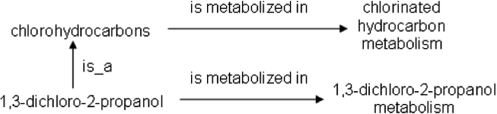Abstract
Motivation: Existing projects that focus on the semiautomatic addition of links between existing terms in the Open Biomedical Ontologies can take advantage of reasoners that can make new inferences between terms that are based on the added formal definitions and that reflect nonalignments between the linked terms. However, these projects require that these definitions be necessary and sufficient, a strong requirement that often does not hold. If such definitions cannot be added, the reasoners cannot point to the nonalignments through the suggestion of new inferences.
Results: We describe a methodology by which we have identified over 1900 instances of nonredundant nonalignments between terms from the Gene Ontology (GO) biological process (BP), cellular component (CC) and molecular function (MF) ontologies, Chemical Entities of Biological Interest (ChEBI) and the Cell Type Ontology (CL). Many of the 39.8% of these nonalignments whose object terms are more atomic than the subject terms are not currently examined in other ontology-enrichment projects due to the fact that the necessary and sufficient conditions required for the inferences are not currently examined. Analysis of the ratios of nonalignments to assertions from which the nonalignments were identified suggests that BP–MF, BP–BP, BP–CL and CC–CC terms are relatively well-aligned, while ChEBI–MF, BP–ChEBI and CC–MF terms are relatively not aligned well. We propose four ways to resolve an identified nonalignment and recommend an analogous implementation of our methodology in ontology-enrichment tools to identify types of nonalignments that are currently not detected.
Availability: The nonalignments discussed in this article may be viewed at http://compbio.uchsc.edu/Hunter_lab/Bada/nonalignments_2008_03_06.html. Code for the generation of these nonalignments is available upon request.
Contact: mike.bada@uchsc.edu
1 INTRODUCTION
Several efforts in recent years have focused on the semiautomatic addition of links between existing terms in the Open Biomedical Ontologies (OBOs) through the creation of formal definitions of these terms using more atomic terms, a process to which we refer as ontology enrichment. Of note, the Gene Ontology Next Generation (GONG) project first used the description-logic-based language DAML+OIL to formally define 250 Gene Ontology (GO) metabolism terms using MeSH terms (Wroe et al., 2003), and later OWL to formally define a much larger number of GO metabolism, binding and transport terms again using MeSH terms (Aranguren, 2004); this project has since evolved into the more general Biological Ontology Next Generation (BONG), which currently exists as a plugin to the Protege ontology editor. The Obol effort uses a series of Prolog production rules that can be used to decompose a given matching GO term into an Aristotelean genus (category) and one or more differentiae (necessary and sufficient conditions that differentiate the term from other terms of the same genus); the Gene Ontology Consortium is currently using Obol to generate Aristotelean definitions of OBO terms that refer to other OBO terms (Mungall, 2004). In our frame-based Protege ontology-enrichment effort, we have created over 9600 assertions linking terms in the GO (The Gene Ontology Consortium, 2000), Chemical Entities of Biological Interest (ChEBI) ontology (Degtyarenko, 2003), and the Cell Type Ontology (CL) (Bard et al., 2005); these base assertions have been integrated into this set of ontologies such that each assertion is consistent with all assertions made at more general levels (Bada and Hunter, 2007).
Both GONG and Obol have been able to take advantage of associated reasoners; for the former, an OWL reasoner can be used, while for the latter, the Aristotelean definitions can be imported into OBO-Edit (www.oboedit.org), the primary tool in which OBOs are developed, and its associated reasoner invoked. A great advantage of using such a reasoner is its ability to make new inferences derived from the added formal term definitions. For example, in the second published GONG study, using the newly added formal defintions for the GO molecular function (MF) terms neurotransmitter binding and glutamate binding (which use the MeSH terms Neurotransmitters and Glutamates, respectively), the OWL reasoner inferred that neurotransmitter binding subsumes glutamate binding, a link absent at that point in GO. However, both GONG/BONG and Obol/OBO-Edit require that these definitions use necessary and sufficient conditions in order for these inferences to be made. This is a strong requirement that does not hold bidirectionally in many, if not most cases: it is necessary and sufficient that catecholamine transport is a transport that results in the directed movement of a catecholamine. However, the semantics of OWL or OBO say that, for an existential restriction expressed for a subject class A linking it to an object class B via property p, each instance of A must have at least one value from B for p. Since we cannot say that every catecholamine takes part in a catecholamine-transport process, it is not even possible to make this a necessary assertion. Consequently, using terms from these two terminologies that have been linked, these new subsumptive inferences can only be made between subject terms for which necessary and sufficient definitions can be created (e.g. substance-transport terms) and not with the object terms (e.g. the substances that are being transported) used in these definitions.
The inferences that are made by these reasoners point to what we call nonalignments—subsets of terms that are linked (other than via is_a), but that are not aligned in that the terms of one side of the links are linked by subsumption while the terms of the other side are not. (The nonalignments we identify all consist of subject terms that are subsumptively linked and object terms that are not subsumptively linked.) For example, as can be seen in Figure 1, we have linked the ChEBI term chlorohydrocarbons to the GO term chlorinated hydrocarbon metabolism and also the ChEBI term 1,3-dichloro-2-propanol to the GO term 1,3-dichloro-2-propanol metabolism. These pairs of terms are not aligned in that 1,3-dichloro-2-propanol is subsumed by chlorohydrocarbons in ChEBI, but 1,3-dichloro-2-propanol metabolism is not subsumed by chlorinated hydrocarbon metabolism in GO. We expect the two sides to be aligned in that if 1,3-dichloro-2-propanol is indeed a kind of chlorohydrocarbon (as represented in ChEBI), then it should be metabolized in a kind of chlorinated-hydrocarbon metabolism—but 1,3-dichloro-2-propanol metabolism is not a kind of chlorinated-hydrocarbon metabolism (as represented in GO). In the nonalignments we identify, if the more specific subject entity (e.g. 1,3-dichloro-2-propanol) is indeed a kind of the more general subject entity (e.g. chlorohydrocarbons), then the assertion made for the more specific subject entity (e.g. that 1,3-dichloro-2-propanol can be metabolized in a 1,3-dichloro-2-propanol-metabolism process) should be subsumed by the assertion made for the more general subject entity (e.g. that a chlorohydrocarbon can be metabolized in a chlorinated-hydrocarbon-metabolism process).
Fig. 1.
The relationships between a pair of terms from ChEBI and another pair of terms from the GO BP ontology, the analysis of which an ontology nonalignment has been identified. Specifically, 1,3-dichloro-2-propanol is subsumed by chlorohydrocarbons in the former, but 1,3-dichloro-2-propanol metabolism is not subsumed by chlorinated hydrocarbon metabolism in the latter. This nonalignment was identified by analyzing the respective object classes of is metabolized in at the levels of 1,3-dichloro-2-propanol and of chlorohydrocarbons.
In this example, with necessary and sufficient definitions of chlorinated hydrocarbon metabolism and 1,3-dichloro-2-propanol metabolism in terms of chlorohydrocarbons and 1,3-dichloro-2-propanol, respectively, these reasoners would point to this nonalignment through the suggestion of an is_a link from 1,3-dichloro-2-propanol metabolism to chlorinated hydrocarbon metabolism. However, if instead 1,3-dichloro-2-propanol was not subsumed by chlorohydrocarbons and 1,3-dichloro-2-propanol metabolism was subsumed by chlorinated hydrocarbon metabolism, these reasoners would not be able to suggest an is_a link from 1,3-dichloro-2-propanol to chlorohydrocarbons, because the required necessary and sufficient definitions of 1,3-dichloro-2-propanol and chlorohydrocarbons in terms of 1,3-dichloro-2-propanol metabolism and chlorinated hydrocarbon metabolism, respectively, could not be created using these terms in an ontologically valid way. This is not a fault of OWL or of Aristotelean formalism; these representational systems have strict semantics, to which ontologists should adhere when making assertions. It is just that reasoners relying solely on necessary and sufficient definitions will likely miss many of these nonalignments because ontologically valid definitions cannot be created, and it is desirable that as many of these nonalignments as possible be rectified.
We have implemented our ontology-enrichment project in Protege-Frames (mainly because this is part of a larger frame-based effort). There is no associated reasoner to Protege-Frames, so we implemented a simple reasoning system to ensure the global consistency of the added assertions in our set of integrated ontologies. It is this same reasoning system we use here to discover nonalignments in the constituent ontologies through structural analysis of the assertions we added in our previous work (Bada and Hunter, 2007). Reasoning over these assertions, we were able to discover nearly 1700 instances of nonredundant nonalignments, 39.8% of which likely could not be identified via suggested inferences by OWL or OBO-Edit reasoners due to the fact that the required necessary and sufficient definitions could not be created in an ontologically valid way using these terms of the linked ontologies. We propose that those nonalignments for which such inferences cannot be made by these reasoners also be examined to increase consistency among the linked ontologies.
2 METHODS
The method by which we ensure the global consistency of the set of assertions to the ontologies is through an analysis of the object classes of the properties of the classes. Specifically, this analysis relies on the fact that the object expression (here, an object class or union of object classes) of a property at a given class level must be subsumed by the object expression of the property at higher (i.e. more general) class levels. Furthermore, the object expression of a given property must be subsumed by the object expression at higher property levels. Put more simply, object expressions should monotonically narrow as one descends to more specific classes and slots. In order for each assertion to be consistent with each assertion made at more general levels, any object class of a property at a given class level that was not subsumed by an object class at a higher class and/or property level such that these conditions were satisfied was appropriately propagated up the class and/or slot hierarchies. The full details of this procedure can be read in the initial publication of our OBO-enrichment work (Bada and Hunter, 2007).
Our methodology for discovering ontology nonalignments follows from this global consistency enforcement. For each base assertion (represented as a triple of a subject class, property and object class), each of the class's direct superclasses is checked to see if it is within the domain of the property. If so, it is checked if at least one of the object classes of the property of the superclass subsumes the object class of the property of the base assertion. If there is no such subsuming class, this is a nonalignment between the subject and object classes of the two assertions. If there is such a subsuming class at the level of this direct superclass, the same examination is performed for each of its direct superclasses. This continues recursively until either all direct superclasses are outside of the domain of the given property or a root of the ontology is reached.
This can be made clearer with a simple but real example. Consider the base assertion 1,3-dichloro-2-propanol is metabolized in 1,3-dichloro-2-propanol metabolism, which states that 1,3-dichloro-2-propanol can be metabolized in a 1,3-dichloro-2-propanol-metabolism process. The sole direct superclass of 1,3-dichloro-2-propanol-chlorohydrocarbons is obtained. It is checked that chlorohydrocarbons is within the domain of the slot is metabolized in, which is the case. The set of allowed classes of is metabolized in at the level of chlorohydrocarbons is then obtained, which is the single class chlorinated hydrocarbon metabolism (which indicates that a chlorohydrocarbon can be metabolized in a chlorinated-hydrocarbon-metabolism process). The set of allowed classes at the superclass level (the one-member set chlorinated hydrocarbon metabolism) should subsume the set of allowed classes at the base-assertion level (the one-member set 1,3-dichloro-2-propanol metabolism). However, it does not; this is thus a nonalignment. Figure 1 illustrates this example.
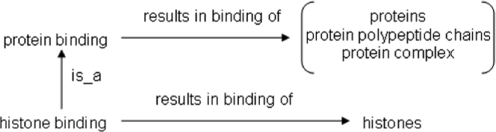
For each discovered nonalignment, we extracted four entities into which the nonalignment can be distilled: the subject class of the base assertion, the superclass of this subject class at the level of which the nonalignment was found, the object class of the base assertion (i.e. the allowed class of the assertion), and the set of object classes at the level of the superclass (i.e. the set of allowed classes for the slot at the level of the superclass). There is only one object class for each base assertion, while there can be more than one object class at the level of the superclass, since monotonicity as one travels down the class hierarchy is preserved as long as an object class of a property of a class is subsumed by at least one object class of the property of the superclass. Figure 2 illustrates another real example where the set of allowed classes at the level of the supeclass has more than one member. In this example, the set of object classes for results in binding of at the level of protein binding was assigned the set [proteins, protein polypeptide chains, protein complex]. Such a multiply membered set of object classes is represented as a union of classes, so this assertion indicates that a protein-binding process can result in the binding of either a protein, a protein polypeptide chain, or a protein complex. (This was done because the definition of protein binding is ‘interacting selectively with a protein or protein complex’.) However, relatively few terms so far have been assigned multiple allowed classes as in this example, so this is currently an exceptional case.
Fig. 2.
The relationships between terms from the GO BP ontology and ChEBI and the GO CC ontology, the analysis of which an ontology nonalignment has been identified. Specifically, histone binding is subsumed by protein binding in the former, but histones is not subsumed by proteins, protein polypeptide chains or protein complex in the latter. This nonalignment was identified by analyzing the respective object classes of results in binding of at the levels of histone binding and of protein binding.
Each stored nonalignment represented by the four summarizing entities was written out to a text file in the following format:
subject class of base assertion -> superclass of subject class
object class of base assertion !-> object-class set at level of superclass
This neatly summarizes the nonalignment by stating that the subject class of the base assertion is subsumed by the superclass, but the object class of the base assertion is not subsumed by any of the object classes at the level of the superclass. Thus, the nonalignment illustrated in Figure 1 is represented as:
1,3-dichloro-2-propanol -> chlorohydrocarbons
1,3-dichloro-2-propanol metabolism !-> chlorinated hydrocarbon metabolism
Such a representation makes clear the essence of the nonalignment—that 1,3-dichloro-2-propanol is subsumed by chlorohydrocarbons (in ChEBI), but 1,3-dichloro-2-propanol metabolism is not subsumed by chlorinated hydrocarbon metabolism (in the GO biological process (BP) ontology).
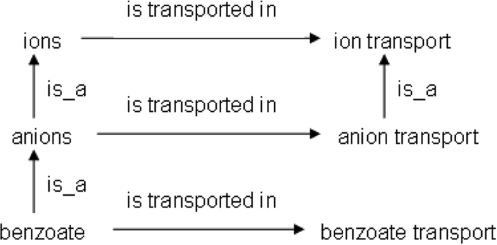
Due to the extensive multiple inheritance of the component ontologies, it is possible to discover redundant nonalignments or even the same nonalignment more than once. Only nonredundant nonalignments were stored and exported, as examining redundant nonalignments to assess whether there are true semantic discrepancies entails additional, unnecessary effort and biases statistics. Two nonalignments are redundant if the resolution of the one also results in the resolution of the other. Consider the following two nonalignments:
benzoate -> anions
benzoate transport !-> anion transport
benzoate -> ions
benzoate transport !-> ion transport
These two nonalignments are redundant with respect to one another. If the first nonalignment was resolved by adding an is_a link from benzoate transport to anion transport, the second nonalignment would also be resolved since this link addition would result in the implication that benzoate transport is a type of ion transport; thus, the second nonalignment would also be resolved. In cases of redundancy, we have kept the more specific nonalignment; thus, for the example above, only the first nonalignment was stored. The relevant relationships between the terms of these two nonalignments are illustrated in Figure 3.
Fig. 3.
The relationships between terms from ChEBI and the GO BP ontology, the analysis of which two redundant ontology nonalignments were identified. Specifically, benzoate is subsumed by anions in the former, but benzoate transport is not subsumed by anion transport in the latter. Also, benzoate is subsumed by ions in the former, but benzoate transport is not subsumed by ion transport in the latter.
The March 6, 2008 versions of GO, ChEBI and CL were used for this study. These base ontologies were previously enriched with 10 270 additional assertions linking the component terms using 50 specific relationships detailed in the initial publication of our OBO-enrichment work. It is important to note that although this study relies upon the links we created in our previously published ontology-enrichment work, our methodology for nonalignment identification is not limited by the specific relationships we chose to use. (The quality of the nonalignments, however, is dependent on the quality of the links that the methodology analyzes.) In fact, we have recently generated nonalignments based on links created by members of the OBO Consortium and have begun a discussion of ways of managing these nonalignments.
3 RESULTS
Using this methodology resulted in a total of 1938 nonredundant nonalignments within the set of GO, ChEBI and CL; this set of nonalignments can be examined at http://compbio.uchsc.edu/Hunter_lab/Bada/nonalignments_2008_03_06.html. To better characterize their distribution, we clustered the nonalignments according to the ontologies that were the sources of the subject and object terms of the nonalignments. For example, the nonalignment illustrated in Figure 1 is a ChEBI-to-BP nonalignment, since the subject terms (1,3-dichloro-2-propanol and chlorohydrocarbons) are from ChEBI and the object terms (1,3-dichloro-2-propanol metabolism and chlorinated hydrocarbon metabolism) are from the GO BP ontology. There is a slight complication in that the two sets of object terms of a nonalignment may be from different ontologies, but this is rare. In such a case, the object term of the base assertion is used for the classification of the nonalignment.
Table 1 lists the number of assertions and nonredundant nonalignments for each directed pairwise combination of ontologies for which there is at least one corresponding assertion. For example, there are 2710 total added assertions from a GO BP term to another GO BP term, and 94 nonredundant nonalignments were identified from these assertions. The numbers of nonalignments are largely symmetric. The biggest discrepancy is that between the 598 nonalignments identified from the BP-to-ChEBI assertions and the 1022 nonalignments identified from the ChEBI-to-BP assertions.
Table 1.
Numbers of assertions and nonredundant alignments for each directed combination of ontologies for which there is at least one added assertion
| Ontology to ontology | Assertions | Nonalignments |
|---|---|---|
| GO BP to GO BP | 2710 | 94 |
| GO BP to GO CC | 156 | 17 |
| GO BP to ChEBI | 3022 | 598 |
| GO BP to CL | 117 | 5 |
| GO BP to GO MF | 65 | 3 |
| GO CC to GO BP | 156 | 19 |
| GO CC to GO CC | 154 | 10 |
| GO CC to GO MF | 32 | 3 |
| ChEBI to GO BP | 3022 | 1022 |
| ChEBI to GO MF | 242 | 79 |
| CL to GO BP | 117 | 10 |
| GO MF to GO BP | 65 | 0 |
| GO MF to GO CC | 32 | 9 |
| GO MF to ChEBI | 242 | 69 |
Table 2 lists the numbers of assertions and nonredundant nonalignments and the ratio of nonalignments to assertions for each undirected pairwise combination of ontologies for which there is at least one corresponding assertion. The lowest ratios of nonalignments to assertions are those between BP terms and MF terms (0.02), between BP terms and BP terms (0.034), between BP terms and CL terms (0.064) and between cellular component (CC) terms and CC terms (0.065). This suggests that terms within these pairs of ontologies are relatively well-aligned. The highest ratios of nonalignments to assertions are those between ChEBI terms and MF terms (0.306), between BP terms and ChEBI terms (0.2680) and between CC terms and MF terms (0.19). This suggests that these pairs of ontologies are relatively not aligned well, which agrees with our empirical observations in our ontology-enrichment work that ChEBI is relatively not aligned well with GO.
Table 2.
Numbers of assertions and nonredundant alignments and the ratio of nonalignments to assertions for each undirected pairwise combination of ontologies for which there is at least one added assertion
| Ontology - ontology | Assertions | Nonalignments | Ratio |
|---|---|---|---|
| GO BP - GO BP | 2798 | 94 | 0.034 |
| GO BP - GO CC | 312 | 36 | 0.12 |
| GO BP - ChEBI | 6044 | 1620 | 0.2680 |
| GO BP - CL | 234 | 15 | 0.064 |
| GO BP - GO MF | 130 | 3 | 0.02 |
| GO CC - GO CC | 154 | 10 | 0.065 |
| GO CC - GO MF | 64 | 12 | 0.19 |
| ChEBI - GO MF | 484 | 148 | 0.306 |
Another way to characterize the nonalignments is whether the subject terms of the nonalignments are the more complex terms or the more atomic terms. For example, in the example illustrated in Figure 1, the subject terms (1,3-dichloro-2-propanol and chlorohydrocarbons) are more atomic than the object terms in that the latter are built up from the former. Conversely, in the example illustrated in Figure 2, the subject terms (protein binding and histone binding) are more complex than the object terms. As will be explained more fully in the next section, this characterization has important implications in that the new inferences made by the GONG/BONG and Obol projects correspond to the first type of nonalignment, in which the subject classes are more atomic, since ontologically valid necessary and sufficient definitions, which are required for these projects, can more easily be constructed in these cases. The second type of nonalignment includes all of the BP-to-CC, BP-to-ChEBI, BP-to-CL, BP-to-MF, MF-to-CC and MF-to-ChEBI nonalignments, while the BP-to-BP and CC-to-CC sets of nonalignments have mixtures of the two types of nonalignments. We have found that 772 (39.8%) of the 1938 nonredundant nonalignments are of the second type, thus showing that our methodology can identify a large number of nonalignments that may be missed by the reasoning methods of the other projects.
4 DISCUSSION
4.1 Evaluation and management of nonalignments
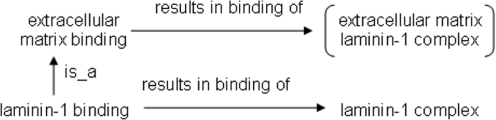
In this study, we have used the term nonalignment to refer to two analogous sets of entities such that one entity is subsumed by the other in the first pair while one entity is not subsumed by the other in the second pair. Upon examination of a given nonalignment, if it is determined that the pairs of entities should be aligned, we term this a discrepancy. Not all nonalignments are discrepancies; Figure 4 illustrates such an example. Here, laminin-1 binding is subsumed by extracellular matrix binding in the GO MF ontology, but laminin-1 complex is not subsumed by extracellular matrix in the GO CC ontology. Even though it is a nonalignment, we believe that this is not a discrepancy in that these pairs of terms should not be aligned; that is, laminin-1 binding is a type of extracellular-matrix binding, but the laminin-1 complex is not a type of extracellular matrix (but rather a component of the extracellular matrix). Nevertheless, we assert that a large majority of the nonalignments we have identified are indeed discrepancies.
Fig. 4.
The relationships between a pair of terms from the GO MF ontology and a pair of terms from the GO cellular-component ontology, the analysis of which an ontology nonalignment has been identified. We assert this is an example of nonalignment that is not a discrepancy in that the subsumption relationship between the subject terms and the lack of a subsumption relationship between the object terms appear to be valid.
If a given nonalignment is assessed to be a discrepancy, there are two ways to resolve it. The first is to add an is_a link from the object term of the base assertion to the object term at the superclass level (or, in the case of multiple object terms at the superclass level, to at least one of the object terms). For example, we assert the nonalignment illustrated in Figure 1 is a discrepancy: according to this model, a chlorohydrocarbon can only be metabolized in a chlorinated-hydrocarbon-metabolism process, but a molecule of 1,3-dichloro-2-propanol, which is a kind of chlorohydrocarbon (according to ChEBI), can only be metabolized in a 1,3-dichloro-2-propanol-metabolism process, which is not a kind of chlorinated-hydrocarbon-metabolism process (according to GO BP). One way to resolve this discrepancy is the addition of an is_a link from 1,3-dichloro-2-propanol metabolism to chlorinated hydrocarbon metabolism. With this addition, a molecule of 1,3-dichloro-2-propanol can be metabolized in a 1,3-dichloro-2-propanol-metabolism process, which is now a more specific kind of chlorinated-hydrocarbon-metabolism process.
The second way to resolve a discrepancy is the removal of the is_a link from the subject term of the base assertion to the subject term at the superclass level. In Figure 1, this corresponds to the removal of the is_a link from 1,3-dichloro-2-propanol to chlorohydrocarbons. With the removal of this link, 1,3-dichloro-2-propanol is no longer a more specific kind of chlorohydrocarbon, which aligns with the fact that a 1,3-dichoro-2-propanol-metabolism process is not a kind of a chlorinated-hydrocarbon-metabolism process.
In the case of a nonalignment that is not a discrepancy, there is still a logical inconsistency, and action should be taken to rectify the inconsistency. A general, automatic solution to such an inconsistency is the propagation of the object class of the base assertion up to the superclass level; this is the type of upward propagation we previously extensively employed in our ontology-enrichment work so as to ensure the global consistency of the ontologies when adding enriching assertions. For example, in Figure 4, we assert that neither of the two steps described in the previous paragraphs should be performed; however, there is still a logical inconsistency in that an extracellular-matrix-binding process results in the binding of an extracellular matrix, but a laminin-1-binding process, which is a kind of extracellular-matrix-binding process (according to GO MF), results in the binding of a laminin-1 complex, which is not an extracellular matrix (according to GO CC). (According to GO CC, laminin-1 complex is transitively part_of extracellular matrix.) The rectification we describe here consists of adding laminin-1 complex as an object class of results in binding of at the level of extracellular matrix binding; this is illustrated in Figure 5. The semantics of this new model are that an extracellular-matrix-binding process results in the binding of an extracellular matrix or a laminin-1 complex, while a laminin-1-binding process further restricts this to a laminin-1 complex.
Fig. 5.
The relationships between terms from the GO MF ontology and cellular-component ontologies in which the nonalignment identified in Figure 4 has been rectified by the propagation of laminin-1 complex. Specifically, laminin-1 complex has been added as an object class of results in binding of at the level of extracellular matrix binding.
A more elegant solution in this example is to instead add the GO CC term extracellular matrix part as an allowed class of results in binding of at the level of extracellular matrix binding; the semantics of this are that an extracellular-matrix-binding-process results in the binding of an extracellular matrix or an extracellular-matrix part, which seems to be a valid definition for extracellular matrix binding. The original nonalignment would be resolved in that laminin-1 complex at the level of laminin-1 binding would be subsumed by extracellular matrix part at the level of extracellular matrix binding. Though this is semantically closer to the definition of extracellular matrix binding, it is also more manual and thus more labor-intensive (which is not to say that it should not be done). Our methodology could be used to either automatically upwardly propagate the specific classes so as to make the ontologies consistent, as described in the previous paragraph, or it could be used to automatically make suggestions to the ontology curators, who would decide to add either the specific terms or more general terms (such as extracellular matrix part).
Of total of 1938, 100 nonredundant nonalignments were randomly selected for an evaluation. Out of these 100, 96 were assessed to be discrepancies; that is, we assert that they should be similarly aligned through the addition or removal of an is_a link, corresponding to the first two types of resolution. The remaining four nonalignments are analogous to the example seen in Figure 4, in which the subject and object terms should not be aligned; rather, the third type of resolution should be undertaken, in which an object term should be added to the higher-level assertion such that the lower-level assertion is subsumed, as seen in Figure 5.
4.2 Comparison to other projects
Both the GONG/BONG and Obol projects have been focusing on creating formal defintions of OBO terms using more atomic OBO terms in necessary and sufficient conditions. These definitions can then be reasoned over (by an OWL reasoner for the former and by the Obol reasoner or the OBO-Edit reasoner for the latter), which can make new inferences using the definitions. However, the reasoner can only make new inferences using the linked terms if ontologically valid necessary and sufficient definitions can be constructed. The type of inferences that can be made largely corresponds to the absent subsumptions in the type of nonalignments in which the subject terms are more atomic than the object terms. Figure 1 is such an example. Necessary and sufficient definitions could be produced for 1,3-dichloro-2-propanol metabolism (as a subclass of metabolism with a results in metabolism of 1,3-dichloro-2-propanol condition) and for chlorinated hydrocarbon metabolism (as a subclass of metabolism with a results in metabolism of chlorohydrocarbons condition). If the associated reasoner reasons over ChEBI and GO (including these added definitions), given that 1,3-dichloro-2-propanol is subsumed by chlorohydrocarbons as in Figure 1, it can infer an is_a link from 1,3-dichloro-2-propanol metabolism to chlorinated hydrocarbon metabolism. This is the same link that is the absent subsumption between the object terms (i.e. that 1,3-dichloro-2-propanol metabolism is not subsumed by chlorinated hydrocarbon metabolism) of the nonalignment described for this example. Thus, these projects could predict analogous inferences for all of our nonalignments in which the subject terms are more atomic than the object terms, so long as ontologically valid necessary and sufficient definitions could be constructed, as was done in this example. Our methodology does not automatically suggest that all object pairs in each identified nonalignment be linked via is_a, as this may not be the correct action to take; it allows the curator to resolve the nonalignment with any of the four methods described in the previous section.
However, these projects likely could not predict new inferences for many if not all of the nonalignments in which the object terms are more atomic than the subject terms presented here, because the required necessary and sufficient definitions likely could not be made in an ontologically valid manner. Figure 6 illustrates such an example. The nonalignment identified here is that aldonate transport is subsumed by hexose transport in GO BP, but aldonates is not subsumed by hexoses in ChEBI. Given necessary and sufficient definitions of hexose transport in terms of hexoses and aldonate transport in terms of aldonates and the fact that aldonate transport is subsumed by hexose transport, a reasoner from one of these projects cannot infer that hexoses subsumes aldonates. In order for the reasoner to infer an is_a link from aldonates to hexoses (which is one way to resolve this nonalignment) from these terms and their definitions, necessary and sufficient definitions for aldonates (perhaps as a subclass of molecular entities and an is transported in aldonate transport condition) and hexoses (perhaps as a subclass of molecular entities and an is transported in hexose transport condition) would have to be created. However, this is too strong a condition, as, for example, an aldonate is not necessarily transported elsewhere; it may be used where it was synthesized. Without these necessary and sufficient definitions, this inference cannot be made.
Fig. 6.
The relationships between a pair of terms from the GO BP ontology and a pair of terms from ChEBI from which a nonalignment was identified. This is an example of a nonalignment that is not currently examined in other ontology-enrichment methodologies, which require necessary and sufficient conditions to make new inferences.
It can be argued that a reasoner in one of these other projects can infer an is_a link between chemicals by creating ontologically valid necessary and sufficient definitions in terms of, for example, parts or functions of these chemicals. However, this presupposes that not only such a more basic ontology but the required specific object terms exist. Such an approach laboriously requires the creation of an entirely new set of assertions, and there may be recursion in that the more basic object terms may not exist in a hierarchical relationship, thus once again preventing the inference of the is_a link between the more composite subject terms. Our approach only requires one set of assertions and their automatically generated inverse assertions and relies on a different kind of reasoning than the deduction used by reasoners in the aforementioned projects. However, we assert that a functionally equivalent methodology could be implemented, e.g. using an OWL API, without the use of explicitly represented inverse assertions.
We have found that 39.8% of the total nonredundant nonalignments identified in this study are those in which the subject terms of the nonalignments are built up from the object terms; these correspond to the instances in which it is difficult to produce the required ontologically valid necessary and sufficient conditions, in which case new inferences by the aforementioned reasoners cannot be made using the linked terms of the ontologies.
Our methodology essentially uses subsumptive analysis of term attributes toward quality assurance of ontologies, a technique which has been used by others in the field. The BERNWARD system reconstructed sets of medical concepts into hierarchies based on five subsumptive principles, but it is different in that it takes into account partonomy in its subsumption without resolution of the type we perform as in Figures 4 and 5 (Bernauer, 1994). In an analysis of UMLS, Cimino (1998) found that the semantic type of 0.5% of concepts was neither the same as nor more specific than the semantic type of their respective parents. In an analysis of the links between diseases and their respective anatomical locations in SNOMED CT, Burgun et al. (2005) looked for differences between sets of disorders associated with all descendants of given anatomical entities and the sets of descendant disorders of the disorders associated with the given anatomical entities. Bodenreider et al. (2007) found that SNOMED CT contained 7226 parent-child pairs in which a role or value present in the parent was not present in the child and 21 799 pairs in which a value of a role present in the parent was not identical or more specific in the child. In addition to being the first subsumptive study of links among OBO terms, ours suggests both fully automatic and semiautomatic solutions to correct the inconsistencies that result upon linking the terms and highlights those that are not currently found by existing reasoning methods in other biomedical ontology-enrichment projects.
We are not calling for the abolition of the use of the OWL, Obol or OBO-Edit reasoners. Rather, we assert that functionality that identifies the type of nonalignments for which inferences cannot be made (due to absence of required necessary and sufficient conditions) can and should be built into ontology-enrichment tools such as BONG. A methodology analogous to ours appears possible through the use of an OWL API through a subsumptive analysis of directly asserted and inherited property-value pairs. Consider Figure 7, in which the nonalignment of Figure 6 has been resolved through the addition of an is_a link from aldonates to hexoses. The links from the subject terms to the object terms can be represented as necessary and sufficient existential (i.e. someValuesFrom) conditions. Comparing the value of results in transport of at the level of aldonate transport (aldonates) to the value of results in transport of at the level of hexose transport (hexoses), it can be determined that the former is subsumed by the latter; thus, there is no inconsistency. Conversely, considering Figure 6, using the same procedure, aldonates is not subsumed by hexoses, which could result in the suggestion of a nonalignment. The same methodology could be used to suggest nonalignments where necessary and sufficient definitions can be made, but this appears unnecessary, since existing reasoners can suggest new inferences for such cases. Moreover, this would require the use of statements for which ontologically valid necessary and sufficient conditions likely could not be made. Thus, the subsumptive inferences made by currently used reasoners and the nonalignments discovered by our methodology are complementary if the OBO curators continue to solely examine those nonalignments indicated by the inferences made by the reasoners using necessary and sufficient definitions.
Fig. 7.
The relationships between a pair of terms from the GO BP ontology and a pair of terms from ChEBI that result from the resolution of the nonalignment of Figure 6 via the addition of an is_a link from aldonates to hexoses.
5 SUMMARY
We have described a methodology by which we have identified over 1900 instances of nonredundant nonalignments between terms from GO, ChEBI and CL. Analysis of the ratios of nonalignments to assertions from which the nonalignments were identified suggests that BP–MF, BP–BP, BP–CL and CC–CC terms are relatively well-aligned, while ChEBI–MF, BP–ChEBI and CCMF terms are relatively not aligned well. We propose that three ways to resolve an identified nonalignment are the addition of an is_a link between the object terms, the removal of an is_a link between the subject terms and the upward propagation of the object term to the superclass level. Many of the 39.8% of these nonalignments in which the object terms are more atomic than the subject terms likely are not currently examined in other ontology-enrichment projects due to the fact that the necessary and sufficient conditions required for the inferences likely could not be added, as they are semantically too strong. We assert that a methodology analogous to ours could be implemented using an OWL API in ontology-enrichment tools in order to identify such nonalignments that are currently not examined.
ACKNOWLEDGEMENTS
Both authors have been supported by NIH grants 5R01 LM008111-2 and 5R01 DE15191-04.
Conflict of Interest: none declared.
REFERENCES
- Aranguren ME. MSc dissertation. Manchester, UK: University of Manchester; 2004. Improving the Structure of the Gene Ontology. [Google Scholar]
- Bada M, Hunter L. J. Biomed. Informat. 2007. Enrichment of OBO Ontologies. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard J, et al. An ontology for cell types. Genome Biol. 2005;6:R21. doi: 10.1186/gb-2005-6-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernauer J. Proceedings of the 1994 Annual Symposium on Computer Applications of Medical Care. DC, USA: Washington; 1994. Subsumption principles underlying medical concept systems and their formal reconstruction. [PMC free article] [PubMed] [Google Scholar]
- Bodenreider O, et al. Investigating subsumption in SNOMED CT: an exploration into large description logic-based biomedical terminologies. Artif. Intell. Med. 2007;39:183–195. doi: 10.1016/j.artmed.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgun A, et al. Proceedings of the 2005 Annual Symposium of the American Medical Informatics Association. DC, USA: Washington; 2005. Classifying diseases with respect to anatomy: a study in SNOMED CT. [PMC free article] [PubMed] [Google Scholar]
- Cimino JJ. Auditing the unified medical language system with semantic methods. J. Am. Med. Informat. Assoc. 1998;5:41–51. doi: 10.1136/jamia.1998.0050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degtyarenko K. Proceedings of the 2003 International Chemical Information Conference. France: Nimes; 2003. Chemical vocabularies and ontologies for bioinformatics. [Google Scholar]
- Mungall CJ. Obol: integrating language and meaning in bio-ontologies. Comp. Funct. Genomics. 2004;5:509–520. doi: 10.1002/cfg.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B, et al. Relations in biomedical ontologies. Genome Biol. 2005;6:R46. doi: 10.1186/gb-2005-6-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroe CJ, et al. Proceedings of the Pacific Symposium on Biocomputing 2003. Hawaii, USA: Lihue; 2003. A Methodology to Migrate the Gene Ontology to a Description Logic Environment Using DAML+OIL. [DOI] [PubMed] [Google Scholar]