Abstract
BACKGROUND
RNA interference offers clinical potential as a therapeutic modality for a variety of diseases; the efficacy of in vivo delivery remains poorly understood. The purpose of our study was to compare and contrast siRNA uptake in vivo utilizing various delivery techniques.
METHODS
DY547- and rhodamine-labeled siRNA was administered to mice by one of four delivery methods: 1) hydrodynamic intravenous (IV) tail vein injection, 2) standard IV tail vein injection, 3) intraperitoneal (IP) injection, and 4) rectal (PR) administration. Mice were sacrificed over a time course; representative tissue samples were collected and analyzed using fluorescent microscopy to determine siRNA uptake.
RESULTS
siRNA uptake was noted in the liver, kidney, pancreas, spleen and bone marrow by both hydrodynamic and standard IV injection. siRNA uptake was detected in the spleen, liver and bone marrow following IP administration. PR administration resulted in siRNA uptake in the spleen, bone marrow and colon.
CONCLUSION
Our results demonstrate differential siRNA uptake depending on delivery technique. Importantly, our results demonstrate the potential of siRNA as a systemic therapeutic option in vivo for selected disease processes.
Keywords: siRNA, hydrodynamic IV injection, delivery methods
INTRODUCTION
RNA interference (RNAi) is a highly conserved, naturally occurring mechanism of gene suppression found in plants, yeast and mammalian cells. Fire et al1 initially identified double-stranded RNA (dsRNA)-mediated gene silencing in nematodes; subsequent reports have demonstrated functional gene silencing in a variety of organisms.2 Since its discovery, RNAi has evolved as a valuable biological tool for gene identification and studying post-transcriptional gene function. Furthermore, RNAi has been proposed as a novel therapeutic modality in the treatment of a variety of inflammatory and neoplastic conditions. Since many diseases are fundamentally gene-based, targeting RNA by RNAi offers great therapeutic potential.
RNAi is mediated by naturally occurring endogenous or synthetic exogenous short interfering RNAs (siRNAs) which typically consist of 19- to 28-nucleotide duplexes with 2- to 4-nucleotide overhangs at the 3′ ends.3-5 Initially, long dsRNA is recognized and cleaved by the RNase III enzyme Dicer into 21- to 24-nucleotide duplexes of siRNA.3-5 The antisense siRNA strand is incorporated into the RNA-induced silencing complex (RISC) which then targets complementary mRNA sequences in the cytoplasm. mRNA is then cleaved by RNase at sites not bound by siRNA thus preventing protein translation.
RNAi therapy offers a number of unique features over traditional chemical inhibitors including a high degree of sequence specificity, high efficacy and the potential to target a multitude of genes.4, 5 However, one major hurdle to the clinical use of siRNA-based therapies has been its delivery to target cells. Although unprotected, or naked siRNA is able to penetrate cell membranes when injected locally, it is rapidly degraded by plasma nucleases and excreted by the kidney with systemic in vivo delivery. Recent studies have focused on modifying siRNA for tissue delivery including the use of viral and bacterial vectors, nanoparticle technology and liposomal envelopes. Additional concerns have been raised with regards to off-target and nonspecific side effects with the use of systemic siRNA therapy. Gene silencing in non-target tissues has the potential to perturb normal cellular functions leading to undesirable side effects. Most studies using siRNA to date have focused on local delivery to target organs. Reich et al6 demonstrated effective siRNA targeting of vascular endothelial growth factor (VEGF) in the retina as a potential treatment for age-related macular degeneration. Zhang et al7 and Bitko et al8 in separate studies reported effective inhibition of the respiratory syncytial virus (RSV) by intranasal siRNA delivery. Finally, Dorn et al9 demonstrated effective treatment of chronic neuropathic pain by intrathecal administration of siRNA. Despite these reports of successful site-specific effects of siRNA, few investigators have reported systemic delivery of siRNA in vivo.
McCaffrey et al10 first reported the use of hydrodynamic IV tail vein injections for the delivery of siRNA to the liver. Subsequently, our laboratory was among the first to report effective siRNA delivery and gene silencing in vivo utilizing this technique11, 12. However, little is known regarding the efficacy of other siRNA delivery methods. Therefore the purpose of our current study was to compare and contrast siRNA uptake in vivo utilizing one of four delivery methods: 1) hydrodynamic IV tail vein injection, 2) standard IV tail vein injection, 3) intraperitoneal (IP) injection and 4) rectal administration.
MATERIALS AND METHODS
siRNA, reagents and antibodies
siSTABLE in vivo SMARTpool siRNA and regular SMARTpool reagents for DY547-labeled non-target control (NTC) and p85α siRNA duplexes were designed and synthesized by Customer SMARTpool siRNA Design from Dharmacon (Lafayette, CO). siSTABLE in vivo duplex is chemically modified to extend siRNA stability in vivo. TransIT In Vivo Gene Delivery System for p85α was purchased from Mirus (Madison, WI). DOTAP liposomal transfection reagent was obtained from Roche Diagnostics (Indianapolis, IN). Hoescht stain and all other molecular biology grade reagents were purchased from Sigma-Aldrich (St. Louis, MO).
Animals and experimental design
Female Swiss-Webster mice (4-6 wks old; 20-25 grams) were obtained from Harlan-Sprague Dawley (Indianapolis, IN) and housed in an environment with controlled temperature (22°C), humidity, and a 12 h light/dark cycle. The mice were fed standard chow (Formula Chow 5008; Purina Mills, St. Louis, MO) and tap water ad libitum and allowed to acclimate for 1 wk. All studies were approved by the Institutional Animal Care and Use Committee of UTMB.
Animals were randomized into 20 groups to receive DY547-labeled NTC (2 mice/group) or Rhodamine-labeled p85α (3 mice/group) siSTABLE siRNA (20μg/mouse) mixed with DOTAP liposomal transfection reagent via one of four delivery methods: 1) hydrodynamic IV tail vein (1000μl), 2) standard IV tail vein (200μl), 3) IP injection (200μl) or 4) administration per rectum (PR) (100μl). Control mice (1 mouse/group) were treated with Hanks Buffered Saline mixed with DOTAP and delivered by one of the four delivery methods. Briefly, for hydrodynamic and standard IV injection, the lateral tail vein was cannulated with a 27 gauge needle as we have previously described.12 IP injections were administered by injecting siRNA into the right lower abdominal compartment to avoid organ injury. PR administration was accomplished by using a 25 gauge angiocatheter into the rectum. Mice were sacrificed at 4 and 24 h after DY547-labeled NTC siRNA injection and 24 h after Rhodamine-labeled p85α siRNA injection. Brain, heart, lung, liver, spleen, stomach, pancreas, small intestine, colon, adrenals, kidney, bone marrow and skin were immediately removed and flash frozen in liquid nitrogen. Tissues were stored at −80°C and analyzed within 1wk.
Tissue processing
Tissues were embedded in Optimal Cutting Temperature compound (OCT; Tissue Tek) and sections (5μm) were cut from the blocks and immediately fixed in cold 70% ethanol. Slides were then stained with Hoescht stain (1:5000 dilution) and washed in PBS. Tissue samples were then examined with an Olympus BX51 microscope (Olympus, Central Valley, PA) and images were processed using Olympus DP Manager and DP Controller software (Olympus, Central Valley, PA). Image fluorescence was assessed using Adobe Photoshop® v.8 software. Briefly, 3 separate images from each animal were merged and photographed from randomly selected tissue sections. Three images were then randomly selected and analyzed by red-channel pixel counts using imaging software. The number of red pixels was averaged for each tissue and delivery method and compared to control images of the same tissue. siRNA uptake was quantified as fold-change as compared to control treated animals.
RESULTS
siRNA tissue uptake following hydrodynamic intravenous (IV) injection
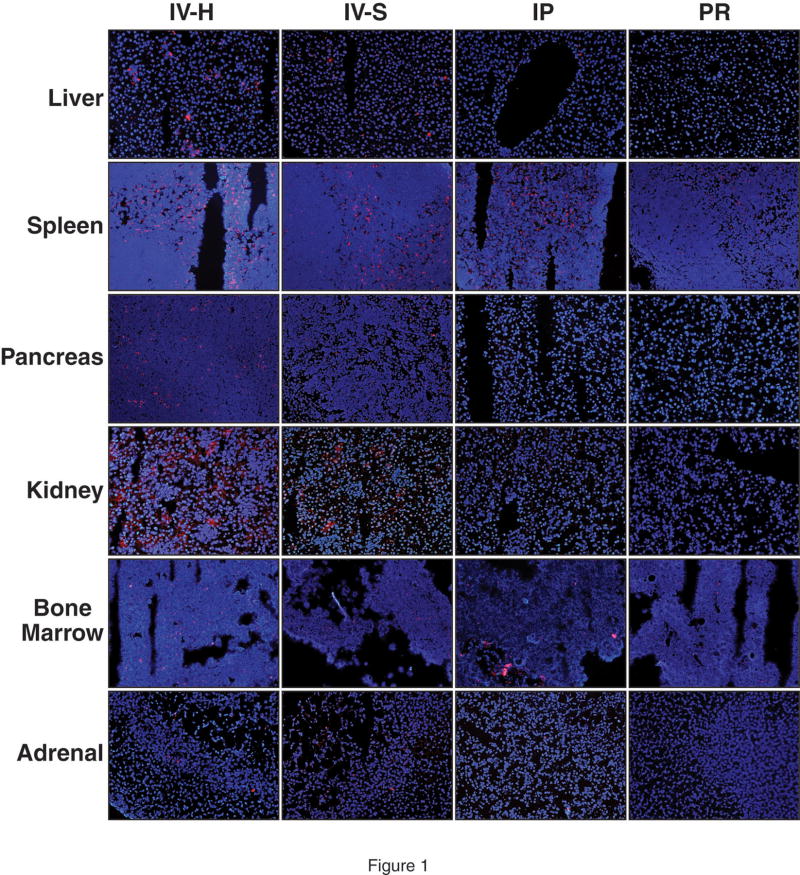
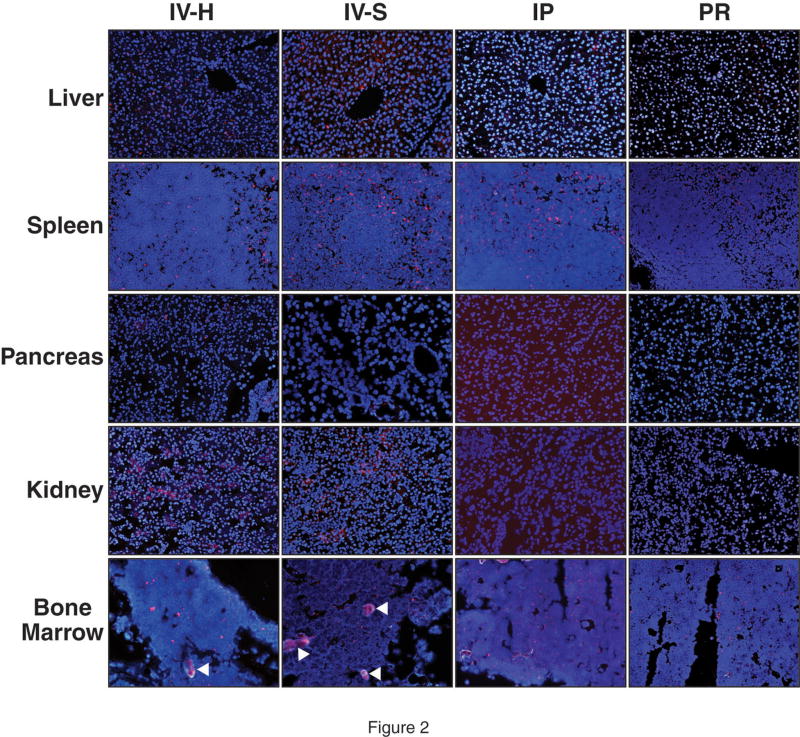
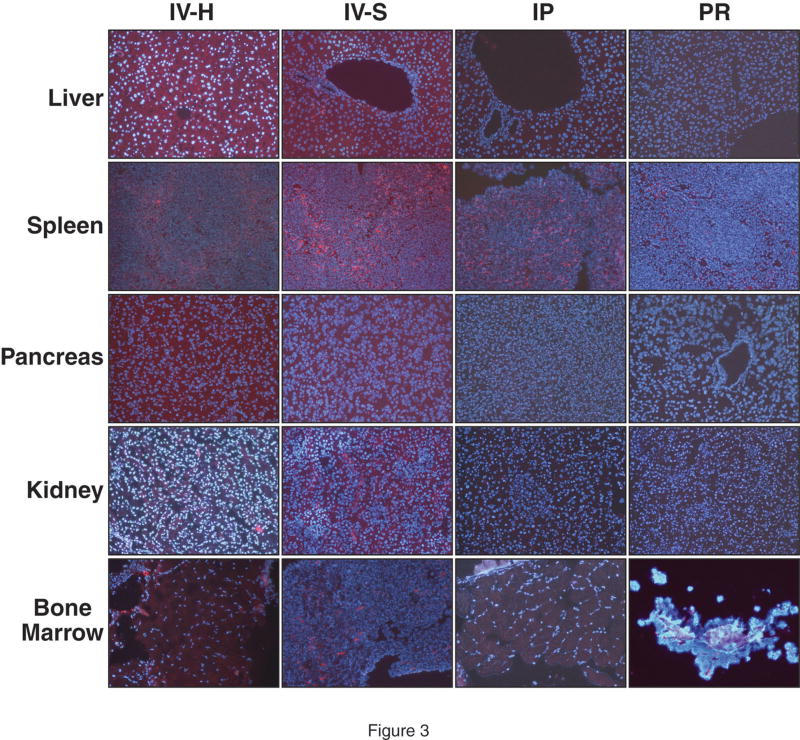
At 4h following hydrodynamic IV tail vein injection of DY547-labeled NTC, siRNA uptake was noted in the liver, spleen, pancreas, kidney, adrenal gland and bone marrow (Fig. 1). A strong fluorescent signal was noted in the liver, spleen and bone marrow whereas the pancreas and kidney showed weaker uptake. siRNA uptake in the liver was localized in a periportal distribution; there was diffuse distribution in the spleen and bone marrow. A weak fluorescent signal was detected in the adrenal gland, mainly localized in the cortex, when compared to control injections (Fig. 1). Table 1 summarizes the fold change in fluorescence as compared to control tissues. At 24h, DY547-labeled NTC siRNA uptake was again detected in the liver, spleen, kidney and bone marrow. No fluorescent signal was detect in the pancreas and adrenal gland at 24h (Fig. 2). Table 2 summarizes the fold change in fluorescence at 24h as compared to control tissues. Rhodamine-labeled p85α siRNA was utilized to further assess siRNA uptake in tissues at 24h and to allow for determination of protein suppression. Similar to DY547-labeled NTC siRNA, rhodamine-labeled p85α siRNA uptake was again detected in the liver, spleen, kidney and bone marrow (Fig. 3). Unlike DY547-labeled NTC treatment, rhodamine-labeled p85α siRNA uptake was detected in the pancreas at 24h. No uptake was noted in the brain, heart, lungs, stomach, small bowel, adrenal, colon or skin at either 4 or 24h following DY547-labeled NTC or rhodamine-labeled p85α siRNA treatment. Furthermore, no fluorescent signal was noted in any organ following control treatment.
Figure 1. Representative tissue uptake of DY547-labeled NTC siRNA at 4h compared by delivery method.
siRNA uptake, as represented by DY547 fluorescence (red), was analyzed 4h following delivery by one of four techniques. Cell nuclei (blue) were counterstained with Hoescht stain. Tissues samples from 13 different organ systems were collected at the time of animal sacrifice, flash frozen and frozen sectioned. Tissues were analyzed using fluorescent microscopy utilizing the same microscope settings for each set of tissues. Representative images of the liver, spleen, pancreas, kidney, bone marrow and adrenal are shown here (magnification = 200x for all images). Strong DY547 fluorescence was noted in the spleen by hydrodynamic, standard and intraperitoneal injection. Fluorescence was noted in kidney and adrenal tissue following both IV techniques only. Images were analyzed by the number of red channel pixels detected using Adobe Photoshop® v.8 software; images were compared to control treated tissues. (IV-H = hydrodynamic IV injection; IV-S = standard IV injection; IP = intraperitoneal administration; PR = rectal administration).
Table 1.
Comparison of DY547-labeled NTC siRNA tissue uptake based on fluorescent intensity at 4h as compared to control treated tissues.
| IV-Hydro | IV-Std. | IP | PR | |
|---|---|---|---|---|
| Liver | ++ | + | + | − |
| Spleen | +++ | ++ | +++ | + |
| Pancreas | + | + | − | − |
| Kidney | ++ | ++ | − | − |
| Bone Marrow | ++ | + | + | + |
| Adrenal gland | + | + | − | − |
(− = Absent uptake; + = ≥1.5 – 2 fold change as compared to control; ++ = ≥2 – 3 fold change; +++ = ≥3 fold change)
Figure 2. Representative tissue uptake of DY547-labeled NTC siRNA at 24h compared by delivery method.
siRNA uptake, as represented by DY547 fluorescence (red), was analyzed 24h following delivery by one of four techniques. Cell nuclei (blue) were counterstained with Hoescht stain. Tissues samples were again collected at the time of animal sacrifice, flash frozen and frozen sectioned. Tissues were analyzed using fluorescent microscopy utilizing the same microscope settings for each set of tissues. Representative images of the liver, spleen, pancreas, kidney and bone marrow are shown here (magnification = 200x for all images). DY547 fluorescence was detected in the liver and spleen with all four techniques. The liver showed moderate DY547 fluorescence in the liver with hydrodynamic and standard IV injection; weak uptake was noted with IP and PR administration. DY547 fluorescence was strongest in the spleen with each delivery method. The white arrowheads denote artifact/autofluorescence in the bone marrow images. Images were analyzed by the number of red channel pixels detected using Adobe Photoshop® v.8 software; images were compared to control treated tissues. (IV-H = hydrodynamic IV injection; IV-S = standard IV injection; IP = intraperitoneal administration; PR = rectal administration).
Table 2.
Comparison of DY547-labeled NTC siRNA tissue uptake based on fluorescent intensity at 24h as compared to control treated tissues.
| IV-Hydro | IV-Std. | IP | PR | |
|---|---|---|---|---|
| Liver | ++ | ++ | + | + |
| Spleen | +++ | ++ | ++ | ++ |
| Pancreas | − | − | − | − |
| Kidney | ++ | + | − | − |
| Bone Marrow | + | + | + | + |
(− = Absent uptake; + = ≥1.5 – 2 fold change as compared to control; ++ = ≥2 – 3 fold change; +++ = ≥3 fold change)
Figure 3. Representative tissue uptake of rhodamine-labeled p85α siRNA at 24h compared by delivery method.
siRNA uptake, as represented by rhodamine-fluorescence (red), was analyzed 24h following delivery by one of four techniques. Cell nuclei (blue) were counterstained with Hoescht stain. Tissues were collected and processed as previously described. Representative images of the liver, spleen, pancreas, kidney and bone marrow are shown here (magnification = 200x for all images). Rhodamine fluorescence was detected in the liver, spleen, pancreas, kidney and bone marrow following hydrodynamic and standard IV injection. Rhodamine fluorescence was detected in the liver and spleen following intraperitoneal injection. Minimal to no rhodamine fluorescence was detected in control tissues. (IV-H = hydrodynamic IV injection; IV-S = standard IV injection; IP = intraperitoneal administration; PR = rectal administration).
siRNA tissue uptake following standard IV tail vein injection
Similar to hydrodynamic IV injection, DY547-labeled siRNA uptake was noted in the liver, spleen, kidney and bone marrow at 4h; however, the overall signal was weaker (Fig. 1). Minimal DY547-labeled siRNA uptake was detected in the pancreas and adrenal. At 24h following DY547-labeled NTC siRNA injection, a fluorescent signal was still present in the liver and spleen; however, the fluorescent signal was diminished in the kidney and bone marrow (Fig. 2). Minimal to no uptake was noted in the pancreas at 24h. Rhodamine-labeled p85α siRNA fluorescence was detected in the liver, spleen, pancreas, kidney and bone marrow at 24h (Fig. 3). No siRNA signal was detected in other tissues or after control treatment.
siRNA tissue uptake following intraperitoneal (IP) injection
DY547-labeled NTC siRNA uptake was noted in the liver, spleen and bone marrow at 4h after IP injection (Fig. 1). The signal in the liver and bone marrow was weak however. At 24h, the fluorescent signal in the spleen remained strong and the signal in the bone marrow showed increased intensity (Fig. 2). The signal in the liver at 24h was minimal, but still present compared to control treatment. Similarly, rhodamine-labeled p85α siRNA uptake was noted in the liver, spleen and bone marrow (Fig. 3). The fluorescent signal in the liver was much stronger following rhodamine-labeled p85α treatment. No siRNA signal was detected in the remaining tissues or after control treatment.
siRNA tissue uptake following rectal (PR) administration
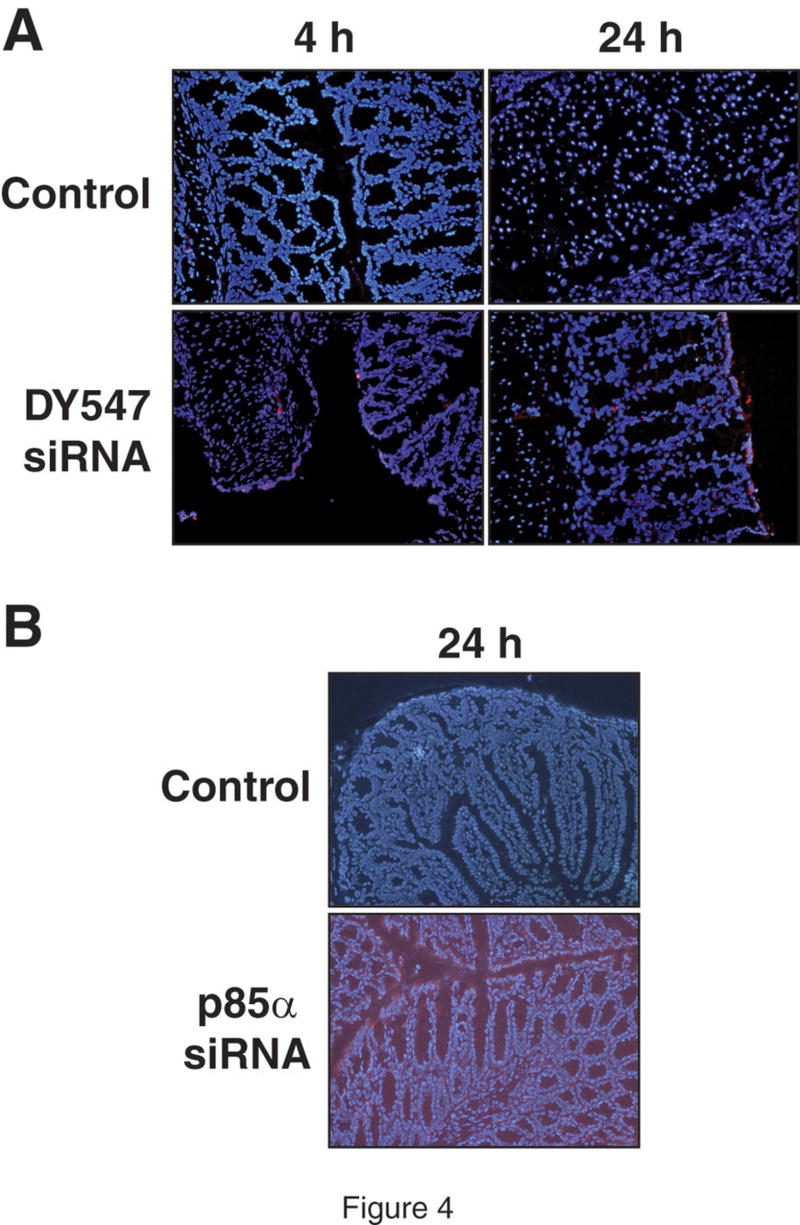
Rectal administration of DY547-labeled NTC siRNA at 4h demonstrated uptake in the spleen and minimal uptake in the bone marrow (Fig. 1) and colon (Fig. 4). At 24h, siRNA uptake was again detected in the spleen, bone marrow (Fig. 2) and the colon (Fig. 4) with a stronger fluorescent signal. Additionally, minimal siRNA uptake was detected in the liver at 24h. Weak rhodamine-labeled p85α siRNA uptake was detected in the spleen (Fig. 3) and colon (Fig. 4) at 24h. No siRNA uptake was noted in the remaining tissues or after control treatment.
Figure 4. siRNA uptake in the colon by rectal (PR) administration.
(A) Representative photographs of colon tissue at 4 and 24h following rectal administration of DY547-labeled NTC siRNA (magnification = 200x for all images). DY547 fluorescence (red) is noted in the colon at 4 and 24h as compared to control treatment (top row). Control tissue (no siRNA; DOTPA and HBS solution only) showed minimal to no fluorescence. Cell nuclei were counterstained with Hoescht stain and appear blue. Tissues were collected and processed as previously described. Images were analyzed by the number of red channel pixels detected using Adobe Photoshop® v.8 software; images were compared to control treated tissues. (B) Representative photographs of colon tissue at 24h following rectal administration of rhodamine-labeled p85α siRNA (magnification = 200x). Rhodamine fluorescence is noted in the colon in the epithelium as well as the crypts. No fluorescence was noted in the control treatment (no siRNA; DOTAP and HBS solution only). Cell nuclei (blue) were counterstaining with Hoescht stain.
DISCUSSION
RNA interference offers great clinical potential as a therapeutic modality especially for a variety of inflammatory and neoplastic conditions. However, one major obstacle remains effective systemic delivery. When administered into the bloodstream, siRNA is rapidly degraded by plasma nucleases and excreted by the kidneys.4 Efforts have focused on modifying siRNA for systemic tissue delivery utilizing various vectors and chemical modification. In our current study, we have utilized siRNA encapsulated in the cationic liposomal agent DOTAP to enhance tissue delivery. Sorensen et al15 previously demonstrated effective suppression of TNFα in vivo using DOTAP encapsulated siRNA. Higher levels of siRNA were noted in ovarian tumors when DOTAP was used compared to naked (unmodified) siRNA.16 Undoubtedly, the type of vehicle utilized for siRNA delivery will affect tissue uptake and will require further refinement before clinical use.
The aim of in vivo systemic delivery is to target specific tissues wile minimizing “off-target” or unintended tissue side effects. We have demonstrated effective delivery of DOTAP encapsulated siRNA to the murine liver, spleen, pancreas, kidney, adrenals and bone marrow by the use of hydrodynamic IV tail vein injections. Our laboratory has also utilized this method to deliver siRNA targeting the phosphatidylinositol 3-kinase (PI3K) pathway in the pancreas and in colon cancers.11, 12 Effective knockdown of gene expression by siRNA has been demonstrated in the murine liver and lung utilizing this delivery method.17-19 Hydrodynamic IV injections require large volumes administered at high pressure over a short period of time. One major criticism of this delivery method is the difficulty in applying this technique to human use. The large volume required and the rapidity of the injection could potentially result in volume overload.
Similar to our results with hydrodynamic IV administration, standard IV injection of siRNA resulted in uptake in the liver, spleen, pancreas, kidney, bone marrow and adrenals. However, the fluorescent signal was weaker in these tissues when compared to hydrodynamic IV injection. Previous studies have demonstrated success with standard IV injection of siRNA with tissue distribution noted in the liver, kidney, lung endothelium and jejunum.20-22 While we did not detect siRNA uptake in the lung or small intestine following standard IV injections, our results corroborate these finding in the liver and kidney. Our results, demonstrating effective uptake in the pancreas, are also consistent with the literature. Bradley et al23 recently reported successful in vivo delivery of siRNA to the pancreas by standard IV tail vein injections. To our knowledge, other investigators have not reported siRNA uptake in the adrenals or bone marrow by standard IV injection. Despite the weaker siRNA uptake with standard IV administration, these results suggest that this technique is still effective and may offer a potential route for systemic therapeutic use.
Few previous reports have focused on siRNA delivery via the intraperitoneal route. We have demonstrated strong siRNA uptake by the spleen and weak uptake by the liver and bone marrow by this method. Urban-Klein et al24 previously reported radioactive-labeled siRNA uptake in murine muscle and tumor tissue after IP injection; little uptake was noted in the liver or kidney. Landen et al16 demonstrated siRNA uptake in vivo in ovarian tumor tissue, liver, kidney, heart and lung tissue after IP delivery. However, these studies utilized a polyethylenimine/siRNA complex and the neutral liposomal carrier agent (DOPC), respectively, which may account for differences in tissue uptake noted in these studies. Apart from the noted uptake in the bone marrow, we did not observe any uptake in extra-abdominal organs by IP administration. IP administration may therefore be a potential strategy for treatment of intra-abdominal pathologies.
Similar to previous studies of local delivery, we have shown uptake of siRNA in the colon by rectal administration. Additionally, we noted uptake in the spleen and bone marrow with this method. There was minimal uptake of DY547-labeled NTC siRNA in the colon at 4h; the fluorescent signal was more intense at 24h. The spleen showed moderate uptake as early as 4h by PR administration and the signal remained strong at 24h suggesting rapid uptake by the spleen using a variety of systemic delivery methods. Zhang et al25 recently reported the effective use of PR-administered siRNA targeting TNFα in a murine colitis model. One potential problem with this delivery method is the volume of injection and the time of tissue exposure. More frequent administration may be required to achieve satisfactory colonic cell uptake by siRNA. Furthermore, utilizing different liposomal envelope carriers may also achieve greater tissue uptake. Similar to IP administration, the PR route demonstrated no significant extra-abdominal tissue delivery suggesting that it may be a potential therapeutic option for inflammatory and neoplastic conditions of the colon.
In summary, DY547-labeled NTC and rhodamine-labeled p85α siRNA uptake was noted in the liver, pancreas, spleen, kidney, adrenal and bone marrow following hydrodynamic and standard IV tail vein injection whereas IP injection showed strong uptake in the spleen and weaker uptake in the liver and bone marrow. PR administration resulted in uptake in the spleen with weak siRNA uptake in the colon and bone marrow. We detected minimal to no uptake in the brain, heart, lungs, stomach, small bowel or skin by any of the four methods tested. Importantly, our results demonstrate differential siRNA uptake in various organs depending on the delivery technique. These analyses are critical prior to the consideration of clinical studies utilizing systemic siRNA as treatment. With the development of more effective reagents to facilitate tissue uptake, it is anticipated that treatment strategies utilizing siRNA to knockdown gene expression will be an effective clinical option for certain disease processes.
Acknowledgments
The authors would like to thank Karen Martin for manuscript preparation and Linda Muehlberger and Rudy Salcedo for their assistance with specimen sectioning. This work was supported by grants RO1CA104748, RO1DK48498, PO1DK35608, and T32DK07639 from the National Institutes of Health and a Jeane B. Kempner Scholar award (to SDL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–8. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 3.Li CX, Parker A, Menocal E, Xiang S, Borodyansky L, Fruehauf JH. Delivery of RNA interference. Cell Cycle. 2006;5:2103–9. doi: 10.4161/cc.5.18.3192. [DOI] [PubMed] [Google Scholar]
- 4.Rychahou PG, Jackson LN, Farrow BJ, Evers BM. RNA interference: mechanisms of action and therapeutic consideration. Surgery. 2006;140:719–25. doi: 10.1016/j.surg.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Uprichard SL. The therapeutic potential of RNA interference. FEBS Lett. 2005;579:5996–6007. doi: 10.1016/j.febslet.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reich SJ, Fosnot J, Kuroki A, Tang W, Yang X, Maguire AM, et al. Small interfering RNA (siRNA) targeting VEGF effectively inhibits ocular neovascularization in a mouse model. Mol Vis. 2003;9:210–6. [PubMed] [Google Scholar]
- 7.Zhang W, Yang H, Kong X, Mohapatra S, San Juan-Vergara H, Hellermann G, et al. Inhibition of respiratory syncytial virus infection with intranasal siRNA nanoparticles targeting the viral NS1 gene. Nat Med. 2005;11:56–62. doi: 10.1038/nm1174. [DOI] [PubMed] [Google Scholar]
- 8.Bitko V, Musiyenko A, Shulyayeva O, Barik S. Inhibition of respiratory viruses by nasally administered siRNA. Nat Med. 2005;11:50–5. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- 9.Dorn G, Patel S, Wotherspoon G, Hemmings-Mieszczak M, Barclay J, Natt FJ, et al. siRNA relieves chronic neuropathic pain. Nucleic Acids Res. 2004;32:e49. doi: 10.1093/nar/gnh044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. RNA interference in adult mice. Nature. 2002;418:38–9. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- 11.Rychahou PG, Jackson LN, Silva SR, Rajaraman S, Evers BM. Targeted molecular therapy of the PI3K pathway: therapeutic significance of PI3K subunit targeting in colorectal carcinoma. Ann Surg. 2006;243:833–42. doi: 10.1097/01.sla.0000220040.66012.a9. discussion 43-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe H, Saito H, Rychahou PG, Uchida T, Evers BM. Aging is associated with decreased pancreatic acinar cell regeneration and phosphatidylinositol 3-kinase/Akt activation. Gastroenterology. 2005;128:1391–404. doi: 10.1053/j.gastro.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 13.Wang Q, Li N, Wang X, Kim MM, Evers BM. Augmentation of sodium butyrate-induced apoptosis by phosphatidylinositol 3′-kinase inhibition in the KM20 human colon cancer cell line. Clin Cancer Res. 2002;8:1940–7. [PubMed] [Google Scholar]
- 14.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 15.Sorensen DR, Leirdal M, Sioud M. Gene silencing by systemic delivery of synthetic siRNAs in adult mice. J Mol Biol. 2003;327:761–6. doi: 10.1016/s0022-2836(03)00181-5. [DOI] [PubMed] [Google Scholar]
- 16.Landen CN, Jr, Chavez-Reyes A, Bucana C, Schmandt R, Deavers MT, Lopez-Berestein G, et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65:6910–8. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- 17.Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–7. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- 18.Tompkins SM, Lo CY, Tumpey TM, Epstein SL. Protection against lethal influenza virus challenge by RNA interference in vivo. Proc Natl Acad Sci U S A. 2004;101:8682–6. doi: 10.1073/pnas.0402630101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zender L, Hutker S, Liedtke C, Tillmann HL, Zender S, Mundt B, et al. Caspase 8 small interfering RNA prevents acute liver failure in mice. Proc Natl Acad Sci U S A. 2003;100:7797–802. doi: 10.1073/pnas.1330920100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braasch DA, Paroo Z, Constantinescu A, Ren G, Oz OK, Mason RP, et al. Biodistribution of phosphodiester and phosphorothioate siRNA. Bioorg Med Chem Lett. 2004;14:1139–43. doi: 10.1016/j.bmcl.2003.12.074. [DOI] [PubMed] [Google Scholar]
- 21.Miyawaki-Shimizu K, Predescu D, Shimizu J, Broman M, Predescu S, Malik AB. siRNA-induced caveolin-1 knockdown in mice increases lung vascular permeability via the junctional pathway. Am J Physiol Lung Cell Mol Physiol. 2006;290:L405–13. doi: 10.1152/ajplung.00292.2005. [DOI] [PubMed] [Google Scholar]
- 22.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–8. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 23.Bradley SP, Kowalik TF, Rastellini C, da Costa MA, Bloomenthal AB, Cicalese L, et al. Successful incorporation of short-interfering RNA into islet cells by in situ perfusion. Transplant Proc. 2005;37:233–6. doi: 10.1016/j.transproceed.2004.12.181. [DOI] [PubMed] [Google Scholar]
- 24.Urban-Klein B, Werth S, Abuharbeid S, Czubayko F, Aigner A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther. 2005;12:461–6. doi: 10.1038/sj.gt.3302425. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Cristofaro P, Silbermann R, Pusch O, Boden D, Konkin T, et al. Engineering mucosal RNA interference in vivo. Mol Ther. 2006;14:336–42. doi: 10.1016/j.ymthe.2006.04.001. [DOI] [PubMed] [Google Scholar]