006;119:2635–2641. doi: 10.1242/jcs.03053. [DOI] [PubMed] [Google Scholar]
- 54.Buratti E., Baralle M., Baralle F.E. Defective splicing, disease and therapy: searching for master checkpoints in exon definition. Nucleic Acids Res. 2006;34:3494–3510. doi: 10.1093/nar/gkl498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krawczak M., Thomas N.S., Hundrieser B., Mort M., Wittig M., Hampe J., Cooper D.N. Single base-pair substitutions in exon–intron junctions of human genes: nature, distribution, and consequences for mRNA splicing. Hum. Mutat. 2007;28:150–158. doi: 10.1002/humu.20400. [DOI] [PubMed] [Google Scholar]
- 56.Baud E., Catilina P., Bignon Y.J. p16 involvement in primary bladder tumors: analysis of deletions and mutations. Int. J. Oncol. 1999;14:441–445. doi: 10.3892/ijo.14.3.441. [DOI] [PubMed] [Google Scholar]
- 57.Orlow I., Lacombe L., Hannon G.J., Serrano M., Pellicer I., Dalbagni G., Reuter V.E., Zhang Z.F., Beach D., Cordon-Cardo C. Deletion of the p16 and p15 genes in human bladder tumors. J. Natl Cancer Inst. 1995;87:1524–1529. doi: 10.1093/jnci/87.20.1524. [DOI] [PubMed] [Google Scholar]
- 58.Greenman C., Stephens P., Smith R., Dalgliesh G.L., Hunter C., Bignell G., Davies H., Teague J., Butler A., Stevens C., et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cliby W., Ritland S., Hartmann L., Dodson M., Halling K.C., Keeney G., Podratz K.C., Jenkins R.B. Human epithelial ovarian cancer allelotype. Cancer Res. 1993;53:2393–2398. [PubMed] [Google Scholar]
- 60.Devlin J., Elder P.A., Gabra H., Steel C.M., Knowles M.A. High frequency of chromosome 9 deletion in ovarian cancer: evidence for three tumour-suppressor loci. Br. J. Cancer. 1996;73:420–423. doi: 10.1038/bjc.1996.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Girard L., Zochbauer-Muller S., Virmani A.K., Gazdar A.F., Minna J.D. Genome-wide allelotyping of lung cancer identifies new regions of allelic loss, differences between small cell lung cancer and non-small cell lung cancer, and loci clustering. Cancer Res. 2000;60:4894–4906. [PubMed] [Google Scholar]
- 62.Lo K.W., Teo P.M., Hui A.B., To K.F., Tsang Y.S., Chan S.Y., Mak K.F., Lee J.C., Huang D.P. High resolution allelotype of microdissected primary nasopharyngeal carcinoma. Cancer Res. 2000;60:3348–3353. [PubMed] [Google Scholar]
- 63.Okada S., Tsuda H., Takarabe T., Yoshikawa H., Taketani Y., Hirohashi S. Allelotype analysis of common epithelial ovarian cancers with special reference to comparison between clear cell adenocarcinoma with other histological types. Jpn. J. Cancer Res. 2002;93:798–806. doi: 10.1111/j.1349-7006.2002.tb01322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suzuki K., Ogura T., Yokose T., Nagai K., Mukai K., Kodama T., Nishiwaki Y., Esumi H. Loss of heterozygosity in the tuberous sclerosis gene associated regions in adenocarcinoma of the lung accompanied by multiple atypical adenomatous hyperplasia. Int. J. Cancer. 1998;79:384–389. doi: 10.1002/(sici)1097-0215(19980821)79:4<384::aid-ijc13>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 65.Wistuba I.I., Maitra A., Carrasco R., Tang M., Troncoso P., Minna J.D., Gazdar A.F. High resolution chromosome 3p, 8p, 9q and 22q allelotyping analysis in the pathogenesis of gallbladder carcinoma. Br. J. Cancer. 2002;87:432–440. doi: 10.1038/sj.bjc.6600490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parry L., Maynard J.H., Patel A., Clifford S.C., Morrissey C., Maher E.R., Cheadle J.P., Sampson J.R. Analysis of the TSC1 and TSC2 genes in sporadic renal cell carcinomas. Br. J. Cancer. 2001;85:1226–1230. doi: 10.1054/bjoc.2001.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parry L., Maynard J.H., Patel A., Hodges A.K., von Deimling A., Sampson J.R., Cheadle J.P. Molecular analysis of the TSC1 and TSC2 tumour suppressor genes in sporadic glial and glioneuronal tumours. Hum. Genet. 2000;107:350–356. doi: 10.1007/s004390000390. [DOI] [PubMed] [Google Scholar]
- 68.Takamochi K., Ogura T., Suzuki K., Kawasaki H., Kurashima Y., Yokose T., Ochiai A., Nagai K., Nishiwaki Y., Esumi H. Loss of heterozygosity on chromosomes 9q and 16p in atypical adenomatous hyperplasia concomitant with adenocarcinoma of the lung. Am. J. Pathol. 2001;159:1941–1948. doi: 10.1016/S0002-9440(10)63041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takamochi K., Ogura T., Yokose T., Ochiai A., Nagai K., Nishiwaki Y., Suzuki K., Esumi H. Molecular analysis of the TSC1 gene in adenocarcinoma of the lung. Lung Cancer. 2004;46:271–281. doi: 10.1016/j.lungcan.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 70.Fang Y., Guan X., Guo Y., Sham J., Deng M., Liang Q., Li H., Zhang H., Zhou H., Trent J. Analysis of genetic alterations in primary nasopharyngeal carcinoma by comparative genomic hybridization. Genes Chromosomes Cancer. 2001;30:254–260. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1086>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 71.Nakayama K., Konno M., Kanzaki A., Morikawa T., Miyashita H., Fujioka T., Uchida T., Miyazaki K., Takao S., Aikou T., et al. Allelotype analysis of gallbladder carcinoma associated with anomalous junction of pancreaticobiliary duct. Cancer Lett. 2001;166:135–141. doi: 10.1016/s0304-3835(01)00436-0. [DOI] [PubMed] [Google Scholar]
- 72.Sarkar S., Julicher K.P., Burger M.S., Della Valle V., Larsen C.J., Yeager T.R., Grossman T.B., Nickells R.W., Protzel C., Jarrard D.F., et al. Different combinations of genetic/epigenetic alterations inactivate the p53 and pRb pathways in invasive human bladder cancers. Cancer Res. 2000;60:3862–3871. [PubMed] [Google Scholar]
- 73.Yeager T.R., DeVries S., Jarrard D.F., Kao C., Nakada S.Y., Moon T.D., Bruskewitz R., Stadler W.M., Meisner L.F., Gilchrist K.W., et al. Overcoming cellular senescence in human cancer pathogenesis. Genes Dev. 1998;12:163–174. doi: 10.1101/gad.12.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bean M.A., Pees H., Fogh J.E., Grabstald H., Oettgen H.F. Cytotoxicity of lymphocytes from patients with cancer of the urinary bladder: detection by a 3-H-proline microcytotoxicity test. Int. J. Cancer. 1974;14:186–197. doi: 10.1002/ijc.2910140207. [DOI] [PubMed] [Google Scholar]
- 75.Southgate J., Hutton K.A., Thomas D.F., Trejdosiewicz L.K. Normal human urothelial cells in vitro: proliferation and induction of stratification. Lab. Invest. 1994;71:583–594. [PubMed] [Google Scholar]
- 76.Chapman E.J., Hurst C.D., Pitt E., Chambers P., Aveyard J.S., Knowles M.A. Expression of hTERT immortalises normal human urothelial cells without inactivation of the p16/Rb pathway. Oncogene. 2006;25:5037–5045. doi: 10.1038/sj.onc.1209513. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.