Abstract
Hepatic resection is associated with rapid proliferation and regeneration of the remnant liver. Phosphatidylinositol 3-kinase (PI3K), composed of a p85α regulatory and a p110α catalytic subunit, participates in multiple cellular processes, including cell growth and survival; however, the role of PI3K in liver regeneration has not been clearly delineated. In this study, we used the potent PI3K inhibitor, wortmannin, and small-interfering RNA (siRNA) targeting the p85α and p110α subunits to determine if total or selective PI3K inhibition would abrogate the proliferative response of the liver after partial hepatectomy in mice. Hepatic resection is associated with an induction in PI3K activity; total PI3K blockade with wortmannin, and selective inhibition of p85α or p110α with siRNA resulted in a significant decrease in hepatocyte proliferation, especially at the earliest timepoints. Fewer macrophages and Kupffer cells were present in the regenerating liver of mice treated with wortmannin or siRNA to p85α or p110α, as reflected by a paucity of F4/80-positive cells. Additionally, PI3K inhibition led to an aberrant architecture in the regenerating hepatocytes characterized by vacuolization, lipid deposition, and glycogen accumulation; these changes were not noted in the sham livers. Our data demonstrate that PI3K/Akt pathway activation plays a critical role in the early regenerative response of the liver after resection; inhibition of this pathway markedly abrogates the normal hepatic regenerative response, most likely by inhibiting macrophage infiltration and cytokine elaboration and thus hepatocyte priming for replication.
Keywords: Liver regeneration, hepatectomy, phosphatidylinositol 3-kinase (PI3K), p85α, p110α, wortmannin, Kupffer cell, macrophage, autophagy
Partial hepatectomy results in the compensatory hypertrophy and hyperplasia of the remaining lobes of the liver to replace the loss of functional mass. In a mouse model of 70% hepatectomy, the original liver mass is restored approximately 7–10 days following resection with a peak in DNA synthesis at approximately 42 hours, while in humans, restoration of liver mass is complete in approximately 3 months following lobectomy, with a peak in DNA synthesis at 7–10 days (12–14, 22). In rodents, the cell type responsible for this proliferation is almost exclusively hepatocytes; in humans, a two-tier system composed of hepatocytes and progenitor (stem) cells contribute to the gain in mass (12, 14, 29, 41). A complex network of signaling pathways leads to successful liver regeneration: a cytokine pathway, responsible for hepatocyte priming; a growth factor pathway, responsible for cell cycle progression; and poorly understood pathways linking metabolic signals with DNA replication (14, 28).
Cytokines, including tumor necrosis factor-alpha (TNF-α) and interleukin 6 (IL-6), are elaborated by non-parenchymal cells such as Kupffer cells and sinusoidal epithelial cells following hepatectomy (10, 14, 24, 26). They are responsible for the initial priming of hepatocytes, with the transition of quiescent hepatocytes from the G0 to G1 stage of the cell cycle. In the absence of cytokines, hepatocytes are minimally responsive to growth factor stimulation, illustrating the importance of this initial priming reaction (14). Upon priming of hepatocytes, growth factor stimulation leads to G1 to S phase transition and cell cycle progression. Three factors of major importance are hepatocyte growth factor (HGF), produced by nonparenchymal cells of the liver (14); transforming growth factor-alpha (TGF-α), produced by hepatocytes (14, 22); and epidermal growth factor (EGF), the major source of which is the salivary glands in rodents (22). HGF, TGF-α, and EGF signaling occurs through receptor tyrosine kinases which, in the activated state, associate with cytosolic proteins rich in Src-homologies (SH-2 and SH-3), such as phosphatidylinositol 3-kinase (PI3K) (4, 22). Downstream signaling leads to the activation of mitogen activated protein kinase (MAPK) cascades, ultimately leading to the regulation of cell cycle and growth (4). While much is known about the cytokines responsible for hepatocyte priming and the growth factors responsible for stimulating cell cycle progression, the complex set of reactions responsible for linking these signaling pathways has yet to be delineated.
PI3K is a ubiquitous protein kinase family involved in signal transduction through receptor tyrosine kinases or G-protein-coupled receptors; examples of known PI3K receptor ligands include TNF-α (34, 35), IL-6 (9), HGF (33), EGF (22, 32), and TGF-α (22). Class IA PI3K is composed of a regulatory p85 (α or β) and a catalytic p110 (α, β, or γ ) subunit (8, 32, 40). When its receptor is activated by growth factors or cytokines, PI3K catalyzes the production of phosphatidylinositol-3,4,5-triphosphate, recruiting a subset of signal proteins with pleckstrin homology domains to the plasma membrane, ultimately leading to their phosphorylation (40, 44). These proteins include phosphoinositide-dependent kinase 1 and Akt, also known as protein kinase B (43). Phosphorylation of Akt leads to the subsequent phosphorylation of downstream targets that affect cell growth and survival (44), as well as membrane ruffling, cell migration, and actin cytoskeletal rearrangement important for leukocyte migration and phagocytosis (8, 32). While Akt has been shown to play an important role in the compensatory recovery of liver mass following resection by regulating hepatocyte hypertrophy (17), little is known regarding the contribution of PI3K subunits to liver regeneration. Interestingly, genetic deletion of all isoforms of p85α in mice leads to perinatal death secondary to extensive hepatocyte necrosis and chylous ascites (15), suggesting a role for the p85α subunit in early liver development.
We have previously demonstrated that pancreatic regeneration is dependent on PI3K/Akt activation (44). This is consistent with findings that PI3K activation: (i) is important for immediate remodeling and cell survival following myocardial infarction (18), (ii) mediates proliferative signals in intestinal epithelial cells in vitro and in vivo (40), and (iii) modulates vascular regeneration following insult (31). Given the role of the PI3K pathway in the regeneration of other organs, we hypothesized that PI3K plays an important role in hepatic regeneration following resection. Here, we demonstrate that the PI3K pathway is activated following hepatectomy, especially at the earliest timepoints. Next, using the pharmacologic PI3K inhibitor, wortmannin (3), or small interfering RNA (siRNA)(37) directed to the p85α regulatory or p110α catalytic subunits, we demonstrate that PI3K signaling is required for early hepatic regeneration following hepatectomy. Our results are the first to define a role for PI3K activation in liver regeneration; decreased PI3K activity, either via total PI3K blockade with wortmannin or selective blockade with p85α or p110α siRNA, attenuates the proliferative response of the liver following hepatectomy.
MATERIALS AND METHODS
Materials
pik3ca (p85α), pik3r1 (p110α), and non-targeting control (NTC) siSTABLE in vivo SMARTpool siRNA duplexes and DY547-labeled NTC and p85α siRNA duplexes were designed and synthesized by Customer SMARTpool siRNA Design from Dharmacon (Lafayette, CO). siSTABLE in vivo duplex is chemically modified to extend siRNA stability in vivo compared with unmodified siRNA. 5-0 silk sutures were purchased from Ethicon (Somerville, NJ). DOTAP liposomal transfection reagent was purchased from Roche (Indianapolis, IN). Wortmannin, bromodeoxyuridine (BrdU), phenol-chloroform-isoamylalcohol (25:24:1), carbon tetrachloride (CCl4), corn oil, mouse monoclonal anti-BrdU antibody, and Hoescht stain were purchased from Sigma-Aldrich (St. Louis, MO). Rabbit monoclonal anti-phospho-Akt (Ser473), anti-p110α, anti-cyclin D1, and anti-p38 MAPK were purchased from Cell Signaling (Beverly, MA). Mouse monoclonal anti-p85α jantibody was purchased from Upstate (Charlottesville, VA). Goat monoclonal anti-cyclophilin B and rabbit monoclonal anti-phospho-Stat3 (Ser727) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse monoclonal anti-F4/80 antibody was purchased from Abcam (Cambridge, MA). Immuno-Blot polyvinylidene difluoride (PVDF) membranes were from Bio-Rad (Hercules, CA), and X-ray film was purchased from Eastman Kodak (Rochester, NY). The enhanced chemiluminescence (ECL) system for Western immunoblot analysis was from Amersham Biosciences (Arlington Heights, IL). TNFα and IL-6 enzyme-linked immunosorbent assay kits were purchased from R&D Systems (Abingdon, Oxon, UK).
Animals
Female Swiss-Webster mice (aged 4–6wks; ~25g) were purchased from Harlan Sprague Dawley (Indianapolis, IN). Mice were housed in an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC)-approved facility under a standard 12h light-dark cycle, fed standard chow (Formula Chow 5008; Purina Mills, St. Louis, MO) and tap water ad libitum and allowed to acclimate for one week. All studies were approved by the Institutional Animal Care and Use Committee of UTMB.
Murine hepatectomy
A model of 70% hepatectomy as originally described by Higgins and Anderson (19) was used in this study. Midline laparotomy was performed, and right medial, left medial, and left lateral lobes were removed; care was taken to preserve the gallbladder. Sham-operated mice underwent an identical exposure and liver manipulation. Operating times and halothane exposure were identical for mice undergoing hepatectomy or sham operation.
Experimental design
(i) Female Swiss-Webster mice (n=60) were randomized to either 70% hepatectomy or sham operation and then further subdivided to receive either vehicle (5% ethanol i.p. bid) or wortmannin (0.75mg/kg in 5% ethanol i.p. bid), which was administered approximately 6h prior to surgery and every 12h thereafter. Mice were sacrificed over a time course after operation. To measure DNA synthesis, BrdU was given i.p. 3h prior to sacrifice. Livers were weighed (wet and dry); portions of liver were extracted for DNA, RNA, or protein, or used for histologic assessment. BrdU was quantitated by a novel dot blot procedure (42). (ii) Mice (n=60) were randomized as above and further subdivided to receive either NTC or p85α siSTABLE siRNA (20μg i.v.), administered 24h prior to surgery and every 24h thereafter; DOTAP liposomal transfection reagent was used for all transfections. Mice were sacrificed over a time course after operation. Analysis of tissues was performed as described above. (iii) a. Mice (n=90) were randomized as above and further subdivided to receive either NTC, p110α, or p85α siSTABLE siRNA (20μg i.v.) administered 24h prior to surgery and every 24h thereafter. Mice were sacrificed over a time course after operation. Analysis of tissues was as above. b. The study was repeated as in (iiia), but mice were not pre-treated with siRNA 24h prior to surgery; instead, to allow priming to occur without alteration, mice were given siRNA 6h following operation, and every 24h thereafter. c. The study was repeated as in (iiia), but mice were sacrificed at earlier time points (2, 4, 6, and 12h) following surgery.
Tissue processing, immunohistochemistry, and BrdU dot blot
Upon sacrifice, liver samples were immediately placed in 10% neutral buffered formalin for 24 h, followed by 70% EtOH for 24h. Samples were then paraffin-embedded, sectioned, and stained with hematoxylin and eosin (H&E) or Periodic Acid Schiff (PAS). Additionally, frozen samples were obtained for sectioning and Oil Red O (ORO) staining. IHC was performed on paraffin-embedded samples as previously described (38). Sections (5pm) were cut from paraffin blocks, then deparaffinized in xylene and rehydrated in descending ethanol series. Protein staining was performed using DAKO EnVision Kit (Dako Corp., Carpinteria, CA). Sections were incubated overnight at 4°C with monoclonal antibodies diluted in 0.05M Tris-HCL+1% BSA against F4/80 (1:100), BrdU (1:1000), or PCNA (1:2000). After exposure to secondary antibody, peroxidase substrate DAB was added for staining. All sections were counterstained with hematoxylin and observed by light microscopy. For negative controls, primary antibody was omitted from the above protocol. Additionally, BrdU dot blot was performed as previously described (42).
Protein preparation and Western immunoblot
Western blotting was performed as previously described (20). Tissues were lysed with TNN buffer using a tissue grinder, then placed on ice for 30 m. Lysates were clarified by centrifugation (10,000×g for 30min at 4°C) and protein concentrations determined using the method of Bradford (6). Briefly, total protein (60μg) was resolved on a 10% Nu-PAGE Bis-Tris gel and transferred to PVDF membranes. Filters were incubated overnight at 4°C in blotting solution, followed by a 1 h incubation with primary antibodies. Filters were washed and incubated with horseradish peroxidase-conjugated secondary antibodies for 1h. After three additional washes, the immune complexes were visualized by ECL detection.
Tissue cytokine ELISA
Frozen liver samples were pulverized by mortar and pestle in liquid nitrogen. Protein was then extracted using methods previously described (25, 36). Ice-cold protein extraction buffer, consisting of 0.1% Igepal CA-630 nonionic detergent in PBS with a protease inhibitor cocktail tablet (Complete Mini), was added to pulverized tissue (50μl to 10mg of tissue) and vortexed. The resultant homogenate was centrifuged and supernatant collected for use in colorimetric ELISA for TNF-α or IL-6. Supernatant (50μl) was added to assay diluent (1:1 dilution) in a 96-well plate, and the assays were performed according to the respective protocols. Experiments were performed in duplicate to assure reproducibility.
siRNA delivery study with F4/80 colocalization
A delivery study utilizing DY547-labeled NTC and p85α siRNA duplexes was performed in our laboratory to determine the distribution of labeled siRNA with various delivery methods (23). Tissues from this study were utilized to determine siRNA delivery to the liver; to further determine delivery to Kupffer cells and macrophages, additional staining with F4/80 was performed. Samples were then examined with an Olympus BX51 microscope (Olympus, Central Valley, PA). Three separate images were captured from each field examined, with blue representing nuclear staining, red representing DY547-labeled siRNA, and green representing F4/80 staining.
Statistical analysis
Remnant liver weight as percent body weight was analyzed using analysis of variance for a two-factor factorial experiment. The two factors were assigned as operation (hepatectomy and sham) and siRNA or wortmannin (present and absent). Effects and interaction were assessed at the 0.05 and the 0.15 levels, respectively, of significance. Fisher’s least significant difference procedure was used for multiple comparisons with Bonferroni adjustment for the number of comparisons. All statistical computations were conducted using the SAS® system, Release 9.1.
RESULTS
PI3K inhibition with wortmannin decreases hepatic regeneration following partial hepatectomy
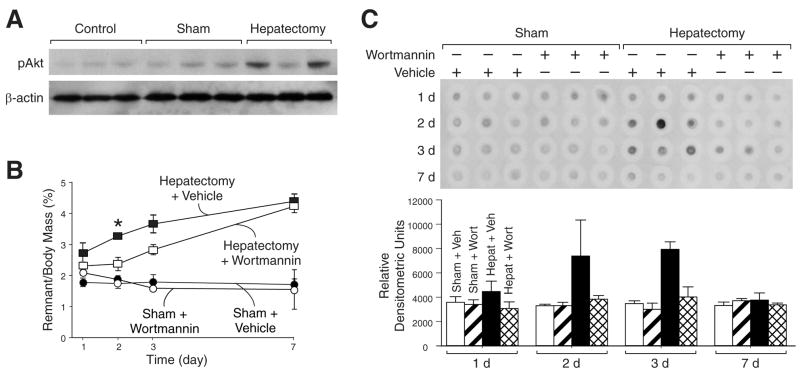
To confirm that PI3K activity is increased following hepatectomy, female Swiss-Webster mice were randomized to receive either 70% hepatectomy (n=3) or sham (n=3) operation; unoperated control mice (n=3) were sacrificed for comparison. Mice were sacrificed 48h following resection, liver was extracted for protein, and Western blot analysis of pAkt was performed. Increased PI3K activity, as reflected by an increase in pAkt expression 48 h following operation (Fig. 1A), was noted. Next, to determine the effects of total PI3K blockade on liver regeneration, female Swiss-Webster mice were randomized to receive either 70% partial hepatectomy or sham operation and then further subdivided to receive the potent PI3K inhibitor, wortmannin, or vehicle via i.p. injection every 12h. Following hepatectomy, a gradual increase in hepatic remnant wet weight was noted in mice treated with vehicle. Remnant liver was approximately 1.8% of body mass at the time of operation, 2.6% after 24h, 3.2% at 48h, 3.8% at 72h, and 4.2% by day 7 (Fig. 1B). Wortmannin treatment decreased liver regeneration relative to vehicle-treated mice, especially at the earliest timepoints. A number of parameters, including coat and skin integrity, color of mucus membranes, daily weight, and quality of stool, were assessed daily. In all groups and timepoints examined, sham mice treated with wortmannin did not manifest side effects; there was no statistically significant weight loss between groups. There was, however, a statistically significant weight loss in mice treated with wortmannin undergoing liver resection and sacrifice 24h later, although this was not noted in mice sacrificed at longer timepoints (data not shown).
Figure 1. Upregulation of PI3K activity following hepatectomy.
(A) Mice (n=3 per group) underwent sham operation or hepatectomy; control mice did not undergo operation. Mice were sacrificed 48h following operation and Western blot analysis was performed to determine pAkt expression; β-actin served as a loading control. (B) Mice (n=60) were randomized to either 70% hepatectomy or sham operation and further subdivided to receive vehicle or wortmannin. Mice were sacrificed over a time course after operation, and wet remnant weight as a percent of body mass was determined as an estimate of liver regeneration. (C) To measure DNA synthesis, BrdU was given ip 3h prior to sacrifice, and DNA dot blot was performed with BrdU probe; densitometric analysis was performed and shown below the blot.
To further confirm that the decrease in hepatic regeneration is due to reduced hepatocyte proliferation, BrdU incorporation was compared in mice treated with wortmannin or vehicle by DNA dot-blot (Fig. 1C). Mice treated with vehicle demonstrated an increase in DNA synthesis at 48h and 72h following hepatectomy; wortmannin significantly inhibited DNA synthesis at these early time points.
siRNA directed to the p85α regulatory and p110α catalytic subunits decreases hepatic regeneration following partial hepatectomy
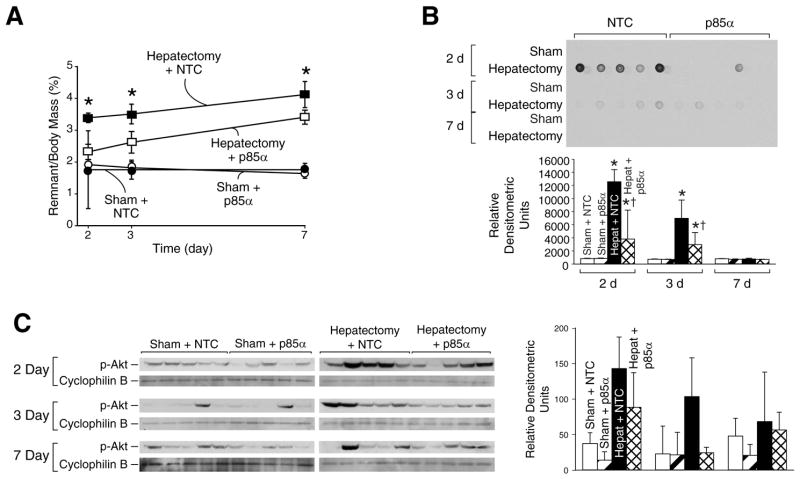
To determine the effects of selective p85α subunit blockade on liver regeneration, female Swiss-Webster mice were randomized to receive either 70% partial hepatectomy or sham operation and further subdivided to receive either NTC siSTABLE siRNA or p85α siSTABLE siRNA via hydrodynamic tail vein injection every 24h. Mice were killed 48h, 72h, and 7d following operation, and wet liver remnant weight was determined (Fig. 2A). Following hepatectomy, mice treated with NTC siRNA demonstrated the expected increase in hepatic remnant wet weight (1.8% of body mass at the time of operation, 3.3% at 48h, 3.5% at 72h, and 4.0% by 7d). Mice treated with p85α siRNA demonstrated a significant decrease in liver regeneration relative to vehicle-treated mice at all timepoints. Liver mass in p85α siRNA-treated mice increased from approximately 1.8% of body mass at the time of operation, to 2.3% at 48h, 2.6% at 72h, and 3.2% at 7d. Body mass of mice treated with p85α siRNA following hepatectomy decreased to a similar extent as mice treated with NTC siRNA; there were no systemic toxic effects of treatment with p85α siRNA in mice undergoing sham operation or hepatectomy (data not shown).
Figure 2. Selective inhibition of the p85α regulatory subunit leads to decreased regeneration and decreased F4/80 positive cells.
(A) Mice (n=60) were randomized to either 70% hepatectomy or sham operation and then further subdivided to receive either NTC or p85α siSTABLE siRNA; mice were sacrificed over a time course after operation, and wet remnant weight as a percent of body mass was determined as in figure 1B. (B) To measure DNA synthesis, BrdU was given ip 3h prior to sacrifice, and DNA dot blot was performed as in figure 1C; densitometric analysis was performed and shown below the blot. (C) Western blot analysis of liver tissue lysates was performed as described in the Materials and Methods to determine phosphorylation and activation of Akt following sham operation or hepatectomy, and to compare pAkt expression in mice treated with either NTC or p85α siRNA; cyclophilin B served as a loading control; densitometric analysis of the Western blot is shown in the right panel.
To further confirm that the decrease in hepatic regeneration is due to reduced hepatocyte proliferation, BrdU incorporation was compared in mice treated with NTC or p85α siRNA by DNA dot blot (Fig. 2B). Mice treated with NTC siRNA demonstrated an expected increase in DNA synthesis at 48 h and 72 h following hepatectomy; this induction of DNA synthesis was inhibited by p85α siRNA.
To confirm that treatment with p85α siRNA led to a decrease in PI3K activation following hepatectomy, Western blot analysis of protein isolates was performed for pAkt (Fig. 2C) with densitometric analysis (Fig. 2D). Following hepatectomy, mice treated with p85α siRNA showed decreased pAkt expression at 48 h and 72 h relative to mice treated with NTC siRNA.
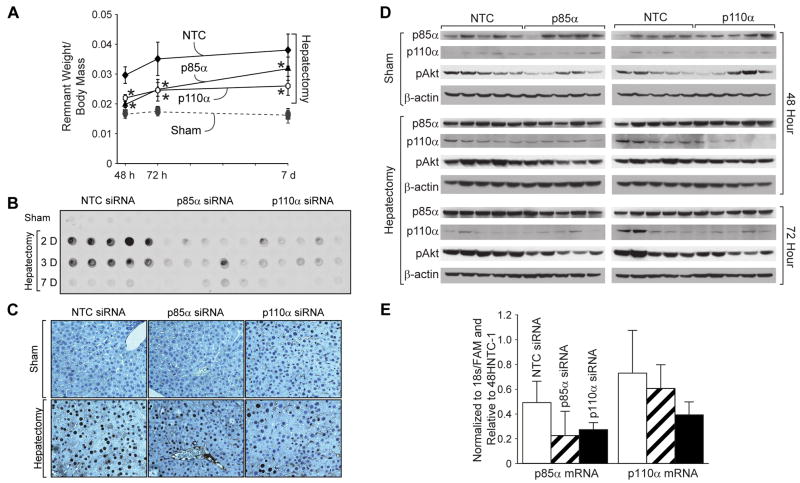
Next, to determine the effects of selective p110α subunit blockade on liver regeneration and to confirm our previous results using p85α siRNA, female Swiss-Webster mice were randomized to receive either 70% partial hepatectomy or sham operation; they were further subdivided to receive either NTC siSTABLE siRNA, p85α siSTABLE siRNA, or p110α siSTABLE siRNA via hydrodynamic tail vein injection every 24h. Mice were killed 48h, 72h, and 7d following operation, and wet liver remnant weight was measured (Fig. 3A). As previously described, following hepatectomy, mice treated with NTC siRNA demonstrated the expected gradual increase in hepatic remnant wet weight. However, liver regeneration was significantly inhibited in p110α siRNA-treated mice. Results for p85α siRNA were identical to those previously shown (data not shown).
Figure 3. Selective inhibition of the p85α regulatory or the p110α catalytic subunit leads to decreased regeneration following liver resection.
(A) Mice (n=90) were randomized to either 70% hepatectomy or sham operation and then further subdivided to receive either NTC, p85α or p110α siSTABLE siRNA; mice were sacrificed over a time course after operation, and wet remnant weight as a percent of body mass was determined as in figures 1B and 2A. (B) To measure DNA synthesis, BrdU was given ip 3h prior to sacrifice, and DNA dot blot was performed as in figures 1C and 2B. (C) IHC was performed with anti-BrdU antibody to confirm decreased staining in p85α- and p110α-siRNA treated mice relative to NTC-treated mice. (D) Western blot analysis of liver tissue lysates was performed as described in the Materials and Methods to determine p85α, p110α and pAkt expression in mice treated with either NTC, p85α or p110α siRNA who underwent sham operation or hepatectomy; β-actin served as a loading control. (E) Real time quantitative PCR was performed on RNA isolated from livers at the 48h timepoint to determine p85α and p110α subunit knockdown. Results were normalized to 18s/FAM.
To again confirm that the decrease in hepatic regeneration is due to reduced hepatocyte proliferation, BrdU incorporation was compared by DNA dot blot (Fig. 3B). Mice treated with NTC siRNA demonstrated an expected increase in DNA synthesis at 48h and 72h following hepatectomy, as previously described; however, mice treated with p110α siRNA did not experience a similar increase. BrdU IHC was also performed to confirm dot blot results; representative staining patterns are presented in Figure 3C.
Additionally, we performed Western blot analysis to assess p110α and pAkt expression (Fig. 3D). There appeared to be a slight decrease in p110α and pAkt protein expression in mice undergoing sham operation or hepatectomy treated with p110α siRNA relative to mice treated with NTC siRNA. To confirm this finding, real time quantitative PCR (RT-PCR) was performed to assess p85α and p110α mRNA expression following siRNA treatment (Fig. 3E). While there appeared to be decreased p85α mRNA in mice treated with p85α siRNA, and decreased p110α mRNA in mice treated with p110α siRNA, this difference was not statistically significant; this suggested the localization of siRNA to only a subpopulation of cells without uptake by the majority of hepatocytes.
Lastly, to determine if p85α and p110α siRNA administration led to decreased liver regeneration by inhibiting the essential priming reaction, we repeated the experiment above without siRNA pre-treatment; instead, mice were given NTC, p85α, or p110α siRNA via hydrodynamic tail vein injection 6 h following sham operation or hepatectomy to allow for the completion of the priming response. Interestingly, there was no statistically significant difference in liver regeneration between mice treated with NTC, p85α, or p110α siRNA (data not shown). This suggests an important role for PI3K in the early response to liver resection, including the initiation of the priming response.
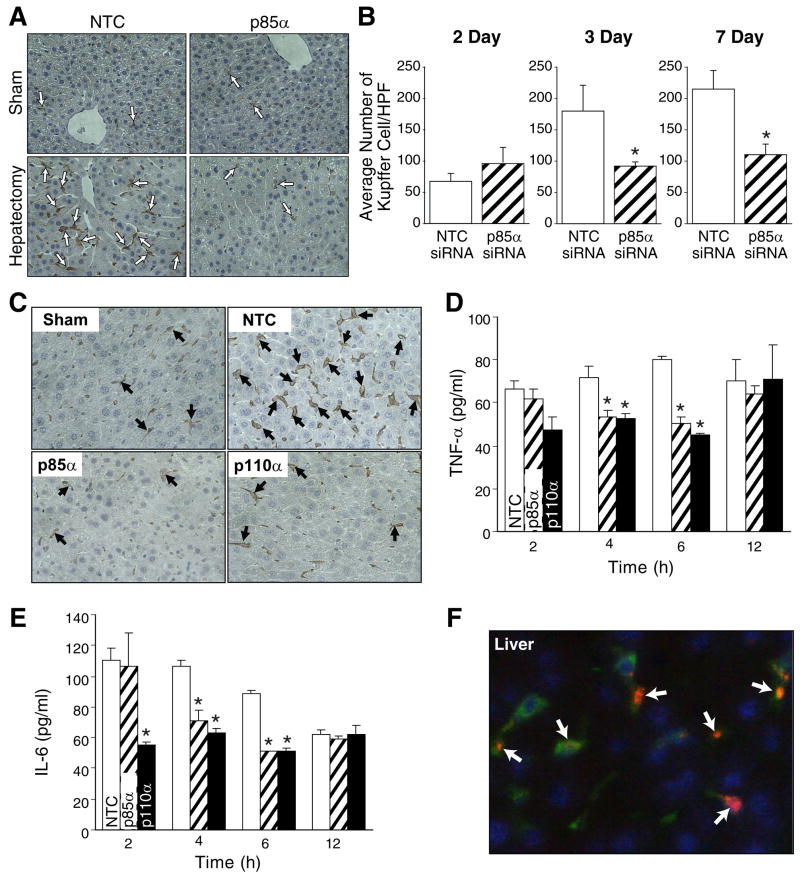
Selective inhibition of the p85α regulatory or the p110α catalytic subunit decreases macrophage infiltration and Kupffer cell hyperplasia/hypertrophy within the regenerating liver with a concomitant decrease in cytokine expression
Circulating macrophages and resident Kupffer cells elaborate cytokines that prime hepatocytes, leading to the transition from quiescence to growth factor-sensitive growth and division. Given the known importance of the PI3K pathway in macrophage migration and cytokine production (7, 32, 35, 43), we next performed F4/80 IHC staining on liver samples from mice undergoing sham operation or liver resection treated with NTC- or p85α siRNA (Fig. 4A). Following hepatectomy, mice treated with NTC siRNA demonstrated an increase in F4/80 positive cells (67±13 at 48h; 180±41 at 72h; and 214±30 at 7d) concentrated around the central vein and scattered throughout the parenchyma. However, mice treated with p85α siRNA showed no increase in F4/80 positive cells (95±25 at 48h; 92±6 at 72h; and 109±16 at 7d) over the same time course (Fig. 4B).
Figure 4. Selective inhibition of the p85α regulatory or the p110α catalytic subunit decreases macrophage infiltration and Kupffer cell hyperplasia/hypertrophy within the regenerating liver; this is associated with decreased tissue cytokine expression.
(A) IHC for F4/80 was performed on mice undergoing sham operation or liver resection; representative sections are shown from the 72h timepoint. Arrows indicate F4/80 staining cells. Magnification: x200. (B) Slides stained for F4/80 were presented to a pathologist in a blinded fashion, and number of positive cells per 8 high power fields were counted. Bars represent average and SD of 8 fields; asterisks indicate statistically significant changes. *=P≤0.01 relative to NTC siRNA-treated mice. (C) To confirm our findings, IHC for F4/80 was performed on mice undergoing sham operation or liver resection treated with NTC, p85α, or p110α siRNA; representative sections are shown from the 72 h timepoint. Arrows indicate F4/80 staining cells. Magnification: x400. To determine if the reduced number of F4/80 staining cells present in liver of p85α or p110α siRNA-treated mice translated to decreased tissue cytokine expression, tissue cytokine ELISAs were performed for TNF-α (D) or IL-6 (E) at 2, 4, 6, and 12h time points as described in the Materials and Methods. Results are expressed as pg/ml. *=P≤0.01 relative to NTC siRNA-treated mice. (F) siRNA delivery to the liver was confirmed by injection of mice with DY547-labeled p85α siRNA; additional staining with anti-F4/80 antibody was performed to determine colocalization of F4/80 with siRNA (arrows). Representative sections are shown for liver from a mouse treated with onetime tail vein injection of labeled siRNA sacrificed 24h following injection. Red = DY547-labeled siRNA; green = F4/80; blue = Hoescht nuclear stain.
To confirm our findings that macrophage migration and Kupffer cell hyperplasia was decreased in mice treated with PI3K inhibitors, we next performed F4/80 IHC staining on liver samples from mice undergoing sham operation or liver resection treated with NTC, p85α, or p110α siRNA (Fig. 4C). Following hepatectomy, mice treated with NTC siRNA again demonstrated a gradual increase in F4/80 positive cells concentrated around the central vein and scattered throughout the parenchyma. However, mice treated with p85α or p110α siRNA showed no increase in F4/80 positive cells over the same time course. To determine whether the decreased number of F4/80 positive cells, coupled with PI3K inhibition, was associated with decreased cytokine expression, we repeated the hepatectomy study with sacrifice at earlier time points (eg, 2, 4, 6, and 12h) and performed tissue cytokine analysis for TNF-α (Fig. 4D) and IL-6 (Fig. 4E) on protein lysates from mice undergoing sham operation or hepatectomy and treatment with NTC, p85α, or p110α siRNA. We found statistically significantly higher levels of both TNF-α and IL-6 in the regenerating livers of mice treated with NTC siRNA at 48h and 72h following hepatectomy relative to p85α and p110α siRNA treated mice.
In an effort to determine whether siRNA was indeed taken up by macrophages, and to determine if siRNA was also delivered to hepatocytes, we performed an siRNA delivery study with DY547-labeled NTC and p85α siRNA (23). Additionally, we stained with F4/80 to determine if Kupffer cells and macrophages expressed the siRNA. Figure 4F represents liver samples harvested from a mouse treated with p85α DY547-labeled siRNA, delivered via tail vein injection, 24h following injection. Surprisingly, there was very little delivery of siRNA to the hepatocytes, but there appeared to be high concentrations of labeled siRNA within the Kupffer cells and macrophages present within the liver. Similarly, there was significant uptake of siRNA by the bone marrow, including the majority of F4/80 cells present (data not shown). Therefore, we hypothesize that the lack of significant p85α and p110α knockdown as detected by Western blot and RT-PCR is due to a relatively selective delivery of siRNA to Kupffer cells, which comprise the minority of the hepatic cellular mass.
Selective inhibition of the p85α regulatory or the p110α catalytic subunit leads to vacuolation and increased lipid and glycogen deposition within the liver after partial hepatectomy
To determine if architectural changes occurred in the livers after treatment, H&E staining was performed on liver samples from mice undergoing sham operation or liver resection treated with NTC, p85α, or p110α siRNA (Fig. 5A). We identified a widespread vacuolar appearance in the livers of mice undergoing liver resection that had been treated with p85α or p110α siRNA, in contrast to the normal appearance of the regenerating liver in NTC siRNA-treated mice (Fig. 5A, right panels); this appearance has been previously termed panlobular hydropic vacuolization, and has been described in association with inhibition of hepatocyte priming following liver resection (21, 30). Notably, the livers of sham-operated mice treated with p85α or p110α siRNA lacked this histological finding (Fig. 5A, left panels). To further investigate the architectural changes found in these mice, we performed ORO staining for lipid (Fig. 5B) and PAS staining for glycogen (Fig. 5C), and found widespread lipid and glycogen deposition, respectively, throughout the livers of mice undergoing liver resection and treated with p85α or p110α siRNA relative to NTC-treated mice (Fig. 5B and 5C, right panels). Again, we found no change in lipid or glycogen deposition in sham-operated mice treated with siRNAs (Fig. 5B and 5C, left panels).
Figure 5. Selective inhibition of the p85α regulatory or the p110α catalytic subunit conferred a vacuolar appearance to the regenerating liver, with increased glycogen and lipid deposition within hepatocytes.
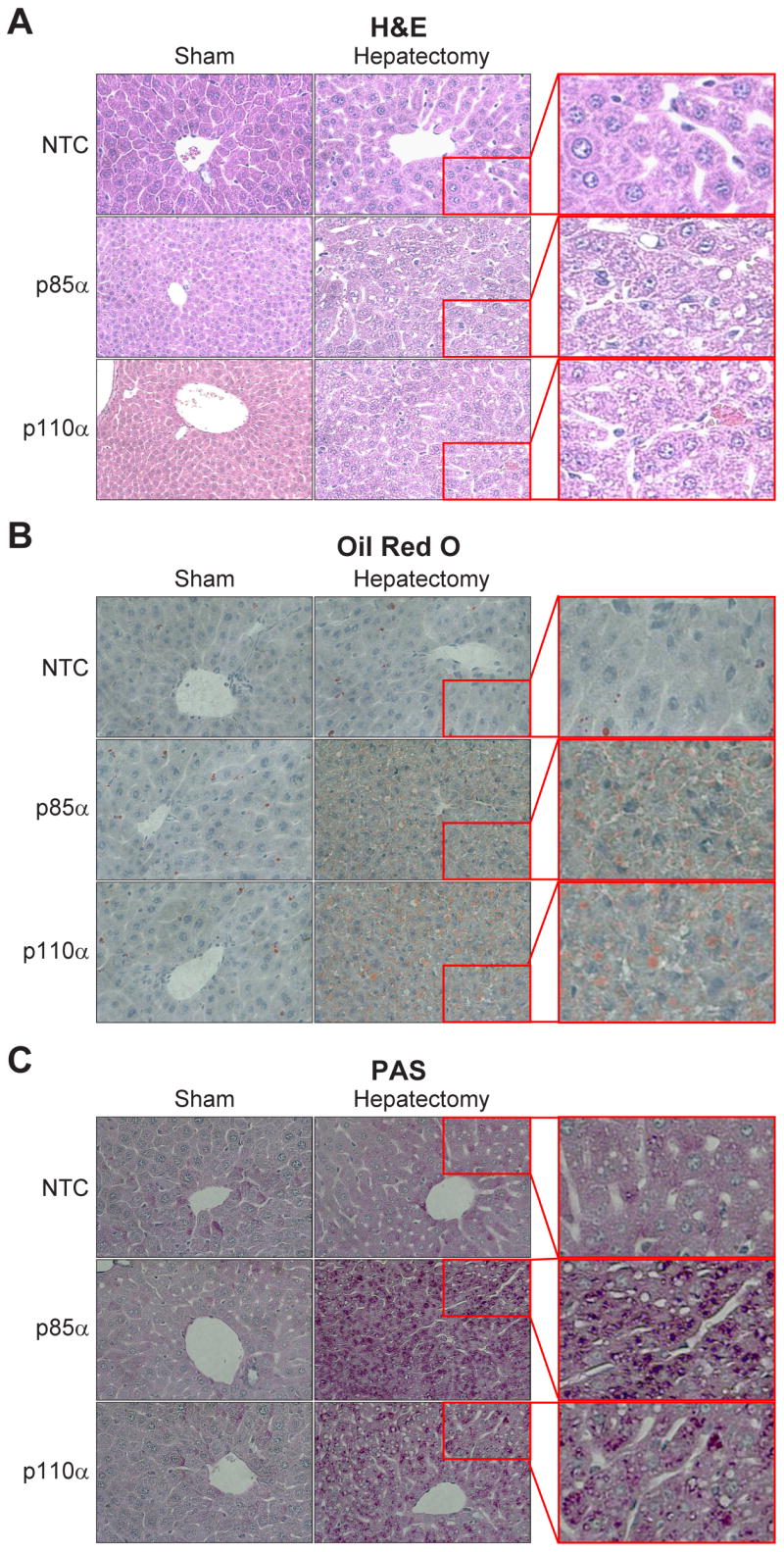
(A) H&E staining was performed on formalin-fixed, paraffin embedded liver samples from mice undergoing sham operation or hepatectomy. Representative sections are shown for NTC-, p85α-, and p110α siRNA-treated mice. Right panels show enlarged views of insets from the hepatectomy groups. (B) Oil Red O (ORO) staining was performed on frozen-sectioned liver samples from mice undergoing sham operation or hepatectomy to determine distribution of lipid storage. Representative sections are shown for NTC-, p85α-, and p110α siRNA-treated mice. Right panels show enlarged views of insets from the hepatectomy groups. (C) PAS staining was performed on paraffin-embedded sections of liver samples from mice undergoing sham operation or hepatectomy to determine distribution of glycogen storage. Representative sections are shown for NTC-, p85α-, and p110α siRNA-treated mice. Right panels show enlarged views of insets from the hepatectomy groups.
DISCUSSION
Class I PI3Ks are heterodimers composed of a regulatory (p85) and catalytic (p110) subunit; the regulatory p85 subunit is essential for the stability of the p110 catalytic subunit and for its recruitment to activated receptor tyrosine kinases or G-protein-coupled receptors. Although PI3K has been shown to be important for the proliferation of various cell types, including intestinal epithelial cells, pancreatic acinar cells, and vascular endothelial cells (31, 40, 44), and important for leukocyte trafficking, signaling, and phagocytosis (7), its role in hepatic regeneration is not known. In our present study, we demonstrate three major findings: (1) PI3K is activated following partial hepatectomy in mice, especially at the earliest timepoints; (2) total PI3K inhibition with the pharmacologic inhibitor wortmannin, or selective inhibition of the PI3K subunits p85α and p110α using siRNA, markedly decreases the regenerative response of the liver after resection, and leads to vacuolar architectural changes with glycogen and lipid deposition; and (3) total or selective PI3K inhibition leads to decreased Kupffer cells and infiltrating macrophages, with concomitant decrease in cytokine expression, within the regenerating liver.
The recruitment of leukocytes to sites of inflammation or regeneration relies on their ability to recognize chemoattractants, reorganize their cytoskeleton and membrane structure in response to such stimulants, undergo transendothelial cell migration, and interact with adhesion molecules expressed at sites of inflammation (35). The PI3K pathway plays a pivotal role in each step of this process, and studies have suggested differential roles for the various catalytic subunits of class I PI3Ks based on cell type. One study found that p110α inhibition leads to complete inhibition of macrophage colony stimulating factor (M-CSF)-mediated DNA synthesis in a macrophage cell line, and p110β or p110δ isoform inhibition leads to impaired lamellipodia formation and macrophage migration (43). Another study utilizing p85α−/− murine bone-marrow derived macrophages concluded that the p85α subunit is required for the regulation of multiple actin-based functions in these cells, including adhesion, migration, wound healing, and phagocytosis (32). A third study found that p110γ or p110δ deficiency results in defective selectin-mediated adhesion of neutrophils, and a failure of accumulation of these leukocytes in an acute lung injury model (35). Thus, PI3K-mediated regulation of leukocyte activation and migration is complex and still poorly understood. In our present study, we identified preferential uptake of labeled siRNA into the reticuloendothelial system, including Kupffer cells and F4/80+ cells within the bone marrow, and hypothesize that total or selective PI3K inhibition leads to decreased Kupffer cell hyperplasia and hypertrophy, as well as decreased mobilization of bone marrow progenitor cells and circulating macrophages into the regenerating liver.
Circulating macrophages and resident Kupffer cells, along with the cytokines they produce, play a critical role in hepatic regeneration following resection. Aldeguer et al (2) utilized a murine model of bone marrow transplantation coupled with 70% hepatectomy to determine the contribution of bone-marrow derived cells to liver regeneration and found that complete replacement of IL-6+/+ bone marrow into IL-6−/− irradiated mice restored regeneration after partial hepatectomy. However, complete replacement of IL-6−/− bone marrow into IL-6+/+ irradiated mice significantly inhibited liver regeneration; regeneration can be restored in this case by administration of IL-6. Similarly, experiments utilizing GFP-labeled bone marrow cells have demonstrated an important role for the bone marrow in repopulation of the resected liver, where a majority of cells (70%) commit to an endothelial cell lineage and others (28%) to a Kupffer cell lineage (16). Other studies have used models of Kupffer cell depletion (liposome-encapsulated dichlormethylene-diphosphonate [Cl2MDP], gadolinium chloride [GdCl3], or pentoxifylline) to demonstrate that a >90% reduction in Kupffer cells leads to significantly impaired liver regeneration following 70% hepatectomy in mice or rats; this response is attributed to a significant decrease in TNFα production (5, 11, 39). Additionally, utilizing the GdCl3 model of Kupffer cell depletion, another group confirmed that Kupffer cell depletion significantly decreased liver regeneration and abolished hepatic expression of IL-6, HGF, and TNFα (27); similarly, the administration of TNFα antibody to mice following hepatectomy significantly inhibited regeneration (1). Given the known importance of IL-6 and TNFα to the priming of quiescent hepatocytes to enter the cell cycle, it is clear that macrophages and Kupffer cells are central to the early regenerative response of the liver, and that the bone marrow does contribute, in part, to the increase in Kupffer cell number following hepatectomy. In our current study, we found that there were significantly fewer F4/80 positive cells present in the regenerating livers of mice treated with total or selective PI3K inhibitors; this correlated with a decrease in cytokine production, as reflected by decreased tissue IL-6 and TNFα cytokine levels. Therefore, we propose that PI3K inhibition leads not only to decreased Kupffer cells and macrophages within the regenerating liver, but secretory dysfunction of Kupffer cells and macrophages present, resulting in decreased cytokine production and reduced hepatocyte priming. This leads to decreased liver regeneration in wortmannin, p85α, or p110α siRNA-treated mice following resection.
In addition to its role in leukocyte trafficking, signaling, and phagocytosis, class I PI3Ks play an important role in cell survival and proliferation. The growth factors HGF, EGF, and TGFa, responsible for the progression of hepatocytes from the G1 to S phase of the cell cycle after priming by IL-6 and TNFα, signal through receptor tyrosine kinases which, in the activated state, associate with cytosolic proteins rich in Src-homologies, such as PI3K (4, 22). Downstream signaling leads to the activation of MAPK cascades, ultimately leading to cell proliferation (4). Therefore, it can be assumed that PI3K inhibition in this setting would lead to altered growth factor signaling; alternate pathways likely compensate over time, such that late liver regeneration may be possible. Given its solubility and wide volume of distribution, treatment with wortmannin is likely to inhibit all cell types present within the liver (3), and likely alters hepatocyte PI3K response to growth factors and cytokines in addition to its action on macrophages and Kupffer cells. However, we did not identify significantly higher attenuation of regeneration in wortmannin-treated mice relative to mice treated with p85α or p110α siRNA. Thus, we propose that decreased hepatocyte priming via decreased cytokine production is the primary mechanism of regeneration attenuation with PI3K inhibition.
We conclude that the PI3K pathway is an important early regulator of hepatic regeneration following partial hepatectomy. We suggest a dual mechanism of action in this case: (1) PI3K is important for the migration of macrophages to the site of regeneration; inhibition leads to decreased IL-6 and TNFα expression and, ultimately, a lack in the priming mechanism central to early regeneration; this appears to be the central mechanism by which PI3K inhibition leads to attenuation of regeneration; and (2) PI3K plays an important role in HGF, EGF, and TGFα-induced hepatocyte survival, DNA synthesis, and cell division.
Acknowledgments
The authors would like to thank Junji Ueda, Hiroaki Watanabe, Cornelius Elferink, and Kathleen O’Connor for helpful comments and advice; Arwen Stelter for technical assistance; Karen Martin for manuscript preparation; and Tatsuo Uchida for statistical analysis.
GRANTS This work was supported by grants R01DK48498, R37AG10885, P01DK35608, and T32DK07639 from the National Institutes of Health.
Footnotes
This paper was presented, in part, at the annual meetings of the American College of Surgeons (October 8-12, 2006, Chicago, IL) and the American Surgical Congress (February 7-10, 2007, Phoenix, AZ).
DISCLOSURES We have nothing to disclose.
References
- 1.Akerman P, Cote P, Yang SQ, McClain C, Nelson S, Bagby GJ, Diehl AM. Antibodies to tumor necrosis factor-alpha inhibit liver regeneration after partial hepatectomy. Am J Physiol. 1992;263:G579–585. doi: 10.1152/ajpgi.1992.263.4.G579. [DOI] [PubMed] [Google Scholar]
- 2.Aldeguer X, Debonera F, Shaked A, Krasinkas AM, Gelman AE, Que X, Zamir GA, Hiroyasu S, Kovalovich KK, Taub R, Olthoff KM. Interleukin-6 from intrahepatic cells of bone marrow origin is required for normal murine liver regeneration. Hepatology. 2002;35:40–48. doi: 10.1053/jhep.2002.30081. [DOI] [PubMed] [Google Scholar]
- 3.Arcaro A, Wymann MP. Wortmannin is a potent phosphatidylinositol 3-kinase inhibitor: the role of phosphatidylinositol 3,4,5-trisphosphate in neutrophil responses. Biochem J. 1993;296 ( Pt 2):297–301. doi: 10.1042/bj2960297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borowiak M, Garratt AN, Wustefeld T, Strehle M, Trautwein C, Birchmeier C. Met provides essential signals for liver regeneration. Proc Natl Acad Sci U S A. 2004;101:10608–10613. doi: 10.1073/pnas.0403412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulton RA, Alison MR, Golding M, Selden C, Hodgson HJ. Augmentation of the early phase of liver regeneration after 70% partial hepatectomy in rats following selective Kupffer cell depletion. J Hepatol. 1998;29:271–280. doi: 10.1016/s0168-8278(98)80013-5. [DOI] [PubMed] [Google Scholar]
- 6.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Camps M, Ruckle T, Ji H, Ardissone V, Rintelen F, Shaw J, Ferrandi C, Chabert C, Gillieron C, Francon B, Martin T, Gretener D, Perrin D, Leroy D, Vitte PA, Hirsch E, Wymann MP, Cirillo R, Schwarz MK, Rommel C. Blockade of PI3Kgamma suppresses joint inflammation and damage in mouse models of rheumatoid arthritis. Nat Med. 2005;11:936–943. doi: 10.1038/nm1284. [DOI] [PubMed] [Google Scholar]
- 8.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 9.Desmots F, Rissel M, Gilot D, Lagadic-Gossmann D, Morel F, Guguen-Guillouzo C, Guillouzo A, Loyer P. Pro-inflammatory cytokines tumor necrosis factor alpha and interleukin-6 and survival factor epidermal growth factor positively regulate the murine GSTA4 enzyme in hepatocytes. J Biol Chem. 2002;277:17892–17900. doi: 10.1074/jbc.M112351200. [DOI] [PubMed] [Google Scholar]
- 10.Diehl A. The role of cytokines in hepatic regeneration. Curr Opinion Gastro. 1997;13:525–533. [Google Scholar]
- 11.Duffield JS, Forbes SJ, Constandinou CM, Clay S, Partolina M, Vuthoori S, Wu S, Lang R, Iredale JP. Selective depletion of macrophages reveals distinct, opposing roles during liver injury and repair. J Clin Invest. 2005;115:56–65. doi: 10.1172/JCI22675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fausto N. Liver regeneration and repair: hepatocytes, progenitor cells, and stem cells. Hepatology. 2004;39:1477–1487. doi: 10.1002/hep.20214. [DOI] [PubMed] [Google Scholar]
- 13.Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology. 2006;43:S45–53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- 14.Fausto N, Riehle KJ. Mechanisms of liver regeneration and their clinical implications. J Hepatobiliary Pancreat Surg. 2005;12:181–189. doi: 10.1007/s00534-005-0979-y. [DOI] [PubMed] [Google Scholar]
- 15.Fruman DA, Mauvais-Jarvis F, Pollard DA, Yballe CM, Brazil D, Bronson RT, Kahn CR, Cantley LC. Hypoglycaemia, liver necrosis and perinatal death in mice lacking all isoforms of phosphoinositide 3-kinase p85 alpha. Nat Genet. 2000;26:379–382. doi: 10.1038/81715. [DOI] [PubMed] [Google Scholar]
- 16.Fujii H, Hirose T, Oe S, Yasuchika K, Azuma H, Fujikawa T, Nagao M, Yamaoka Y. Contribution of bone marrow cells to liver regeneration after partial hepatectomy in mice. J Hepatol. 2002;36:653–659. doi: 10.1016/s0168-8278(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 17.Haga S, Ogawa W, Inoue H, Terui K, Ogino T, Igarashi R, Takeda K, Akira S, Enosawa S, Furukawa H, Todo S, Ozaki M. Compensatory recovery of liver mass by Akt-mediated hepatocellular hypertrophy in liver-specific STAT3-deficient mice. J Hepatol. 2005;43:799–807. doi: 10.1016/j.jhep.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 18.Hausenloy DJ, Yellon DM. Survival kinases in ischemic preconditioning and postconditioning. Cardiovasc Res. 2006;70:240–253. doi: 10.1016/j.cardiores.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 19.Higgins G, Anderson R. Experimental pathology of the liver: restoration of liver of the white rat following partial surgical removal. Arch Pathol. 1931;365:179–183. [Google Scholar]
- 20.Jackson LN, Li J, Chen LA, Townsend CM, Evers BM. Overexpression of wild-type PKD2 leads to increased proliferation and invasion of BON endocrine cells. Biochem Biophys Res Commun. 2006;348:945–949. doi: 10.1016/j.bbrc.2006.07.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanel GC, Korula J. Atlas of liver pathology. Edinburgh: Elsevier Saunders; 2005. [Google Scholar]
- 22.Koniaris LG, McKillop IH, Schwartz SI, Zimmers TA. Liver regeneration. J Am Coll Surg. 2003;197:634–659. doi: 10.1016/S1072-7515(03)00374-0. [DOI] [PubMed] [Google Scholar]
- 23.Larson SD, Jackson LN, Chen LA, Rychahou PG, Evers BM. Effectiveness of siRNA uptake in target tissues by various delivery methods. Surgery. 2007;142:262–269. doi: 10.1016/j.surg.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malik R, Selden C, Hodgson H. The role of non-parenchymal cells in liver growth. Semin Cell Dev Biol. 2002;13:425–431. doi: 10.1016/s1084952102001301. [DOI] [PubMed] [Google Scholar]
- 25.Matalka KZ, Tutunji MF, Abu-Baker M, Abu Baker Y. Measurement of protein cytokines in tissue extracts by enzyme-linked immunosorbent assays: application to lipopolysaccharide-induced differential milieu of cytokines. Neuro Endocrinol Lett. 2005;26:231–236. [PubMed] [Google Scholar]
- 26.Mawet E, Shiratori Y, Hikiba Y, Takada H, Yoshida H, Okano K, Komatsu Y, Matsumura M, Niwa Y, Omata M. Cytokine-induced neutrophil chemoattractant release from hepatocytes is modulated by Kupffer cells. Hepatology. 1996;23:353–358. doi: 10.1053/jhep.1996.v23.pm0008591863. [DOI] [PubMed] [Google Scholar]
- 27.Meijer C, Wiezer MJ, Diehl AM, Schouten HJ, Schouten HJ, Meijer S, van Rooijen N, van Lambalgen AA, Dijkstra CD, van Leeuwen PA. Kupffer cell depletion by CI2MDP-liposomes alters hepatic cytokine expression and delays liver regeneration after partial hepatectomy. Liver. 2000;20:66–77. doi: 10.1034/j.1600-0676.2000.020001066.x. [DOI] [PubMed] [Google Scholar]
- 28.Michalopoulos GK. Liver regeneration: molecular mechanisms of growth control. Faseb J. 1990;4:176–187. [PubMed] [Google Scholar]
- 29.Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell KA, Lockhart CA, Huang G, Elferink CJ. Sustained aryl hydrocarbon receptor activity attenuates liver regeneration. Mol Pharmacol. 2006;70:163–170. doi: 10.1124/mol.106.023465. [DOI] [PubMed] [Google Scholar]
- 31.Miyashita K, Itoh H, Sawada N, Fukunaga Y, Sone M, Yamahara K, Yurugi-Kobayashi T, Park K, Nakao K. Adrenomedullin provokes endothelial Akt activation and promotes vascular regeneration both in vitro and in vivo. FEBS Lett. 2003;544:86–92. doi: 10.1016/s0014-5793(03)00484-8. [DOI] [PubMed] [Google Scholar]
- 32.Munugalavadla V, Borneo J, Ingram D, Kapur R. p85α subunit of class IA PI-3 kinase is crucial for macrophage growth and migration. Blood. 2005;106:103–109. doi: 10.1182/blood-2004-10-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okano J, Shiota G, Matsumoto K, Yasui S, Kurimasa A, Hisatome I, Steinberg P, Murawaki Y. Hepatocyte growth factor exerts a proliferative effect on oval cells through the PI3K/AKT signaling pathway. Biochem Biophys Res Commun. 2003;309:298–304. doi: 10.1016/j.bbrc.2003.04.002. [DOI] [PubMed] [Google Scholar]
- 34.Osawa Y, Banno Y, Nagaki M, Brenner DA, Naiki T, Nozawa Y, Nakashima S, Moriwaki H. TNF-alpha-induced sphingosine 1-phosphate inhibits apoptosis through a phosphatidylinositol 3-kinase/Akt pathway in human hepatocytes. J Immunol. 2001;167:173–180. doi: 10.4049/jimmunol.167.1.173. [DOI] [PubMed] [Google Scholar]
- 35.Puri KD, Doggett TA, Huang CY, Douangpanya J, Hayflick JS, Turner M, Penninger J, Diacovo TG. The role of endothelial PI3Kgamma activity in neutrophil trafficking. Blood. 2005;106:150–157. doi: 10.1182/blood-2005-01-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosengren S, Firestein GS, Boyle DL. Measurement of inflammatory biomarkers in synovial tissue extracts by enzyme-linked immunosorbent assay. Clin Diagn Lab Immunol. 2003;10:1002–1010. doi: 10.1128/CDLI.10.6.1002-1010.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rychahou PG, Jackson LN, Farrow BJ, Evers BM. RNA interference: mechanisms of action and therapeutic consideration. Surgery. 2006;140:719–725. doi: 10.1016/j.surg.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 38.Rychahou PG, Jackson LN, Silva SR, Rajaraman S, Evers BM. Targeted molecular therapy of the PI3K pathway: therapeutic significance of PI3K subunit targeting in colorectal carcinoma. Ann Surg. 2006;243:833–842. doi: 10.1097/01.sla.0000220040.66012.a9. discussion 843–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Selzner N, Selzner M, Odermatt B, Tian Y, Van Rooijen N, Clavien PA. ICAM-1 triggers liver regeneration through leukocyte recruitment and Kupffer cell-dependent release of TNF-alpha/IL-6 in mice. Gastroenterology. 2003;124:692–700. doi: 10.1053/gast.2003.50098. [DOI] [PubMed] [Google Scholar]
- 40.Sheng H, Shao J, Townsend CM, Jr, Evers BM. Phosphatidylinositol 3-kinase mediates proliferative signals in intestinal epithelial cells. Gut. 2003;52:1472–1478. doi: 10.1136/gut.52.10.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thorgeirsson SS. Hepatic stem cells in liver regeneration. Faseb J. 1996;10:1249–1256. [PubMed] [Google Scholar]
- 42.Ueda J, Saito H, Watanabe H, Evers BM. Novel and quantitative DNA dot-blotting method for assessment of in vivo proliferation. Am J Physiol Gastrointest Liver Physiol. 2005;288:G842–847. doi: 10.1152/ajpgi.00463.2004. [DOI] [PubMed] [Google Scholar]
- 43.Vanhaesebroeck B, Jones GE, Allen WE, Zicha D, Hooshmand-Rad R, Sawyer C, Wells C, Waterfield MD, Ridley AJ. Distinct PI(3)Ks mediate mitogenic signalling and cell migration in macrophages. Nat Cell Biol. 1999;1:69–71. doi: 10.1038/9045. [DOI] [PubMed] [Google Scholar]
- 44.Watanabe H, Saito H, Rychahou PG, Uchida T, Evers BM. Aging is associated with decreased pancreatic acinar cell regeneration and phosphatidylinositol 3-kinase/Akt activation. Gastroenterology. 2005;128:1391–1404. doi: 10.1053/j.gastro.2005.03.016. [DOI] [PubMed] [Google Scholar]