Abstract
We have previously demonstrated that haplotypes of three single nucleotide polymorphisms (SNPs) within the first intron of the OCA2 gene are extremely strongly associated with variation in human eye color. In the present work, we describe additional fine association mapping of eye color SNPs in the intergenic region upstream of OCA2 and within the neighboring HERC2 (hect domain and RLD2) gene. We screened an additional 92 SNPs in 300–3000 European individuals and found that a single SNP in intron 86 of HERC2, rs12913832, predicted eye color significantly better (ordinal logistic regression R2 = 0.68, association LOD = 444) than our previous best OCA2 haplotype. Comparison of sequence alignments of multiple species showed that this SNP lies in the center of a short highly conserved sequence and that the blue-eye-associated allele (frequency 78%) breaks up this conserved sequence, part of which forms a consensus binding site for the helicase-like transcription factor (HLTF). We were also able to demonstrate the OCA2 R419Q, rs1800407, coding SNP acts as a penetrance modifier of this new HERC2 SNP for eye color, and somewhat independently, of melanoma risk. We conclude that the conserved region around rs12913832 represents a regulatory region controlling constitutive expression of OCA2 and that the C allele at rs12913832 leads to decreased expression of OCA2, particularly within iris melanocytes, which we postulate to be the ultimate cause of blue eye color.
Introduction
Human eye color is a polymorphic phenotype under strong genetic control.1 The gene responsible for oculocutaneous albinism type II (OCA2)2–4 has hitherto seemed the best candidate to explain the genetic linkage of blue-brown eye (BEY2/EYCL3 [MIM 227220]) and brown hair (HCL3 [MIM 601800]) color to chromosome 15q11.2–q12.5–11 OCA2 is the human homolog of the mouse pink-eyed dilution gene (p).12 The human OCA2 gene is divided into 24 exons covering >345 kbp of DNA; 23 of these exons span the 836 amino acid coding region, with exon 1 representing exclusively a noncoding 5′UTR.13 The resulting gene product, the P protein, is an integral membrane protein containing 12 transmembrane spanning regions that helps regulate melanogenesis.14
Many polymorphisms in OCA2 occur in different populations with at least 13 nonsynonymous amino acid substitutions reported.2,13,15–18 Only two of these were found to be present at significant frequency in our previous studies of adolescent twins and family members of mainly European descent from southeast Queensland, Arg305Trp (rs1800401) and Arg419Gln (rs1800407) at 0.05 and 0.09, respectively.18 They exhibited only a minor impact on eye color, leading us to conclude that OCA2 coding alleles account for only a small proportion of the variation in iris pigmentation, at least within fair-skinned populations.
We proceeded therefore to screen tagging SNPs throughout the entire OCA2 locus. The highest association for blue:nonblue (green/hazel or brown) eye color was found with three SNPs in intron 1: rs7495174 T/C, rs6497268 (now rs4778241) G/T, and rs11855019 (now rs4778138) T/C. We found the TGT/TGT diplotype in 62.2% of samples, and this was the major blue-eye genotype, with a frequency of 91% in blue- or green-eyed individuals, compared with only 9.5% in those with brown eyes.18 The position of this major diagnostic haplotype for eye color suggested that differences within the 5′ proximal regulatory control region of OCA2 alter temporospatial expression of the gene and may be responsible for these associations.
To further refine the elements controlling eye color, we have tested for association with haplotype-tagging SNPs proximal to intron 1 of OCA2, which span the intergenic region and encompass the 3′ end of the upstream gene, HERC2. We describe the fine mapping of SNPs that are better predictors of blue-brown eye color than the existing OCA2 intron 1 TGT haplotype block. Moreover, we propose a mechanism whereby a single base change (our best predictive SNP), contained within a highly conserved region of the HERC2 gene intron 86, may abrogate accessibility of the OCA2 chromosomal region by transcription factors necessary for expression of OCA2 in people of different eye color.
Material and Methods
Structure of the Study Population and Pigmentation Characteristics
Adolescent twins and their siblings were recruited for an investigation of genetic and environmental factors contributing to the development of pigmented nevi and were also phenotyped for pigment traits including skin, hair, and eye color. The pigmentation characteristics of the twins were examined on up to three occasions, at 12, 14, and 16 years of age as previously described.19 Subjects were overwhelmingly (>95%) of northern European origin (mainly Anglo-Celtic). Eye color was rated by one research nurse (AE) as blue/gray (1), green/hazel (2), or brown (3) and crossvalidated with the individual's self-report.19
There were 5075 family members in 1100 pedigrees with some phenotypic data, and DNA was available for genotyping for 3839 individuals within 1037 of these pedigrees. Excluding one member of each genotyped MZ twin pair, there were 3011 individuals with complete OCA2 genotype, hair color, eye color, and sex recorded. We analyzed the data collected when the twins were 12 years old, because these are most complete.
We also carried out genotyping of an additional collection of Queensland cutaneous malignant melanoma (CMM) cases and their family members. In brief, the Queensland Familial Melanoma Project ascertained all 10407 CMM cases diagnosed from 1982 to 1990 in the state of Queensland, and a sample of families stratified on strength of family history was drawn.20–23 Phenotypes including hair, skin, and eye color were assessed by questionnaire, and DNA from available cases and selected unaffected members was collected. Only a subset of 96 blue-eyed and 96 brown-eyed individuals from this sample was genotyped for the present study.
OCA2 and HERC2 SNP Genotyping
Genotyping at a set of 58 OCA2 SNPs was described previously.18 We designed assays for another 92 SNPs including 30 within OCA2, 7 SNPs within the intergenic region, and 55 within HERC2, concentrating on 3′ introns/exons. These were selected based on their location within the gene and/or differences in published allele frequencies among the three HapMap populations. They were designed to be assayed in three multiplexes, so SNPs were typed with iPLEX chemistry on a Compact MALDI-TOF Mass Spectrometer (Sequenom, Inc., San Diego, CA) by standard methods.24 In a subset of twins, we also utilized SNPs genotyped as part of a genome-wide association study with the Affymetrix 100K array.
Statistical Methods
We performed standard linear and logistic regression analyses of the data with the R computer package (Version 2.5, R Development Core Team 2007). We used SOLAR 4.0.7 and MENDEL 7.01 to perform individual SNP and haplotypic association analysis correctly allowing for the relatedness within the sample. Some simulation-based p values for association have been generated with the Sib-pair program.18 Long-range haplotypes were inferred with Beagle25 and Haploview 4.26
Results
Association of OCA2-HERC2 SNP Alleles and Haplotypes with Blue Eye Color in an Adolescent Twin Collection
The eye color grade distributions were similar to our earlier reports,18,27 with 46.1% blue/gray, 27.7% green/hazel, and 26.3% brown in the total sample collection. The genotyped subjects were approximately 52% female and 48% male and there were no significant gender differences in eye color distribution.
In our first round of follow-up genotyping of 8 SNPs 5′ of the OCA2 gene, the most strongly associated of the SNPs was rs12913832 (p = 2 × 10−78). Surprisingly, this SNP was 21.1 kb upstream of the OCA2 first exon, in intron 86 of the HERC2 gene (Figure 1). Individuals carrying the C/C genotype had only a 1% probability of having brown eyes. By contrast, T/T carriers had an 80% probability of being brown eyed.
Figure 1.
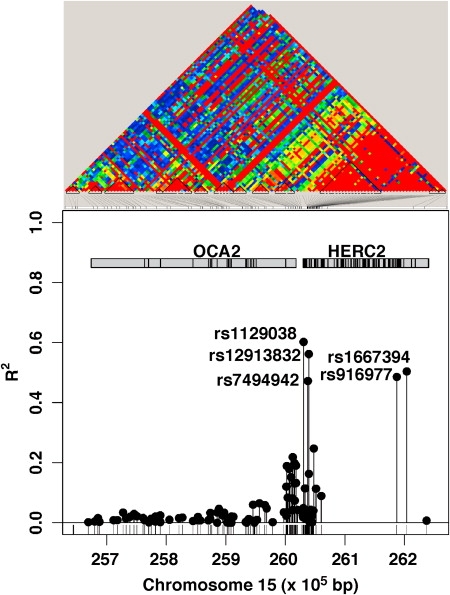
Association of OCA2 and HERC2 SNPs with Eye Color
Association of OCA2 and HERC2 SNPs with eye color (three-point scale) measured as R2 in ordinal logistic regression analysis in the lower panel. The upper panel shows the linkage disequilibrium (r2) between the SNPs.
We were able to show that this SNP gave additional information over that obtained with our previous best predictor, the three SNP haplotype in the first intron of OCA2. A haplotype analysis combining the original three SNPs with rs12913832 showed that the latter “split” the original haplotypes more precisely into eye color groups (see Table 1). Most critically, the C-TGT haplotype has a frequency of 91% in blue-eyed individuals and 37% in brown-eyed individuals, whereas the T-TGT haplotype is present in 0.3% of blue-eyed individuals and 15% of brown-eyed individuals (odds ratio = 122). Confirming this, a multiple ordinal logistic regression including these four SNPs found that the original three SNPs could be dropped from the model without a significant loss of predictive power.
Table 1.
Haplotype Frequencies at HERC2-OCA2 Locus SNPs rs12913832-rs7495174-rs6497268-rs11855019 versus Eye Color
| Haplotype | Total | Blue | Green-Hazel | Brown |
|---|---|---|---|---|
| C-TGT | 0.73547 | 0.91268 | 0.74083 | 0.37359 |
| C-TTC | 0.02627 | 0.02661 | 0.02115 | 0.02377 |
| C-TGC | 0.01031 | 0.01458 | 0.00260 | 0.01140 |
| C-TTT | 0.00768 | 0.01447 | 0.00345 | 0.0 |
| C-CTC | 0.00202 | 0.00115 | 0.00249 | 0.00004 |
| C-CGT | 0.00120 | 0.00131 | 0.00054 | 0.00328 |
| C-CGC | 0.00021 | 0.0 | 0.0 | 0.00204 |
| T-TTT | 0.07598 | 0.01260 | 0.11152 | 0.15577 |
| T-CTC | 0.05431 | 0.00708 | 0.03599 | 0.16711 |
| T-TGT | 0.05224 | 0.00318 | 0.05441 | 0.15851 |
| T-TGC | 0.02482 | 0.00393 | 0.02309 | 0.07032 |
| T-TTC | 0.00830 | 0.00212 | 0.00298 | 0.03270 |
| T-CGT | 0.00059 | 0.0 | 0.00096 | 0.0 |
| T-CTT | 0.00049 | 0.00030 | 0.0 | 0.00146 |
| T-CGC | 0.00011 | 0.0 | 0.0 | 0.0 |
Assay of HERC2 SNP Alleles and Haplotype Association in Individuals Selected for Blue or Brown Eye Color
We then carried out genotyping at an additional 92 SNPs within the OCA2 and HERC2 genes in a subsample of 384 individuals from our collection (192 with blue eyes, 192 with brown eyes), choosing only one person from any family. Only one SNP, rs1129038, was found to slightly more accurately predict eye color than rs12913832 in this small subsample (Figure 1). This was located in the 3′UTR of the HERC2 gene exon 93 and 12.4 kb upstream of the OCA2 first exon. This had similar genotype frequencies to rs12913832 among blue-eyed and brown-eyed individuals. In addition, two more distant SNPs exhibited similar though slightly smaller effects on eye color: rs1667394 and rs916977, in introns 4 and 12 of HERC2, respectively. Further examination of these four eye color SNPs confirmed that they were all in strong linkage disequilibrium with one another (see Table 2). However, it was possible to show that rs1129038 and rs12913832 were more likely to be causative or in strong LD with the causative variant (see Table 3). Specifically, the rs1129038-rs12913832-rs916977-rs1667394 ACGA/ACGA genotype had a blue-eye phenotype whereas the ACGA/GTGA genotypes were brown-eyed (p = 2 × 10−16).
Table 2.
Minor Allele Frequencies and Measures of Intermarker Linkage Disequilibrium for the Four HERC2 Locus SNPs Most Strongly Associated with Eye Color in Our Sample
| SNP | Sequence Position (bp) | MAF | rs1129038 | rs12913832 | rs916977 | rs1667394 |
|---|---|---|---|---|---|---|
| rs1129038 | 26,030,454 | 0.236 (G > A) | 1 | 0.984 | 0.556 | 0.588 |
| rs12913832 | 26,039,213 | 0.220 (C > T) | 0.997 | 1 | 0.546 | 0.580 |
| rs916977 | 26,186,959 | 0.204 (G > A) | 0.989 | 0.978 | 1 | 0.928 |
| rs1667394 | 26,203,777 | 0.214 (A > G) | 0.990 | 0.979 | 1 | 1 |
MAF, minor allele frequencies; r2 above diagonal; D' below diagonal.
Table 3.
Phased Genotypes at Four Key HERC2 Locus SNPs versus Eye Color
| rs1129038 | rs12913832 | rs916977 | rs1667394 | Phaseda | Blue | Brown | |
|---|---|---|---|---|---|---|---|
| 1b | A/A | C/C | G/G | A/A | ACGA/ACGA | 169 | 1 |
| 2b,c,d | A/G | C/T | G/G | A/A | ACGA/GTGA | 1 | 47 |
| 3c,d | A/G | C/T | A/G | A/G | ACGA/GTAG | 10 | 81 |
| 4d | A/G | C/T | G/G | A/G | ACGA/GTGG | 0 | 8 |
| 5 | G/G | T/T | A/A | G/G | GTAG/GTAG | 0 | 18 |
| 6 | G/G | T/T | A/G | A/G | GTAG/GTGA | 0 | 14 |
| 7 | A/A | C/C | A/G | A/G | ACGA/ACAG | 1 | 1 |
| 8 | A/G | C/C | A/G | A/G | ACGA/GCAG | 1 | 1 |
| 9 | A/G | C/C | G/G | A/A | ACGA/GCGA | 1 | 1 |
| 10 | G/G | T/T | A/G | G/G | GTAG/GTGG | 0 | 1 |
| 11 | G/G | T/T | G/G | A/A | GTGA/GTGA | 0 | 3 |
AC phased rs1129038 and 12913832 in bold.
The Fisher exact test comparing counts for row 1 versus 2, p = 2 × 10−16.
Row 2 versus 3, p = 0.097.
Row 2 versus 3 versus 4, p = 0.195.
Assay of HERC2 SNP Alleles and Haplotype Association with Eye Color in an Adolescent Twin Collection
We used the Beagle program25 to impute the most likely haplotypes for our sample of 384 individuals including all genotyped SNPs between R419Q in OCA2 and rs1667394 in HERC2 (129 SNPs). When we tested for an optimum combination of alleles from these haplotypes to best predict eye color by a recursive partitioning method,28 rs1129038 and rs12913832 were selected (Bonferroni-corrected p < 10−48) (Figure 2), along with R419Q (included at a far lower level of statistical significance, p = 0.041).
Figure 2.

SNP Haplotypes Associated with Eye Color
SNP haplotypes associated with blue and nonblue eye color spanning rs11855019 to rs1667394 of the OCA2-HERC2 locus. The SNP bases associated with blue and brown haplotypes are indicated by color.
Based on these results, we elected to genotype only these two SNPs in our entire collection. The total number of individuals genotyped at these key SNPs was 3961. In the total sample, these two SNPs were confirmed to be in almost perfect LD. Of the rs1129038-rs12913832 AC haplotype homozygotes, 99% were nonbrown (blue or green/hazel) in eye color, and 99% of GT haplotype homozygotes were nonblue in eye color (Table 4). These results closely match those expected for the brown eye color BEY2 locus based on segregation analysis. It was not possible by statistical methods to further decide between these two SNPs as the best candidate for a causal variant.
Table 4.
Genotypes with Penetrances Given in Parentheses for HERC2 rs12913832 and OCA2 R419Q versus Eye Color in the Complete Sample Collection
| rs12913832 | R419Q | Blue | Green | Brown |
|---|---|---|---|---|
| C/C | R/R | 1268 (0.71) | 499 (0.28) | 13 (0.01) |
| C/C | R/Q | 125 (0.83) | 25 (0.17) | 1 (0.01) |
| C/C | Q/Q | 1 (1.00) | 0 (0.00) | 0 (0.00) |
| C/T | R/R | 38 (0.05) | 214 (0.29) | 496 (0.66) |
| C/T | R/Q | 36 (0.12) | 176 (0.58) | 91 (0.30) |
| C/T | Q/Q | 0 (0.00) | 9 (0.75) | 3 (0.25) |
| T/T | R/R | 2 (0.02) | 7 (0.06) | 100 (0.92) |
| T/T | R/Q | 0 (0.00) | 12 (0.22) | 42 (0.78) |
| T/T | Q/Q | 0 (0.00) | 5 (0.50) | 5 (0.50) |
We then used multivariate approaches to further dissect effects of other variants within OCA2 via the complete data set. We were able to confirm that the OCA2 R419Q substitution is independently associated with eye color. The most striking effect was seen on the rs12913832∗T/T background, where the penetrance for green/hazel eyes for 419Q/419Q, 419Q/419R, and 419R/419R was then 50%, 21%, and 6%, respectively (p = 0.0003). In a parallel analysis, we examined the effects of these two SNPs on risk of cutaneous malignant melanoma. Interestingly, R419Q was a significant independent risk factor (allelic odds ratio adjusted for the other locus, OR = 1.27, empirical p = 0.0006), as previously found by ourselves and others, and rs12913832 was less strongly associated (OR = 1.15, p = 0.008).
Sequence Alignment of HERC2 Exon 86 to Exon 93 across Species
We examined cross-species similarity in alignment of the 10.4 kb human HERC2 genomic sequence from exon 86 to exon 93 (GenBank NT_010280.17), encompassing the interval between our two peak eye color SNPs. Alignments were first tested by comparing each individual exonic and intronic segment separately with the corresponding mouse Herc2 genomic sequence (GenBank NT_039424.7). Apart from the exonic coding regions, there was only one region of marked similarity found—between the human and mouse intron 86 sequences, as seen in a dot matrix plot (Figure 3A). The GenBank reference sequence for HERC2 was also compared with the Celera sequence (Celera NW_925783) across this 10.4 kb region, and 23 nucleotide differences were apparent in this alignment, notably the rs1129038-rs12913832 SNP haplotype was the brown GT in GenBank and blue AC in the Celera sequences, respectively, consistent with the eye color of one of the individuals used to generate the Celera database.29 The alignment of the conserved region of intron 86, between the two versions of the human 406 bp and corresponding 420 bp mouse sequence, showed more than 77% shared identity (Figure 3B). An additional SNP rs6497271 contained within this region was also identified. To search for additional variants in the immediate vicinity of our key SNPs, we sequenced the regions around rs12913832 and rs1129038 in five individuals with brown eyes and five individuals with blue eyes (690 bp either side of rs12913832 and 600 bp either side of rs1129038; primers available on request). No novel variants were found in these regions.
Figure 3.

Conserved Sequences in HERC Intron 86
(A) Dot-matrix-aligned sequence comparison of intron 86 from human HERC2 (x axis) and mouse Herc2 (y axis) with the MacVector program (MacVector, Inc.).
(B) ClustalW alignment of the sequence similarity found in (A) with the RefSeq and Celera human with the RefSeq mouse sequences. Transcription factor binding sites identified by the MatInspector Program are boxed with SNP changes shown in color.
(C) UCSC Genome Browser53 screen shot showing sequence alignment for seven species and associated phastCONS scores for sequence around rs12913832 of HERC2 intron 86. Sequence is shown in the 3′ to 5′ direction of the upper strand.
An alignment of this interval across multiple species, concentrating especially on the regions close to our two peak SNPs, was also examined with the UCSC browser Multiz alignment track30 and in the Vista Enhancer “Computational Dataset.”31 The latter resource flagged only two regions of HERC2 as strongly conserved and thus likely to contain enhancer elements—portions of intron 2 (which contains no known SNPs) and intron 86 (containing rs12913832 and rs6497271). Although we observed little sequence conservation around rs1129038, there was again striking conservation immediately around rs12913832 (Figure 3C). The Regulatory Potential mammalian conservation scoring method provided by the ESPERR 7 program32 gave a score 10-fold higher than the baseline signal (a level found to predict enhancer activity in 60%–100% of cases when validated by chromatin immunoprecipitation or integration of the putative expression cassette via the Cre-loxP system33).
Analysis of this sequence with the MatInspector Program34 suggests that this may represent a HLTF (SMARCA3) binding site. Tellingly, this consensus sequence is abolished in the rs12913832∗C blue-eyed allele. Other potential transcription factor binding sites relevant to melanocytic cell gene expression identified in this search are two strong MITF sites35 and three LEF1 sites. Notably, rs6497271 is in the third of these LEF1 sites, and presence of the minor allele results in the failure of this motif to be identified in a MatInspector search. rs6497271 had only a 1% minor allele frequency (MAF) within our sample, so it cannot explain the association to the region.
Discussion
There is overwhelming evidence implicating the OCA2 gene region in regulation of human pigmentation. Mutations in the OCA2 gene lead to oculocutaneous albinism. Deletion of the region encompassing the OCA2 gene on chromosome 15 as observed in Prader-Willi and Angelman syndromes is associated with hypopigmentation of the skin, hair, and eyes, and extra copies of this chromosomal region result in generalized hyperpigmentation of the skin. Normal variation in eye color shows strong genetic linkage8 and association18 to markers in the OCA2 locus. In our previous combined segregation-linkage analysis in this twin sample, we estimated the frequencies of a dominant brown eye B allele as 21% and recessive b allele as 79%, which was close to the 26% B and 74% b allele modeled for the US white population by Hasstedt.36 The allele frequencies and penetrances estimated for rs12913832 in our Anglo-Celtic population match these predictions almost exactly. The European population frequencies in Hapmap for this SNP are in agreement (MAF 21%), and the variant is absent from other population groups.37 There is strong and extensive linkage disequilibrium across the OCA2-HERC2 region, as well as marked differences in SNP allele frequencies between different ethnic groups. Voight et al.38 used HapMap data to show that this can be interpreted as a signal of positive genetic selection in Europeans, similar to that seen around other pigmentation loci such as TYRP1, DTNBP1, and SLC24A5. The hitchhiking seen around such loci can decrease the power of association-based fine mapping.
Although rs12913832 lies within a distal intron of HERC2 (intron 86 of a 93 exon encoded gene spanning 211 kb), we do not believe that the HERC2 protein is involved in regulating the pigmentation pathway. Three mouse deletions that map within this distal region of HERC2, which do not include the OCA2 locus, were originally labeled as p gene mutants because they exhibit pigmentation changes that are intermediate to those seen in canonical p mutant homozygotes: dark pink eyes with mottling or partially decreased eumelanin quantities in hair.39–41 Effects on pigmentation are thought to result from sequences within this region of HERC2 controlling expression of OCA2. One of the radiation-induced mutant mouse alleles characterized by Lehman et al.,40 pbs (black eyed, sterile; originally p24H42), involves a deletion of only 8 kb in Herc2, which spans the orthologous region to human HERC2 exons 86–93. Lehman et al. comment in parentheses that “[t]he relatively mild hypopigmentation associated with the pbs deletion apparently reflects an effect of the deletion on expression of the closely linked p gene.” Walkowicz et al.41 describe characterization of the p12DTR, p103G, and p39DSD alleles.43 These all involve deletions or rearrangements of Herc2. Those authors concluded that elements regulating the expression of p probably lie within Herc2.
Recently, strong association between eye color and rs1667394 in HERC2 was reported in Icelandic population,44 but we excluded this as the casual SNP based on haplotype analysis. One of our two most strongly associated SNPs, rs12913832, lies within an evolutionarily conserved 406 bp region. Moreover, database searches of this region revealed transcription factor binding sites for LEF1, HLTF, and MITF. Both LEF1 and MITF are critically important to gene regulation in melanocyte cell development, differentiation, and tissue-specific transcription.45,46 Helicase-Like Transcription Factor (HLTF) is a member of the SWI2/SNF2 family, DNA-dependent chromatin-remodelling ATPases that have been implicated in a wide variety of processes involving the modification of chromatin configuration to allow access of the transcriptional machinery. HLTF is the only member of the family in humans that incorporates a specific DNA recognition site that binds a variety of gene promoters, including PAI-1 and beta-globins (where it seems to be involved in controlling levels of constitutive expression),47,48 uteroglobin (transducing of the effects of prolactin) on uteroglobin,49 and an enhancer region of the myosin light chain locus.50,51 The binding site in intron 86 matches the consensus recognized by the HLTF variant known as RUSH-1-α as determined by CASTing.49
Another SWI/SNF family member, BRG1, has recently been shown to be involved in pigmentation pathways, being recruited by MITF to melanocyte-specific promoter regions to induce changes in chromatin structure at endogenous loci, and thus initiating the process of melanocyte differentiation.52 We therefore suggest that the presence of both MITF and HLTF enhancer sequence elements in the evolutionary conserved region of HERC2 intron 86 is a further pointer that this is a locus control region that determines the expression of the OCA2 gene product.
We have now genotyped most of the reported SNPs in the 3′ end of HERC2 and the 5′ end of OCA2, finding a large number (39 out of 93 or 42% in the second round of genotyping) to be monomorphic in our sample. The blue-eye-color-associated allele must be reasonably common if a single SNP is to explain most of the variation in eye color in European-descended populations. This makes it less likely that another SNP as yet unidentified in the region is the true causative variant. Based on the foregoing, we conclude that the conserved region around rs12913832 represents a regulatory region controlling constitutive expression of OCA2, and that the C allele at rs12913832 leads to decreased expression of OCA2, particularly within iris melanocytes. We speculate that the regulatory mechanism is abrogation of the binding site for HLTF that regulates transcription of the neighboring OCA2 gene. We also confirmed that the common coding variant OCA2∗R419Q acts to modify the penetrance of this locus and that this effect includes modification of the risk of malignant melanoma.
Web Resources
The URLs for data presented herein are as follows:
HapMap, http://www.hapmap.org/
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim
UCSC Genome Browser, http://genome.ucsc.edu/
VISTA Enhancer Browser, http://enhancer.lbl.gov/
Acknowledgments
This work was supported by grants from the National Health and Medical Research Council of Australia (NHMRC 241944, 389875) and the National Cancer Institute (CA88363). The Institute for Molecular Bioscience incorporates the Centre for Functional and Applied Genomics as a Special Research Centre of the Australian Research Council. R.A.S., D.L.D., and G.W.M. are NHMRC Senior Research Fellows. F.P.N.L. was supported by a grant from Capes - Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brazilian Education Ministry. We thank Ann Eldridge and Marlene Grace for data collection, Anjali Henders and Megan Campbell for managing sample processing and preparation, and the twins and their families for participation in the research.
References
- 1.Sturm R.A., Frudakis T.N. Eye colour: portals into pigmentation genes and ancestry. Trends Genet. 2004;20:327–332. doi: 10.1016/j.tig.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Ramsay M., Colman M.A., Stevens G., Zwane E., Kromberg J., Farrall M., Jenkins T. The tyrosinase-positive oculocutaneous albinism locus maps to chromosome 15q11.2-q12. Am. J. Hum. Genet. 1992;51:879–884. [PMC free article] [PubMed] [Google Scholar]
- 3.Gardner J.M., Nakatsu Y., Gondo Y., Lee S., Lyon M.F., King R.A., Brilliant M.H. The mouse pink-eyed dilution gene: association with human Prader-Willi and Angelman syndromes. Science. 1992;257:1121–1124. doi: 10.1126/science.257.5073.1121. [DOI] [PubMed] [Google Scholar]
- 4.Rinchik E.M., Bultman S.J., Horsthemke B., Lee S.T., Strunk K.M., Spritz R.A., Avidano K.M., Jong M.T., Nicholls R.D. A gene for the mouse pink-eyed dilution locus and for human type II oculocutaneous albinism. Nature. 1993;361:72–76. doi: 10.1038/361072a0. [DOI] [PubMed] [Google Scholar]
- 5.Eiberg H., Mohr J. Assignment of genes coding for brown eye colour (BEY2) and brown hair colour (HCL3) on chromosome 15q. Eur. J. Hum. Genet. 1996;4:237–241. doi: 10.1159/000472205. [DOI] [PubMed] [Google Scholar]
- 6.Rebbeck T.R., Kanetsky P.A., Walker A.H., Holmes R., Halpern A.C., Schuchter L.M., Elder D.E., Guerry D. P gene as an inherited biomarker of human eye color. Cancer Epidemiol. Biomarkers Prev. 2002;11:782–784. [PubMed] [Google Scholar]
- 7.Jannot A.S., Meziani R., Bertrand G., Gerard B., Descamps V., Archimbaud A., Picard C., Ollivaud L., Basset-Seguin N., Kerob D. Allele variations in the OCA2 gene (pink-eyed-dilution locus) are associated with genetic susceptibility to melanoma. Eur. J. Hum. Genet. 2005;13:913–920. doi: 10.1038/sj.ejhg.5201415. [DOI] [PubMed] [Google Scholar]
- 8.Zhu G., Evans D.M., Duffy D.L., Montgomery G.W., Medland S.E., Gillespie N.A., Ewen K.R., Jewell M., Liew Y.W., Hayward N.K. A genome scan for eye color in 502 twin families: most variation is due to a QTL on chromosome 15q. Twin Res. 2004;7:197–210. doi: 10.1375/136905204323016186. [DOI] [PubMed] [Google Scholar]
- 9.Posthuma D., Visscher P.M., Willemsen G., Zhu G., Martin N.G., Slagboom P.E., de Geus E.J., Boomsma D.I. Replicated linkage for eye color on 15q using comparative ratings of sibling pairs. Behav. Genet. 2006;36:12–17. doi: 10.1007/s10519-005-9007-x. [DOI] [PubMed] [Google Scholar]
- 10.Frudakis T., Thomas M., Gaskin Z., Venkateswarlu K., Chandra K.S., Ginjupalli S., Gunturi S., Natrajan S., Ponnuswamy V.K., Ponnuswamy K.N. Sequences associated with human iris pigmentation. Genetics. 2003;165:2071–2083. doi: 10.1093/genetics/165.4.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frudakis T., Terravainen T., Thomas M. Multilocus OCA2 genotypes specify human iris colors. Hum. Genet. 2007;122:311–326. doi: 10.1007/s00439-007-0401-8. [DOI] [PubMed] [Google Scholar]
- 12.Brilliant M.H. The mouse p (pink-eyed dilution) and human P genes, oculocutaneous albinism type 2 (OCA2), and melanosomal pH. Pigment Cell Res. 2001;14:86–93. doi: 10.1034/j.1600-0749.2001.140203.x. [DOI] [PubMed] [Google Scholar]
- 13.Lee S.T., Nicholls R.D., Jong M.T., Fukai K., Spritz R.A. Organization and sequence of the human P gene and identification of a new family of transport proteins. Genomics. 1995;26:354–363. doi: 10.1016/0888-7543(95)80220-g. [DOI] [PubMed] [Google Scholar]
- 14.Sturm R.A. A golden age of human pigmentation genetics. Trends Genet. 2006;22:464–468. doi: 10.1016/j.tig.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 15.Oetting W.S., Gardner J.M., Fryer J.P., Ching A., Durham-Pierre D., King R.A., Brilliant M.H. Mutations of the human P gene associated with Type II oculocutaneous albinism (OCA2). Mutations in brief no. 205. Online. Hum. Mutat. 1998;12:434. doi: 10.1002/(SICI)1098-1004(1998)12:6<434::AID-HUMU16>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 16.Kerr R., Stevens G., Manga P., Salm S., John P., Haw T., Ramsay M. Identification of P gene mutations in individuals with oculocutaneous albinism in sub-Saharan Africa. Hum. Mutat. 2000;15:166–172. doi: 10.1002/(SICI)1098-1004(200002)15:2<166::AID-HUMU5>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki T., Miyamura Y., Tomita Y. High frequency of the Ala481Thr mutation of the P gene in the Japanese population. Am. J. Med. Genet. A. 2003;118:402–403. doi: 10.1002/ajmg.a.20044. [DOI] [PubMed] [Google Scholar]
- 18.Duffy D.L., Montgomery G.W., Chen W., Zhao Z.Z., Le L., James M.R., Hayward N.K., Martin N.G., Sturm R.A. A three-single-nucleotide polymorphism haplotype in intron 1 of OCA2 explains most human eye-color variation. Am. J. Hum. Genet. 2007;80:241–252. doi: 10.1086/510885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu G., Duffy D.L., Eldridge A., Grace M., Mayne C., O'Gorman L., Aitken J.F., Neale M.C., Hayward N.K., Green A.C. A major quantitative-trait locus for mole density is linked to the familial melanoma gene CDKN2A: a maximum-likelihood combined linkage and association analysis in twins and their sibs. Am. J. Hum. Genet. 1999;65:483–492. doi: 10.1086/302494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aitken J., Welch J., Duffy D., Milligan A., Green A., Martin N., Hayward N. CDKN2A variants in a population-based sample of Queensland families with melanoma. J. Natl. Cancer Inst. 1999;91:446–452. doi: 10.1093/jnci/91.5.446. [DOI] [PubMed] [Google Scholar]
- 21.Aitken J.F., Green A.C., MacLennan R., Youl P., Martin N.G. The Queensland Familial Melanoma Project: study design and characteristics of participants. Melanoma Res. 1996;6:155–165. doi: 10.1097/00008390-199604000-00011. [DOI] [PubMed] [Google Scholar]
- 22.Box N.F., Duffy D.L., Chen W., Stark M., Martin N.G., Sturm R.A., Hayward N.K. MC1R genotype modifies risk of melanoma in families segregating CDKN2A mutations. Am. J. Hum. Genet. 2001;69:765–773. doi: 10.1086/323412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siskind V., Aitken J., Green A., Martin N. Sun exposure and interaction with family history in risk of melanoma, Queensland, Australia. Int. J. Cancer. 2002;97:90–95. doi: 10.1002/ijc.1563. [DOI] [PubMed] [Google Scholar]
- 24.Zhao Z.Z., Nyholt D.R., Le L., Martin N.G., James M.R., Treloar S.A., Montgomery G.W. KRAS variation and risk of endometriosis. Mol. Hum. Reprod. 2006;11:671–676. doi: 10.1093/molehr/gal078. [DOI] [PubMed] [Google Scholar]
- 25.Browning B.L., Browning S.R. Efficient multilocus association testing for whole genome association studies using localized haplotype clustering. Genet. Epidemiol. 2007;31:365–375. doi: 10.1002/gepi.20216. [DOI] [PubMed] [Google Scholar]
- 26.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 27.Duffy D.L., Box N.F., Chen W., Palmer J.S., Montgomery G.W., James M.R., Hayward N.K., Martin N.G., Sturm R.A. Interactive effects of MC1R and OCA2 on melanoma risk phenotypes. Hum. Mol. Genet. 2004;13:447–461. doi: 10.1093/hmg/ddh043. [DOI] [PubMed] [Google Scholar]
- 28.Hothorn, T., Hornik, K., and Zeileis, A. (2004) Unbiased Recursive Partitioning: A Conditional Inference Framework. Research Report Series/Department of Statistics and Mathematics, WU (Wien, Austria).
- 29.Levy S., Sutton G., Ng P.C., Feuk L., Halpern A.L., Walenz B.P., Axelrod N., Huang J., Kirkness E.F., Denisov G. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prabhakar S., Poulin F., Shoukry M., Afzal V., Rubin E.M., Couronne O., Pennacchio L.A. Close sequence comparisons are sufficient to identify human cis-regulatory elements. Genome Res. 2006;16:855–863. doi: 10.1101/gr.4717506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor J., Tyekucheva S., King D.C., Hardison R.C., Miller W., Chiaromonte F. ESPERR: learning strong and weak signals in genomic sequence alignments to identify functional elements. Genome Res. 2006;16:1596–1604. doi: 10.1101/gr.4537706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H., Zhang Y., Cheng Y., Zhou Y., King D.C., Taylor J., Chiaromonte F., Kasturi J., Petrykowska H., Gibb B. Experimental validation of predicted mammalian erythroid cis-regulatory modules. Genome Res. 2006;16:1480–1492. doi: 10.1101/gr.5353806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 35.Aksan I., Goding C.R. Targeting the microphthalmia basic helix-loop-helix-leucine zipper transcription factor to a subset of E-box elements in vitro and in vivo. Mol. Cell. Biol. 1998;18:6930–6938. doi: 10.1128/mcb.18.12.6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hasstedt S.J. Phenotypic assortative mating in segregation analysis. Genet. Epidemiol. 1995;12:109–127. doi: 10.1002/gepi.1370120202. [DOI] [PubMed] [Google Scholar]
- 37.International HapMap Consortium (IHMC). A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voight B.F., Kudaravalli S., Wen X., Pritchard J.K. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lyon M.F., King T.R., Gondo Y., Gardner J.M., Nakatsu Y., Eicher E.M., Brilliant M.H. Genetic and molecular analysis of recessive alleles at the pink-eyed dilution (p) locus of the mouse. Proc. Natl. Acad. Sci. USA. 1992;89:6968–6972. doi: 10.1073/pnas.89.15.6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lehman A.L., Nakatsu Y., Ching A., Bronson R.T., Oakey R.J., Keiper-Hrynko N., Finger J.N., Durham-Pierre D., Horton D.B., Newton J.M. A very large protein with diverse functional motifs is deficient in rjs (runty, jerky, sterile) mice. Proc. Natl. Acad. Sci. USA. 1998;95:9436–9441. doi: 10.1073/pnas.95.16.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walkowicz M., Ji Y., Ren X., Horsthemke B., Russell L.B., Johnson D., Rinchik E.M., Nicholls R.D., Stubbs L. Molecular characterization of radiation- and chemically induced mutations associated with neuromuscular tremors, runting, juvenile lethality, and sperm defects in jdf2 mice. Mamm. Genome. 1999;10:870–878. doi: 10.1007/s003359901106. [DOI] [PubMed] [Google Scholar]
- 42.Phillips R. A new allele at the p-locus. Mouse News Lett. 1965;32:39. [Google Scholar]
- 43.Russell L.B., Montgomery C.S., Cacheiro N.L., Johnson D.K. Complementation analyses for 45 mutations encompassing the pink-eyed dilution (p) locus of the mouse. Genetics. 1995;141:1547–1562. doi: 10.1093/genetics/141.4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sulem P., Gudbjartsson D.F., Stacey S.N., Helgason A., Rafnar T., Magnusson K.P., Manolescu A., Karason A., Palsson A., Thorleifsson G. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat. Genet. 2007;39:1443–1452. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 45.Levy C., Khaled M., Fisher D.E. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol. Med. 2006;12:406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 46.Larue L., Delmas V. The WNT/Beta-catenin pathway in melanoma. Front. Biosci. 2006;11:733–742. doi: 10.2741/1831. [DOI] [PubMed] [Google Scholar]
- 47.Ding H., Benotmane A.M., Suske G., Collen D., Belayew A. Functional interactions between Sp1 or Sp3 and the helicase-like transcription factor mediate basal expression from the human plasminogen activator inhibitor-1 gene. J. Biol. Chem. 1999;274:19573–19580. doi: 10.1074/jbc.274.28.19573. [DOI] [PubMed] [Google Scholar]
- 48.Mahajan M.C., Weissman S.M. DNA-dependent adenosine triphosphatase (helicaselike transcription factor) activates beta-globin transcription in K562 cells. Blood. 2002;99:348–356. doi: 10.1182/blood.v99.1.348. [DOI] [PubMed] [Google Scholar]
- 49.Hewetson A., Hendrix E.C., Mansharamani M., Lee V.H., Chilton B.S. Identification of the RUSH consensus-binding site by cyclic amplification and selection of targets: demonstration that RUSH mediates the ability of prolactin to augment progesterone-dependent gene expression. Mol. Endocrinol. 2002;16:2101–2112. doi: 10.1210/me.2002-0064. [DOI] [PubMed] [Google Scholar]
- 50.Sheridan P.L., Schorpp M., Voz M.L., Jones K.A. Cloning of an SNF2/SWI2-related protein that binds specifically to the SPH motifs of the SV40 enhancer and to the HIV-1 promoter. J. Biol. Chem. 1995;270:4575–4587. doi: 10.1074/jbc.270.9.4575. [DOI] [PubMed] [Google Scholar]
- 51.Gong X., Kaushal S., Ceccarelli E., Bogdanova N., Neville C., Nguyen T., Clark H., Khatib Z.A., Valentine M., Look A.T. Developmental regulation of Zbu1, a DNA-binding member of the SWI2/SNF2 family. Dev. Biol. 1997;183:166–182. doi: 10.1006/dbio.1996.8486. [DOI] [PubMed] [Google Scholar]
- 52.de la Serna I.L., Ohkawa Y., Higashi C., Dutta C., Osias J., Kommajosyula N., Tachibana T., Imbalzano A.N. The microphthalmia-associated transcription factor requires SWI/SNF enzymes to activate melanocyte-specific genes. J. Biol. Chem. 2006;281:20233–20241. doi: 10.1074/jbc.M512052200. [DOI] [PubMed] [Google Scholar]
- 53.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]


