Abstract
Since the discovery of the first human neocentromere in 1993, these spontaneous, ectopic centromeres have been shown to be an astonishing example of epigenetic change within the genome. Recent research has focused on the role of neocentromeres in evolution and speciation, as well as in disease development and the understanding of the organization and epigenetic maintenance of the centromere. Here, we review recent progress in these areas of research and the significant insights gained.
Main Text
In all eukaryotic organisms, the centromere is the fundamental structure that controls the segregation of genetic material at meiosis and mitosis. With a few exceptions (such as the point centromeres of the budding yeast Saccharomyces cerevisiae and the centromeres of at least one species of Trypanosome1), almost all centromeres are characterized by an accumulation of repetitive satellite DNA, often present in higher-order arrays.
In general, such centromeric repeats are specific to the species and indicate some form of sequence sharing between centromeres. In primates, the repeat motif has been termed alpha-satellite (or alphoid) DNA,2 and in humans, a consensus sequence exists between chromosomes.3 Understandably, therefore, the concept of a close relationship between DNA sequence and centromere formation was compelling in early centromere research.
All this changed, however, with the discovery in 1993 of an ectopic centromere, or neocentromere, formed on a marker chromosome without any alpha-satellite DNA.4 The marker chromosome in question, designated mardel(10), had formed from a de novo rearrangement of chromosome 10 into a ring chromosome containing the normal centromere, and a linear chromosome completely lacking in centromeric alpha-satellite DNA. Nevertheless, this acentric maker chromosome had been rescued by the spontaneous formation of a new centromere at the cytogenetic band 10q25—a euchromatic region of the chromosome arm that had not undergone any rearrangement or sequence change.5,6 This was the discovery of a striking epigenetic phenomenon: the ability of a structure as complex as a centromere to spontaneously form at a seemingly random genomic location was unprecedented.
Such neocentromeres are quite different from the “classical” plant neocentromeres first described by Rhoades and Vilkomerson,7 and they lack fundamental centromere proteins and interact with microtubules in a very different manner to normal centromeres (for review, see Dawe and Hiatt8). In contrast, human neocentromeres have been shown to bind all known essential centromere proteins and behave identically in mitosis and meiosis to their satellite-DNA-based counterparts.
Since the initial discovery in 1993, over ninety cases of neocentromere formation in humans have been described in the literature. These cases, together with research from other organisms, have led not only to a greater understanding of the processes of neocentromere formation itself, but also to important insights into the structure and function of all centromeres and the major role the neocentromere phenomenon plays in karyotype evolution and speciation.
Constitutional Human Neocentromeres
Most of the initial information on neocentromeres has stemmed from human clinical data gathered through cytogenetic screening (Table 1, Figure 1A). In general, neocentric marker chromosomes form when an acentric chromosomal fragment is rescued via the formation of a neocentromere, and these marker chromosomes result from two main classes of chromosomal rearrangement. These are either an inverted duplication (inv dup) of the distal part of a chromosome arm resulting in an unbalanced karyotype (class I), or a balanced chromosomal rearrangement into linear and circular marker chromosomes after an interstitial deletion (class II) (Figure 2).
Table 1.
Ninety-Three Constitutional Neocentromere Cases
| Chromosome and Neocentromere Site | Rearrangement | Karyotype | Mosaicism (Percent Abnormal Cells) |
Reference | |
|---|---|---|---|---|---|
| Fibroblasts | Lymphoblasts | ||||
| 1: | |||||
| 1p32-p36.1 | Interstitial deletion (paracentric) | Balanced | 97 | 100 | Slater et al.106 |
| 1q21-q22 | Supernumerary ring | Trisomy | Constantinou et al.107 (case 1) | ||
| 1q23-q32 | Interstitial deletion (paracentric) | Balanced | 85 | Higgins et al.108 | |
| 1q32-qter | Deletion + inv dup | Trisomy | 100 | Kucerova et al.109 | |
| 1q43-q44 | Supernumerary ring | Trisomy | 70 | 50 | Spiegel et al.110 (case 1) |
| 2: | |||||
| 2p11-p21 | Interstitial deletion (paracentric) | Balanced | 100 | Petit and Fryns111 | |
| 2q35-q36 | Supernumerary ring | Trisomy | 28 | Pietrzak et al.112 | |
| 3: | |||||
| 3p23 | Interstitial deletion (pericentric) | Balanced | 100 | 100 | Maraschio et al.113 |
| 3q21.2-qter | Supernumerary inv dup | Tetrasomy | 0 | 87 | Gimelli et al.114 (case 3) |
| 3q24 | None | Balanced | (Ventura et al.71 (case 2) | ||
| 3q26.1 | Interstitial deletion (pericentric) | Balanced | 100 | 100 | Wandall et al.;14 Ventura et al.71 (case 1) |
| 3q26-qter | Supernumerary inv dup | Tetrasomy | 100 | Batanian et al.115 | |
| 3q26.2-qter | Supernumerary inv dup | Tetrasomy | 2 | 87 | Teshima et al.116 (case 2) |
| 3q26.2-qter | Supernumerary inv dup | Tetrasomy | 57 | Cockwell et al.117 | |
| 3q26.2-qter | Supernumerary inv dup | Tetrasomy | Yu et al.118 | ||
| 3q26.2-qter | Supernumerary inv dup | Tetrasomy | Sullivan et al.119 | ||
| 3q27 | Deletion + inv dup | Trisomy | 100 | Papenhausen et al.120 | |
| 3q27.1-qter | Supernumerary inv dup | Tetrasomy | 30 | 6 | Portnoi et al.121 |
| 3q27.2-qter | Supernumerary inv dup | Tetrasomy | 71 | Teshima et al.116 (case 1) | |
| 3q28-qter | Supernumerary inv dup | Tetrasomy | 100 | Barbi et al.122 | |
| 3q27.3 | Supernumerary inv dup | Tetrasomy | 87 | Gimelli et al.123 | |
| 4: | |||||
| 4q21 | Interstitial deletion (paracentric) | Balanced | 75 | Grimbacher et al.124 | |
| 4q21.2 | Interstitial deletion (pericentric) | Balanced | 100 | Warburton et al.13 | |
| 4q21.3 | None | Balanced | 100 | Amor et al.92 | |
| 5: | |||||
| 5p14-p15.1 | Supernumerary inv dup | Tetrasomy | 19 | Fritz et al.125 | |
| 6: | |||||
| 6q16.2-q22.2 | Interstitial deletion (paracentric) | Balanced | 100 | Qin et al.126 | |
| 6q26 | Supernumerary inv dup | Tetrasomy | 60 | Sala et al.127 | |
| 8: | |||||
| 8p22-pter | Supernumerary inv dup | Trisomy | 50 | 18 | de Pater et al.128 |
| 8p23-pter | Supernumerary inv dup | Tetrasomy | 21 | 28 | Herry et al.129 (case 2) |
| 8p23.1-pter | Supernumerary inv dup | Tetrasomy | 100 | Ohashi et al.130 | |
| 8p23.1-pter | Supernumerary inv dup | Tetrasomy | 25 | Neumann et al.131 | |
| 8p23.2 | Supernumerary inv dup | Tetrasomy | 23–46 | Voullaire et al.11 (case 1) | |
| 8p23.2 | Supernumerary inv dup | Tetrasomy | 53–60 | Voullaire et al.11 (case 2) | |
| 8p distal - pter | Supernumerary inv dup | Tetrasomy | 90 | 100 | Velinov et al.132 |
| 8q23-qter | Supernumerary inv dup | Tetrasomy | 75 | Sulcova et al.133 | |
| 8q23.3-qter | Supernumerary inv dup | Tetrasomy | 75 | Reddy et al.134 | |
| 9: | |||||
| 9p23 | Supernumerary inv dup | Tetrasomy | 100 | Depinet et al.9 (case 5); Satinover et al.135 (case 2) | |
| 9p23 | Deletion + inv dup | Trisomy | 100 | Depinet et al.9 (case 7); Vance et al.;136 Satinover et al.135 (case 1) | |
| 10: | |||||
| 10p14-pter | Supernumerary inv dup | Tetrasomy | 100 | Levy et al.137 | |
| 10q11-q23 | Interstitial deletion (paracentric) | Balanced | 62 | 80 | Depinet et al.9 (case 8) |
| 10q25.2 | Interstitial deletion (pericentric) | Balanced | 100 | 100 | Voullaire et al.;4 Lo et al.4 |
| 11: | |||||
| 11p11.12-11.2 | Interstitial deletion (paracentric) | Balanced | 76 | Chuang et al.15 | |
| 11q22-qter | Deletion + inv dup | Trisomy | 100 | 100 | Depinet et al.9 (case 6) |
| 12: | |||||
| 12p12.3-pter | Supernumerary inv dup | Tetrasomy | 50–57 | Dufke et al.138 | |
| 12p13.31-pter | Supernumerary inv dup | Tetrasomy | 100 | Vermeesch et al.139 | |
| 13: | |||||
| 13q21 | Supernumerary inv dup | Tetrasomy | 49 | Warburton et al.99 (case C) | |
| 13q21 | Deletion + inv dup | Trisomy | 100 | 100 | Morrissette et al.100 |
| 13q21 | Supernumerary inv dup | Tetrasomy | 14 | 20 | Li et al.101 (case 2) |
| 13q21.3 | Deletion + inv dup | Trisomy | 100 | Warburton et al.99 (case A); Cardone et al.34 | |
| 13q21.3 | Interstitial deletion (paracentric) | Balanced | 100 | Knegt et al.;16 Cardone et al.34 | |
| 13q31 | Supernumerary inv dup | Tetrasomy | 60 | Warburton et al.99 (case E) | |
| 13q31 | Supernumerary inv dup | Tetrasomy | 13 | 13 | Barwell et al.102 |
| 13q31-q32 | Interstitial deletion (paracentric) | Balanced | 50–70 | Amor et al.103 | |
| 13q31.3 | Supernumerary inv dup | Tetrasomy | 11 | Tonnies et al.104 | |
| 13q32 | Supernumerary inv dup | Tetrasomy | 98 | 8 | Depinet et al.;9 Warburton et al.99 (case F) |
| 13q32 | Supernumerary inv dup | Tetrasomy | 100 | 100 | Warburton et al.99 (case G) |
| 13q32 | Supernumerary inv dup | Tetrasomy | 74 | 25 | Warburton et al.99 (Case H) |
| 13q32 | Supernumerary inv dup | Tetrasomy | 56 | Li et al.101 (case 1); Alonso et al.33 | |
| 13q32.1 | Deletion + inv dup | Trisomy | 100 | Rivera et al.;140 Warburton et al.99 (case B); Alonso et al.33 | |
| 13q33.1 | Supernumerary inv dup × 2 | Hexasomy | 12–26 | Li et al.101 (case 3); Alonso et al.33 | |
| 13q33.1 | Supernumerary inv dup | Tetrasomy | 88 | Warburton et al.99 (case D); Alonso et al.33 | |
| 14: | |||||
| 14q32.1-qter | Deletion + inv dup | Trisomy | 100 | Magnani et al.;141 Sacchi et al.142 | |
| 15: | |||||
| 15 | ? | ? | 100 | Li et al.143 (case 11) | |
| 15q22 | ? | ? | Constantinou et al.107 (case 2) | ||
| 15q24 | Supernumerary inv dup | Tetrasomy | 80 | Blennow et al.144 (case B) | |
| 15q24-qter | Supernumerary inv dup | Tetrasomy | 66 | 50 | Spiegel et al.110 (case 2) |
| 15q24.1 | Supernumerary inv dup | Tetrasomy | 70 | 11 | Blennow et al.144 (case A); Ventura et al.10 (case 1) |
| 15q24.1-qter | Supernumerary inv dup | Tetrasomy | Tonnies et al.145 | ||
| 15q25 | Supernumerary inv dup | Tetrasomy | 80 | Huang et al.146 | |
| 15q25-qter | Supernumerary inv dup | Tetrasomy | 79 | Van den Enden et al.147 | |
| 15q25-qter | Supernumerary inv dup | Tetrasomy | 100 | Huang et al.148 | |
| 15q25.2 | Supernumerary inv dup | Tetrasomy | Ventura et al.10 (case 2) | ||
| 15q25.3-qter | Supernumerary inv dup | Tetrasomy | 82 | Depinet et al.9 (case 1) | |
| 15q25.3-qter | Supernumerary inv dup | Tetrasomy | 74 | Depinet et al.9 (case 2) | |
| 15q25.3-qter | Supernumerary inv dup | Tetrasomy | 95 | Hu et al.149 | |
| 15q25.3-qter | Supernumerary inv dup | Tetrasomy | 68 | Chen et al.150 | |
| 15q26.1 | Supernumerary inv dup | Tetrasomy | 50–100 | 18 | Rowe et al.151 |
| 15q26.1-qter | Supernumerary inv dup | Tetrasomy | 86 | Depinet et al.9 (case 3) | |
| 15q26.1-qter | Supernumerary inv dup | Tetrasomy | 70 | Mahjoubi et al.152 | |
| 16: | |||||
| 16p | Deletion + iso(16q) | Trisomy 16q | 100 | Tabet et al.153 | |
| 17: | |||||
| 17q22-q23 | Deletion + inv dup | Trisomy | 100 | Ravnan et al.154 | |
| 18: | |||||
| 18 | ? | ? | 64 | Rauch et al.155 (case 4) | |
| 20: | |||||
| 20p12.2 | Deletion + inv dup | Trisomy | 100 | 100 | Lo et al.32; Voullaire et al.156 |
| 21: | |||||
| 21q21.1-qter | Deletion + inv dup | Trisomy and deletion 21 | 100 | Barbi et al.157 | |
| X: | |||||
| Xq12 | Deletion + asymmetric inv dup | Trisomy and deletion X | 82 | Kaiser-Rogers et al.158 | |
| Xp22.31-pter | Supernumerary inv dup | Tetrasomy | 100 | Yu et al.159 | |
| Y: | |||||
| Distal Yp | Interstitial deletion? | Deletion Y | 15 | 15 | Conde et al.160 |
| Yq11.2 | None (or inversion?) | Balanced? | Mosaic | Rivera et al.89 | |
| Yq11.2 | Deletion + inv dup | Disomy and deletion Y | 70 | Floridia et al.161 | |
| Yq11.2 | Deletion + inv dup | Disomy and deletion Y | 70 | Warburton et al.162 | |
| Yq11.2 | Deletion + inv dup | Disomy and deletion Y | 100 | Assumpção et al.163 | |
| Yq12 | Supernumerary Y | Disomy | 5 | Bukvic et al.90 | |
| Yq12 | None | Balanced | 94 | 100 | Tyler-Smith et al.91 |
Figure 1.
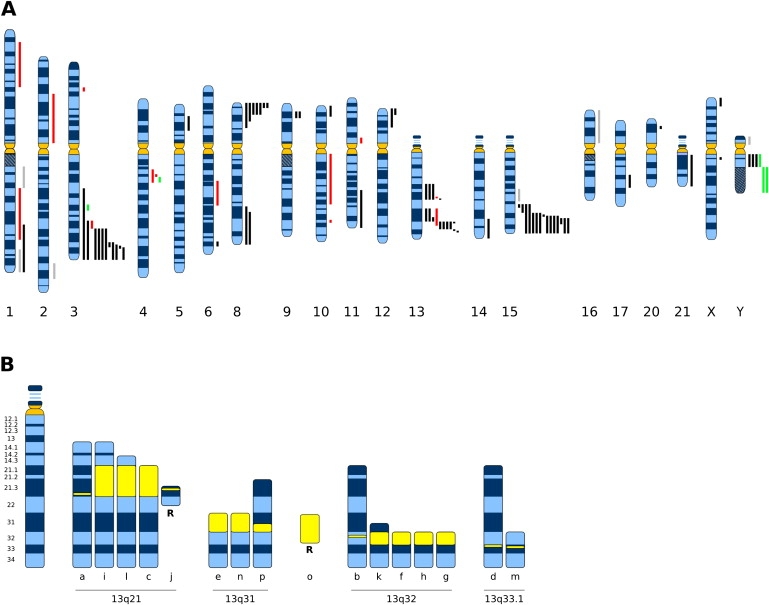
Constitutional Neocentromeres
(A) Sites of constitutive neocentromere formation in the human genome. The known locations of neocentromeres are represented by bars aligned against the chromosome ideograms; black bars represent neocentromere formation on class I marker chromosomes, red bars represent neocentromere formation on class II marker chromosomes, green bars represent sites of centromere repositioning, and gray bars represent unknown chromosomal rearrangements. Adapted from Amor and Choo.75
(B) Neocentromere hotspots on 13q. Sites of neocentromere formation are shown in yellow within the length of the marker chromosomes, with markers grouped by neocentromere formation within cytogenetic bands. All reported neocentromere cases from chromosome 13 are illustrated: a–h are as described by Warburton et al;99 i, Morrissette et al.;100 j, Knegt et al.;16 k–m are cases 1–3, respectively, as described by Li et al.;101 n, Barwell et al.;102 o, Amor et al.;103 and p, Tonnies et al.104 Additional mapping data of a and j are from Cardone et al.34 and b, d, and g from Alonso et al.33 All marker chromosomes are inverted duplications (for the sake of simplicity, the inversion is not illustrated for these chromosomes), with the exception of two ring chromosomes designated “R.”
Figure 2.
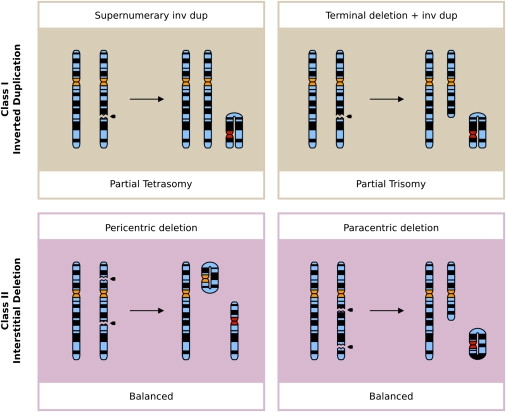
Chromosome Rearrangements after Chromatid Breaks That Are the Common Cause of Neocentromere Formation in Humans
Single, unreplicated chromatids of a homologous chromosome pair are depicted; break points are indicated with arrowheads, and the neocentromeres colored red. The resulting effect on the karyotype is listed underneath each alternative rearrangement.
Of the two main forms of neocentric chromosomal rearrangements, class I marker chromosomes are by far the most commonly reported. These inverted duplicated marker chromosomes represent 74% (67 out of 90 classified cases, Table 1) of neocentric chromosomes. They might be present as either supernumerary inv dup chromosomes (thus making the individual tetrasomic for the region of duplication) or with a deleted chromosome complementary for the region of duplication (thus leading to partial trisomy).
Precisely how these markers form is unknown, but from studies of parental DNA markers, it is clear that they might form at either meiosis or mitosis,9,10 and several mechanisms for their formation have been proposed11 (Figure 3). One clear mechanism involves chromatid breakage at mitosis, leading to an acentric chromosomal fragment that might subsequently segregate with either an intact chromatid or with the complementary broken chromatid (Figure 3A). If the fragment segregates with an intact chromatid, partial tetrasomy will be the result. However, if the fragment segregates with the complementary broken chromatid, the broken chromatid might be saved through telomere restitution and the end result is partial trisomy. In both cases, the inv dup marker forms after cell division and DNA replication by rejoining the broken, replicated ends of the acentric fragment. Partial tetrasomy might also result from a distal U type exchange at meiosis I, with subsequent neocentromere formation within the inverted duplication allowing the rescue of the marker chromosome (Figure 3B). Once the inv dup marker chromosome has been formed, the neocentromere itself can form at any location, although a significant minority of inv dup markers are approximately metacentric (see Epigenetic Mechanisms of Neocentromere Formation for a discussion of the significance of this observation). Out of all class I markers, partial tetrasomy is by far the more frequent occurrence, representing 80% of all cases (51 out of 64 cases, Table 1), which might represent either the extra contribution of meiotic formation to these marker chromosomes and/or the relative rarity of a broken mitotic fragment to segregate with its derivative chromosome. One final point to note regarding the formation of inverted duplicated markers is the presence of a small region of unduplicated genetic material at the duplication and rearrangement boundary of at least some marker chromosomes, thus giving an ABC::BA structure to the marker10 (and therefore resulting in partial trisomy [in partially tetrasomic individuals] or normality [in partially trisomic individuals] for region C). Although such marker structure is suggestive of a meiotic recombination event, analysis of polymorphic markers within two of these chromosomes has demonstrated that both chromosomes originated during mitosis.10 The implications of this structure for the process of inv dup marker chromosome formation are thus unclear.
Figure 3.
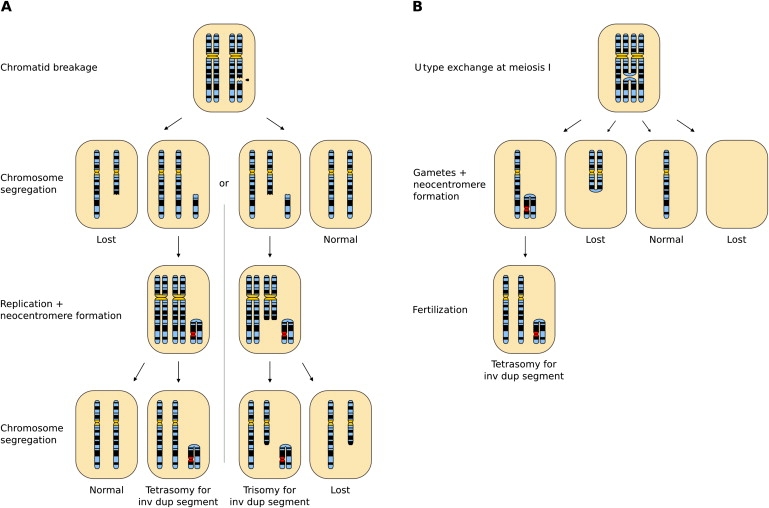
Possible Mechanisms for the Formation of Inverted Duplicated Neocentric Marker Chromosomes
(A) Formation at mitosis. After chromatid breakage, the acentric fragment can segregate in two possible ways. After subsequent replication, the broken ends of the acentric fragment rejoin to create the inverted duplication. Neocentromere formation can occur at this stage or after further rounds of cell division. If the neocentric fragment segregates with its sister chromatid, the result is partial tetrasomy for the duplicated fragment. On the other hand, if the centric fragment of the chromatid segregates with the neocentric fragment, the broken ends of the centric fragment can be stabilized by telomere restitution, and the result is partial trisomy for the duplicated fragment.
(B) Formation at meiosis. An acentric inverted duplicated (inv dup) chromosome is formed through anomalous crossing over during meiosis I and segregates with a normal sister chromatid to yield the gametes depicted. After fertilization, cells will be tetrasomic for the duplicated region. The inv dup marker might form a neocentromere during meiosis (as shown) or during subsequent rounds of mitotic division after fertilization.
Inverted duplicated neocentric marker chromosomes are often present in the individual in mosaic form. This mosaicism might be due to the mechanisms of marker chromosome formation or some intrinsic mitotic instability of the resulting neocentromere, but the selective disadvantage of partial tetrasomy is likely to be a contributing factor in some tissues. The loss of an inv dup marker chromosome from a partially tetrasomic cell will result in a balanced karyotype and is likely to be favored, whereas the loss of the marker chromosome from a partially trisomic cell will result in partial monosomy—a situation generally more deleterious to cell survival. Notably, this is reflected in the clinical data: 82% (42 out of 51 cases, Table 1) of partial tetrasomy caused by an inv dup marker chromosome were found to be mosaic for the marker chromosome, compared to only 15% (2 out of 13 cases, Table 1) of partial trisomy cases.
The second most common form of neocentric marker chromosomes are interstitial deletions (13 out of 90 cases; 14%), whereby a chromosome has been rearranged to form a ring chromosome and a linear marker chromosome, giving rise to a balanced karyotype (Figure 2). The neocentromere can form on either the linear or ring derivative, whichever is left acentric from the initial rearrangement. Precisely how and when this rearrangement occurs is unclear—the general assumption is that this process occurs via the chromosome breaking twice and the ends rejoining,12,13 although an alternative explanation would be looping and homologous recombination within a sister chromatid during meiosis I.
These balanced chromosomal rearrangements are generally marked by the stability of the linear chromosome derivative, and some degree of mosaicism with the ring derivative (as is common with ring chromosomes), regardless of which fragment contains the neocentromere. The phenotype associated with class II rearrangements is thus limited to the disrupted region of the chromosome at or around the breakpoints and the slight aneuploidy of the ring derivative caused through ring behavior. Because such genotypic changes can be relatively minor, it is possible that many such rearrangements have not been detected through clinical screening. Indeed, there are at least three examples of class II neocentric marker chromosomes being detected serendipitously in a phenotypically normal individual: twice where the rearrangement was ascertained in the offspring of an individual14,15 and once where an individual was only discovered to possess a rearrangement due to a high proportion of miscarriages.16
Of the remaining ten classified constitutional neocentromere cases, three were found on supernumerary rings and two on deleted p arm fragments of chromosomes. The final five neocentromeres were in fact not present on marker chromosomes at all, but rather were found on normal chromosomes with the original, alphoid centromere present, but deactivated. These extraordinary cases of centromere repositioning are discussed in greater detail below (see Centromere Repositioning, Karyotype Evolution, and Speciation).
Prevalence of Human Neocentromeres
An important question in understanding neocentromere formation is how frequent the phenomenon is. Clear estimation of this frequency is difficult, but an analysis of the statistics of reported small supernumerary marker chromosomes (sSMCs) in the literature might give an indication of the relative frequency of deleterious neocentromere formation. Such sSMC chromosomes are rearranged small markers often featuring the short arms of the acrocentric chromosomes, with the inverted duplications of chromosomes that feature neocentromeres also grouped within this category. An ongoing compilation of published sSMC cases made available online has cataloged 2480 sSMC cases in the literature, of which 81 feature confirmed or putative neocentromeres—suggesting that neocentromeres represent around 3% of published sSMC cases. Because the novelty of neocentromere cases prompts a greater likelihood of publication, this number might be an overestimate. However, a study of 241 unpublished sSMC cases in 2005 suggested a similar frequency—the study found only three putative occurrences of neocentromere formation,17 suggesting that neocentromeres represent around 1% of total sSMC cases. Considering that sSMCs are found in 0.043% of live births,18 these numbers give us an estimate of neocentromere formation on inverted duplicated chromosomes occurring in approximately 0.0005%–0.0014%, or once in every 70,000–200,000 live births.
However, such an estimate does not give a full picture of neocentromere formation. These studies do not include the incidence of balanced rearrangements (class II neocentromere markers), which, owing to the less severe phenotype associated with such cases, might be under reported in the literature. Furthermore, we can gain no clear idea of the frequency of centromere-repositioning events (see Centromere Repositioning, Karyotype Evolution, and Speciation, below) from these statistics because such rearrangements have only been detected serendipitously—individuals with such rearrangements show no detrimental phenotype at all. The overall picture, then, is that neocentromerization is a rare, but by no means infrequent event.
Cancer
Although neocentromere formation is in general a rare occurrence, certain cancers are associated with the formation of a complex, rearranged chromosome containing a neocentromere. These instances are especially interesting considering the clear somatic nature of neocentromere formation in these tumors and the suggestion that neocentromeres have formed in order to stabilize complex rearranged acentric chromosomes.
The best characterised link between neocentromeres and a specific form of cancer is found in the atypical lipomas and well-differentiated liposarcomas (ALP-WDLPS) class of lipomatous tumors. These cancers are marked by a supernumerary ring chromosome or large marker chromosome devoid of alpha-satellite DNA, comprised primarily of amplified 12q14-15 sequences that contain oncogenes19,20 and possessing a neocentromere at the primary constriction.21,22 This is in direct contrast to other liposarcomas, which contain the same amplified 12q14-15 sequences but on chromosomes with alphoid centromeres.21 Thus, the presence of a marker chromosome with a neocentromere appears to be a defining characteristic of this tumor class.
With the strong link between an amplified sequence and the formation of a neocentromere, it might be expected that the neocentromere was formed from the amplified DNA. This, however, appears not to be the case. Although the neocentric marker chromosomes consist largely of the 12q14-15 region, immuno-fluorescence in situ hybridization (immuno-FISH) showed the kinetochore proteins to be bound to DNA from other regions present in these chromosomes.21 The formation of these marker chromosomes might thus be a multistep process, involving the amplification of 12q14-15 and the capture and/or amplification of a different sequence involved in neocentromere formation.
Precisely why there should be a selective advantage conferred by the formation of a new marker chromosome complete with a neocentromere, as opposed to the amplification of these sequences within an existing chromosome, is unclear. Interestingly, though, similar lipomatous tumors where the region from 12q is amplified on marker chromosomes with alphoid centromeres are aggressive, metastatic tumors, and thus have very different behavior to the ALP-WDLPS tumors—suggesting a difference between alphoid centromeres and neocentromeres in this instance.21 One intriguing possibility would be that these more aggressive lipomatous tumors are derived from ALP-WDLPS tumors where the neocentromere has rapidly evolved into a repetitive, alphoid centromere, in a process similar to centromere repositioning, described below. However, whether there is a link between the aggressive nature of a tumor and the presence of alpha-satellite at a marker centromere is at this stage unclear.
Neocentromeres have also been reported in other types of cancers. The marker chromosomes in one lung carcinoma patient were found to contain a neocentromere associated with an amplification of the 9p23-24 region.23 The fact that neocentromeres have twice been previously reported at 9p23 suggests that amplification of this region might perhaps have facilitated neocentromere formation and stabilized the marker chromosome. Neocentromeres have also been described in two acute myeloid leukemia (AML [MIM 601626]) tumors, although these two cases involved different chromosome rearrangements. In one instance, the neocentromere was present on a standard inverted duplication of 10q,24 and in the other, a complex, rearranged ring chromosome.20
Although neocentromeres have only been described in three forms of cancer thus far, it is possible that the frequency of neocentromere formation in cancers is much higher than the literature suggests, because most cancers are not subjected to karyotype analysis that would detect neocentromere formation. There are also the intriguing reports that two important centromere proteins, CENPA (MIM 117139) and CENPH (MIM 605607), are overexpressed in all colorectal cancer (CRC [MIM 114500]) cell lines and tissues.25,26 Considering the possible ectopic kinetochore formation associated with CENPA overexpression27 described below (see Epigenetic Mechanisms of Neocentromere Formation), it is possible that upregulation of centromere proteins that promote neocentromerization is a common step in the promotion of the genome rearrangements and abnormalities that lead to cancer. Karyotyping of these cancer cell lines known to overexpress centromere proteins would provide a clear indication as to whether a link exists between the two.
Organization of Centromere Proteins
At mitosis, human neocentromeres behave identically to alphoid centromeres, binding all known centromere proteins (with the exception of the alphoid sequence-specific and apparently redundant protein CENPB [MIM 117140]).28,29 The spatial relationships between these proteins in building the inner centromere is of great importance in the understanding of the structure of the centromere, and neocentromeres have proved to be a vital means of examining this problem. A unique research advantage that neocentromeres provide is their lack of repetitive DNA, and this has allowed detailed mapping of the chromatin-binding domains of centromere proteins—something that has been difficult to achieve with normal centromeres.
Centromere Protein CENPA
The fundamental innerkinetochore protein CENPA is the most extensively mapped protein at neocentromeres. This protein, a histone H3 (H3F2 [MIM 142780]) paralogue found only at the nucleosomes of active centromeres,30,31 has been mapped to six neocentromeres at 100–200 kb resolution level via chromatin immunoprecipitation (ChIP) and bacterial artificial chromosome (BAC) DNA array analysis.6,32–34 From these somewhat low-resolution ChIP and array mapping experiments, it is clear that some variability in the size of the CENPA domain exists between neocentromeres (Figure 4A), with the extent covered by CENPA ranging from 218 kb to 464 kb.
Figure 4.
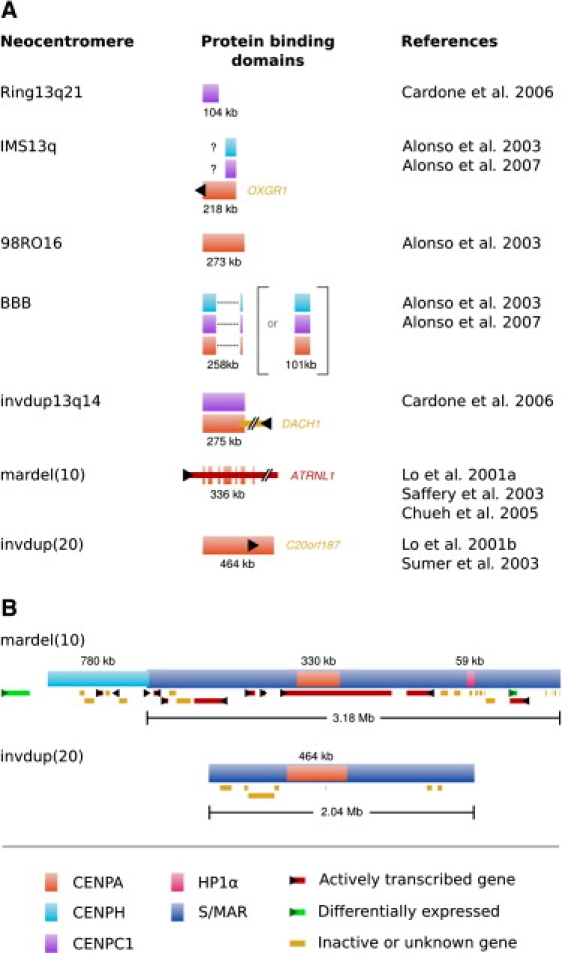
The Size and Distribution of Mapped Protein Domains at Neocentromeres
(A) Innerkinetochore domain organization at seven neocentromeres. The size of the protein-binding domain for each neocentromere is listed. Known protein-coding genes present within these domains are shown (derived from Ensembl release 44). The discontinuous nature of the mardel(10)35 and BBB36 neocentromeres is illustrated. Two possibilities of the layout of the BBB neocentromere are provided—see main text for details. The IMS13q CENPC1 and CENPH domains are marked “? ” to represent an uncertainty as to the size of these domains—the authors of this study suggested that the lower efficiency of ChIP with CENPC1 and CENPH antibodies might prevent the full extent of these domains from being detected.36
(B) Scaffold domains, protein binding domains, and genes present within two mapped neocentromeres. Known protein-coding genes present within these domains are shown (derived from Ensembl release 44), with expression data from cell lines derived from Saffery et al.50 and Wong et al.67 Differentially expressed genes denote two protein-coding genes found to be activated after neocentromere formation.67 (Domain positions are derived from BAC data from Lo et al.,6,32 Alonso et al.,33 Cardone et al.,34 and Saffery et al.50 updated against Ensembl release 44 from the Ensembl project105.)
For two of the neocentromeres, the mardel(10) neocentromere at 10q25 and the BBB neocentromere at 13q33, CENPA has been mapped at higher resolution with an array of polymerase chain reaction (PCR) fragments.35,36 The first study to achieve this used PCR fragments with an average size of 8 kb to map the mardel(10) neocentromere.35 At this resolution, the CENPA domain was shown to comprise seven separate regions interspersed with histone H3 (Figure 4A). The regions of CENPA binding were regularly spaced, with an average of 54 kb peak-to-peak distance (standard deviation: 10 kb). Such a result suggested a similar organization of centromeric chromatin at the neocentromere to that found at normal centromeres, where interspersed regions of CENPA and H3 binding had previously been shown by stretched chromatin fiber studies.37 Taken together, the data suggested a contiguous unit of CENPA at the inner kinetochore plate formed by coiling of the chromatin fiber into a series of stacked loops.35,37,38
Evenly spaced clusters within the CENPA domain were less evident at the smaller BBB neocentromere.36 This study used PCR fragments around 1 kb in size as the basis of the array—providing a higher resolution, but with perhaps a lower signal:noise ratio. The study showed a major CENPA binding domain 88 kb in size, and a second, smaller domain of 13 kb. Curiously, the two domains were separated by a stretch of 157 kb—a distance greater than the size of the two domains combined. It is therefore unlikely that these two domains represent two adjacent coils of chromatin, and it is possible that this represents a malformed neocentromere with CENPA present in a discontinuous domain across the innerkinetochore plate. However, considering that the inv dup chromosome marker carrying this neocentromere was 100% stable in cell culture,33 this scenario seems unlikely. Rather, we would suggest the alternative possibility that this curious distribution of CENPA represents a small inversion of 160 kb in the patient and that the CENPA domain is, in fact, a contiguous unit of 101 kb that merely appears to be discontinuous when mapped back to the consensus sequence of the human genome (Figure 4A) (see Epigenetic Mechanisms of Neocentromere Formation below for a further discussion of the possible implications of this observation.) Within the major CENPA-binding domain, there was some evidence of regular, localized signal troughs—suggesting a possible concordance with the data from the mardel(10) neocentromere, albeit with much smaller chromatin loops. Nevertheless, some CENPA binding was shown to be present by real-time PCR within the trough domains,36 and the implications of this for the structure of the BBB neocentromere remain unclear.
One interesting aspect of the multiple neocentromere mapping results for CENPA is the variability in the size of the CENPA domain. Does this reflect a similar change in the size of the innerkinetochore plate and the primary constriction between neocentromeres? If the physical size of the kinetochore is variable, this might point to the transient nature of neocentromeres and perhaps suggest that such structures only reach maturity upon the subsequent incorporation of satellite DNA. However, an alternative explanation is also possible. Rather than reflecting a variation in the physical size of the kinetochore, the results might instead represent a variation in the size of the loops of the coiled chromatin fiber that form the basis of the CENPA domain (see above). By a variety of methods, the average loop size in human chromosomes has been estimated to be between 30 and 90 kb.39–42 If the seven peaks of CENPA binding at the mardel(10) neocentromere represent an end-to-end distance of six loops (five full loops and two half loops), it could be expected that a CENPA-binding domain made of seven binding peaks could range between 180 and 540 kb. Such a prediction fits well with five of the seven mapped neocentromeres but fails to adequately explain the size of the two smallest and perhaps structurally more primitive neocentromeres.
Although such mapping data provides a substantial amount of information regarding the structure of neocentromeres, it is important to understand whether neocentromeres are truly structurally homologous to alphoid centromeres. In order to answer this question, the physical distribution of CENPA has recently been studied at normal centromeres and at a neocentromere through the use of high-resolution electron microscopy (EM). Through the use of flow-cytometry to isolate individual populations of chromosomes, the distribution of CENPA at the mardel(10) neocentromere could be directly compared to that found at normal alphoid centromeres (O.J.M., A.T.M., and K.H.A.C., unpublished data). The results suggested a surprising similarity in the CENPA-binding-domain size between the two types of centromeres: In both cases, there was no significant difference between the physical size of the CENPA domain or in the proportion of chromatin occupied by CENPA relative to the constriction (Figure 5).
Figure 5.
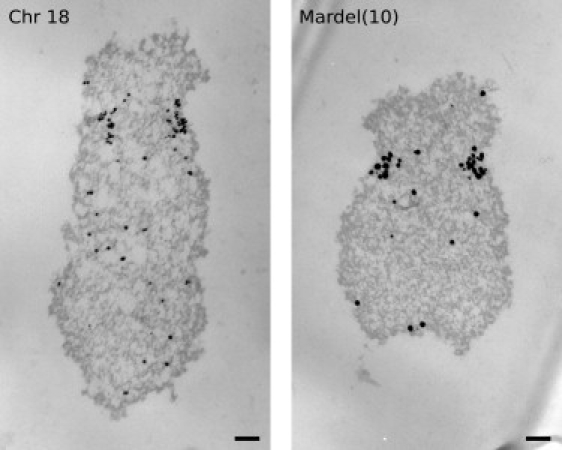
Fine Structural Localization of CENPA at a Human Alphoid Centromere and a Neocentromere
A 45-nm-thick section through each chromosome is shown. Chromosomes were sorted by flow cytometry, fixed in acetone, and labeled with a mouse monoclonal anti-human CENPA primary antibody (MBL) and a Ultrasmall gold anti-mouse secondary antibody (Aurion) before postfixation, embedding, and sectioning. The scale bar represents 200 nm.
In addition to investigation of the physical size of the CENPA-binding domain, the relative amount of CENPA present on neocentromeres has been compared to that found on normal centromeres. Through the use of a cell line expressing a green fluorescent protein (GFP)-CENPA fusion protein and the measurement of the relative levels of fluorescence, two separate neocentromeres on the mardel(10) and invdup(20) chromosomes were shown to bind only one-third the amount of CENPA as most other human centromeres.43 With the EM data above, this suggests that the loading of CENPA at the inner plate is less efficient at neocentromeres. This suggestion is also supported by recent ultra-high-resolution ChIP at a subnucleosome level with an oligonucleotide DNA array, which has demonstrated that at one neocentromere, CENPA nucleosomes are not present in contiguous, uniform blocks, but rather that individual CENPA-containing nucleosomes are interspersed with canonical H3 nucleosomes.36
Precisely why the incorporation of CENPA at neocentromeres should be less efficient, though, is less clear. Interestingly, neocentromeres and the Y chromosome centromere—which also exhibits significantly reduced CENPA binding43—both fail to bind CENPB. Considering that CENPA has been shown to be strongly associated with alpha-satellite sequences containing the CENPB box (as distinct from a second abundant class of centromeric alphoid sequences lacking the CENPB box motif)44 and that the presence of such satellite sequences appears to be essential for de novo human artificial chromosome formation,45–48 it is tempting to speculate a role for CENPB at centromeres in which the protein, although nonessential, significantly enhances the recruitment of CENPA to centromeric regions. Such an observation would also explain why neocentromeres are merely a transient structure in evolution, eventually incorporating satellite DNA in order to facilitate optimal binding of kinetochore proteins (see Centromere Repositioning, Karyotype Evolution, and Speciation, below).
Centromere Proteins CENPC1 and CENPH
Two other functionally essential innerkinetochore proteins, CENPC1 [MIM 117141] and CENPH [MIM 605607], have also been mapped by ChIP-on-chip on multiple neocentromeres. Both proteins are known to interact with CENPA,49 and as a result, the two proteins would be expected to occupy overlapping locations with CENPA on neocentromeres. Indeed, colocalization between CENPC1 and CENPA in ChIP-on-chip studies has been now shown for three neocentromeres,34,36 and, between CENPH and CENPA, on two neocentromeres.36 On one neocentromere there was only partial overlap of the protein binding domains (with CENPC1 and CENPH occupying a subset of the CENPA domain)36 (Figure 4A). However, the authors suggested that this result might merely reflect less efficient ChIP antibodies, and this is perhaps the more likely explanation, considering that precise colocalization of CENPA, CENPC1 and CENPH has been shown with a high-resolution PCR DNA array at the BBB neocentromere.36
Interestingly, no colocalization was seen between CENPA and CENPH on the mardel(10) neocentromere—CENPH was found to be present in a large, 900 kb domain over 1 Mb distant from the CENPA domain50 (Figure 4B). The implications of this last result are unclear, but it might imply a higher-order chromatin folding at this particular neocentromere—which incidentally was the largest of the CENPA- or CENPH-mapped neocentromeres—bringing the two separate regions into closer proximity to enable protein-protein interactions.
The Chromosomal Scaffold
The presence of scaffold or matrix proteins at the cores of condensed, mitotic chromosomes has long been demonstrated.51–56 At alphoid centromeres, the frequency of sites of scaffold attachment increases dramatically,57 suggesting a tighter compaction of centromeric chromatin. The same observation has also been shown for two neocentromeres,50,58 and the lack of repetitive DNA at neocentromeres has allowed the extent of the enhanced scaffold/matrix attachment region (S/MAR) to be defined. In both cases, the enhanced S/MAR domain was found to be much larger than the associated CENPA-binding domain, covering an expanse of 3.2 Mb for the mardel(10) neocentromere and 2.0 Mb for the invdup(20) neocentromere (Figure 4B). Although it is unknown whether this domain represents the physical boundaries of the centromere, it seems logical to suggest that the enhanced S/MAR domain defines the primary constriction considering the clear change in the physical structure of chromatin at the centromere.
Of interest, the size of the CENPA-binding domain at these neocentromeres does not appear to be proportional to the size of the mapped S/MAR domain. In the case of the mardel(10) neocentromere, CENPA occupied only one-tenth of the region of increased scaffold attachment, whereas at the invdup(20) neocentromere CENPA occupied almost one-quarter of the enhanced S/MAR domain (Figure 4B).
In both cases, numerous genes were present within the boundaries of the enhanced scaffold domain, and some of these have been shown to be transcriptionally active (see Transcriptional Competence within Neocentromeres, below).
Heterochromatin Protein HP1α
The organizational distribution of the heterochromatin protein marker HP1α [MIM 604478] has also been studied on a single neocentromere, the mardel(10). Curiously, considering the strong requirement of heterochromatin for sister chromatid cohesion at the centromeres of fission yeast59,60 and vertebrates,61 the protein showed only slight enrichment at the neocentromere compared to the progenitor chromosome 10, at a single BAC position 800 kb distant from the CENPA domain50 (Figure 4B). Although this region represents a relatively small domain of heterochromatin, it is important to note that the chromatin immunoprecipitation study was a comparative one, only measuring the levels of enrichment of the protein. Thus, it is possible that an extant domain of heterochromatin already existed at the normal 10q25.3 region, which has merely been augmented after neocentromere formation.
Transcriptional Competence within Neocentromeres
One of the more fascinating features of neocentromeres is their location within euchromatic, protein-encoding regions of the genome. This is particularly evident in the case of the mardel(10) neocentromere, which has a long gene transcript spanning the entire length of the CENPA-associated domain (Figure 4). In addition, three other neocentromeres similarly have known protein-coding genes within their mapped CENPA-binding domains (Figure 4A)—indicating that of the seven mapped innerkinetochore plates, the majority contain protein-coding genes.
Whether such euchromatic genes could be expressed within the boundaries of the kinetochore chromatin has been a question of considerable interest. The CENPA N-terminal tail lacks a lysine amino acid at residue 4, preventing the methylation marks that denote euchromatin and active genes, and consequently it might be thought that centromeric chromatin was silent by default. It has also been shown that CENPA forms a tighter nucleosome structure62,63 than the histone H3-containing alternative, which might also form a barrier to transcription.
Through comparison of the expression levels of known protein-coding genes within the mardel(10) neocentromere domain to the same genes on the progenitor chromosome 10 from which the neocentric marker chromosome had formed, investigation of this problem has been possible. Surprisingly, ATRNL1, the gene that spans the CENPA-associated domain on the mardel(10) neocentromere, was found to be actively expressed, and the formation of the mardel(10) neocentromere at 10q25 did not significantly change the expression levels of this gene.50 Most recently, this phenomenon of active transcription through a region of CENPA-containing chromatin has also been demonstrated in alpha-satellite-containing human artificial chromosomes, where the CENPA-containing domain was shown to not be restricted to the alpha-satellite repeats but to have spread over the active selective marker gene64 adjacent to these repeats. Transcriptional competence of centromeric chromatin has also been shown for two rice centromeres65,66 (see Centromere Repositioning, Karyotype Evolution, and Speciation, below). From these results, it appears that CENPA-containing chromatin represents no barrier to gene transcription.
Within the enriched S/MAR domain of the mardel(10) neocentromere, there are eight other actively expressed genes that were similarly unaffected by the formation of a neocentromere50 (Figure 4). Indeed, the only differences in gene expression detected after neocentromere formation was the activation of two protein-coding genes on either end of the S/MAR domain,67 where in both instances the genes were only expressed after neocentromere formation (Figure 4). These genes corresponded with regions of hypomethylation at the mardel(10) neocentromere, and their activation might have been induced as a byproduct of the epigenetic remodeling that accompanies neocentromerization (see Epigenetic Mechanisms of Neocentromere Formation, below).
Thus, despite the increased scaffold attachment sites and a corresponding tighter packing of chromatin, gene transcription can continue regardless within the primary constriction and is occasionally even promoted. All the evidence therefore points to the centromeric structure being largely irrelevant to gene transcription and again raises the question as to the purpose of satellite repeats—which have been traditionally linked to genetic silencing—at the eukaryotic centromere (see further discussion in Centromere Repositioning, Karyotype Evolution, and Speciation, below).
Epigenetic Mechanisms of Neocentromere Formation
Neocentromerization
Although much is now known about the structural and functional characteristics of neocentromeres, comparatively little is known about how they actually form. Currently, there are no reports of neocentromeres forming experimentally in human cell lines, and the little information that we have stems from studies undertaken in flies and plants.
The first example of neocentromerization occurring experimentally was from work undertaken in Drosophila, investigating the germline transmission of fragments of noncentromeric DNA after radiation damage.68,69 In an initial study, a small (<300 kb) subtelomeric fragment of a rearranged Drosophila X minichromosome (γ238) was shown to spontaneously form a neocentromere after release by radiation.68 Significantly, the rearrangements within the γ238 minichromosome had placed this subtelomeric fragment very close to the repetitive DNA-based normal centromere of the chromosome. In contrast, when the same subtelomeric fragment was released from a normal X chromosome, where it is separated from the active centromere by 40 Mb of DNA, no neocentromere formation was detected.68 A follow-up study released the same subtelomeric fragment from various sites within the Drosophila genome and found that a neocentromere was only formed when the subtelomeric DNA was located within 20 kb of an active centromere.69 Such results demonstrated that it was a close association to a normal centromere in the γ238 minichromosome that was responsible for conferring neocentromerization potential on the fragment. These studies thus suggested that neocentromere formation—in flies, at least—is reliant upon close-proximity spreading of a centromeric signal from an active centromere. It should be noted, however, that these studies were based upon the neocentromere-forming capacity of a single, short length of subtelomeric DNA. It is possible that other regions of the Drosophila genome might be more predisposed to neocentromere formation without requiring proximity to an active centromere.
A second report of neocentromerization has come from plant chromosomes, and it also describes neocentromeres forming close to the site of active centromeres.70 This study used a gametocidal system in wheat to induce structural changes in added barley chromosomes. Two fragments of barley chromosome 7 were recovered that lacked the original primary constriction and any form of centromeric repeats. Both chromosome derivatives were shown to form telocentric neocentromeres at the pericentric breakpoint from the original chromosome, suggesting that close-proximity spreading of a centromeric signal from the original chromosome was the cause of neocentromere formation.70
Despite the above studies, the potential of centromeric spreading to generate a neocentromere cannot account for the neocentromere cases reported in humans, where there is no record of neocentromeres forming near active centromeres. Considering the chromosomal rearrangements known to result in neocentromere formation, though, this is not entirely surprising. Neocentric inverted duplications generally occur far from the centromere on the distal ends of chromosome arms; balanced chromosomal rearrangements contain one fragment with the centromere and much of the surrounding pericentric DNA. Thus, although it might be possible for the centromeric signal to spread in humans just as in flies, this will rarely, if ever, be seen in vivo. Nevertheless, it is worth noting the balanced neocentromere rearrangement on chromosome 3 described by 14 where the break points for the rearrangement were within 2 Mb either side of the original alphoid centromere.14,71 Even in this scenario, no spreading of the centromeric signal was observed: The neocentromere was formed at 3q26.1, more than 70 Mb distant from the break point, rather than in a region close to the original alphoid centromere.
If the centromere signals do not possess the capability to spread in humans, at least through considerable distances, how then are neocentromeres formed? One possibility is that aberrant incorporation of CENPA at ectopic sites within the genome during DNA replication might lead to the formation of a new centromere at mitosis. It is known that overexpression of CENPA in human and fly cell lines leads to misincorporation of the protein within the chromosome arms.27,72 There has also been the suggestion that CENPA misincorporation is common and that subsequent protease digestion is required for the removal of the protein from the chromosome arms during the normal cell cycle.73,74 Could neocentromerisation, therefore, simply occur via small pockets of CENPA retained on acentric fragments? The only evidence that this might be possible stems from a study in Drosophila, where overexpression of the Drosophila CENPA ortholog CID was shown to occasionally give rise to regions with ectopic kinetochore activity, which were able to successfully bind microtubules at mitosis.27 However, a similar study in humans reported no such kinetochore formation72 despite observing a high degree of CENPA incorporation within the chromosome arms. It seems likely, therefore, that overexpression of the protein might need to be inordinately high to drive the formation of ectopic kinetochores in humans, at least, and that this is unlikely to be a main mechanism of neocentromerization (although it might well play a role in cancer formation—see Cancer, above).
Related to this theory is the idea that CENPA is incorporated in potential neocentromeric sites at high efficiency but is rapidly removed on chromosomes that already possess a centromere by factors acting in cis.75 In this hypothesis, each acentric fragment has a strong potential to form a neocentromere at multiple locations, and each active centromere plays a role in silencing alternative centromeres. Removal of the controlling centromere—by a chromosome rearrangement, break, or epigenetic silencing—would thus allow one of these alternative regions to form a neocentromere. In humans, there is some evidence in support of the idea of centromere silencing in cis: Although functional dicentric X chromosomes with two pairs of active kinetochores can form when the distance between the two centromeres is small, once this distance becomes greater than 12 Mb, two active centromeres are never observed.76 However, this phenomenon might simply result from the instability and loss of chromosomes with a large degree of separation between the two active centromeres—thereby imposing a positive selection for chromosomes that have deactivated one of the centromeres through various loss-of-function mechanisms—rather than from the involvement of cis-acting silencing factors. Furthermore, studies in Drosophila have demonstrated that a satellite-rich region in a chromosome can bind centromere proteins and occasionally form an active centromere, even in the presence of the wild-type active centromere77—an event that suggests a lack of centromeric silencing in cis, or at least that certain regions can escape silencing. (This particularly unusual DNA region, termed bwD, comprises an insertion of over one megabase of satellite DNA.78 Although it possesses the ability to form an active centromere at high frequency after chromosome breakage, the bwD region is not euchromatic and has a peculiar affinity for centromere proteins. It thus should not be confused with neocentromeres, which form at nonrepetitive regions of the genome and do not bind centromere proteins prior to chromosome breakage.)
The above hypotheses imply that a protokinetochore is already present at the site of neocentromere formation before breakage of the chromosome (or silencing of the initial centromere) occurs. An alternative possibility is that the neocentromere signals are acquired after chromosome breakage or rearrangement, via aberrant association of the acentric fragment with centromeric DNA during CENPA loading.69 Considering that loading of CENPA occurs immediately after cell division and before DNA replication,79,80 this would imply that neocentromerization would occur only after the cell has progressed through one round of division with an acentric fragment.
Perhaps most intriguing of all is the suggestion that chromosome rearrangements might be responsible for inducing neocentromere formation, possibly through a change in the epigenetic state of the chromatin after DNA repair.10,81 The evidence for this theory is based upon the high-resolution FISH mapping of two 15q25 neocentromeres on inverted-duplicated marker chromosomes. The mapping showed a close association (to within 500 kb) between the site of neocentromere formation and the duplication and rearrangement boundaries of the marker chromosomes.10 Indeed, a significant proportion of all known inv dup marker chromosomes have a centromere at the approximate position of the site of recombination (see Figure 1B for examples of chromosome 13-derived neocentromeres), suggesting a link between the two events. In this respect, the possibility of a small inversion at the mapped BBB neocentromere (see Organization of Centromere Proteins, above) is particularly interesting, and it is tempting to suggest that the inversion and formation of the neocentromere in this case might be related.
Hotspots of Neocentromere Formation
A clearly recognizable trend from the human clinical cytogenetic data is the clustering of sites of neocentromere formation at chromosomal “hotspots.”75,82 Certain regions of chromosomes—for example, 3q, 8p, 13q, and 15q—seem particularly prone to forming neocentromeres (Figure 1A). In part, this trend must be ameliorated by the recognition that these sites cluster in chromosomes that are inverted duplications and that the survival of individuals with more distal inverted duplications will be favored (as such individuals possess a smaller region of partial trisomy or tetrasomy). It is therefore logical to see a clustering of neocentromeres around the distal ends of chromosomes, which might explain the 8p hotspot in particular. Nevertheless, it is clear that neocentromere formation in certain regions is favored, something that is perhaps best illustrated by the neocentromeres found on chromosome 13. Within the q arm of this chromosome, two clear regions of neocentromere formation, or hotspots, are present within the cytogenetic bands 13q21 and 13q32, and the formation of neocentromeres within these bands does not appear to be related to the length of the marker chromosome (Figure 1B).
Whether neocentromeres form at precisely the same location and underlying genomic DNA sequence within each hotspot has been an important issue in understanding neocentromerization. If this were indeed the case, it would suggest a fundamental relationship between DNA sequence (or a highly localized chromatin environment) and neocentromere formation. This question has been the subject of investigations into the 13q3233 and 13q21 hotspots.34 Both studies mapped the CENPA- or CENPC1-binding domains of several neocentromeres known to be located within these hotspots and found that each binding domain localized to a different region of the same cytogenetic band.33,34
These still rather limited data by themselves are not necessarily proof against a common sequence basis for the primary constriction within these hotspots. The combined neocentromere mapping studies (described in Organization of Centromere Proteins, above) suggest that the exact location of the CENPA domain within the primary constriction (as indicated by the S/MAR domain) might be variable, raising the possibility that the same region of the chromosome forms a primary constriction each time, within which the position of the CENPA domain might change. However, the distance between the 13q neocentromeres investigated in these studies suggests that this scenario is unlikely. In the case of three 13q32 neocentromeres, the CENPA domains of each centromere were found to be located within a stretch of more than 6 Mb of DNA,33 with two domains separated by over 5 Mb; similarly, the two 13q21 neocentromeres were separated by over 3 Mb of DNA. These distances are greater than the size of either previously mapped neocentric constriction50,58 and suggest that the neocentromeres studied have indeed formed at different locations within the cytogenetic bands.
If the exact chromosomal location is not a contributing factor, could there be more general characteristics of neocentromere hotspots that make them conducive to neocentromere formation? One possibility is that neocentromerization is favored in gene-poor regions of the genome.34 It is logical to assume that such regions might be more conducive to subsequent incorporation of alpha-satellite DNA and pericentric heterochromatin, and there are indeed some regions of the genome that have a low gene density and appear to be favored for neocentromere formation (e.g., 9p23). However, there are also apparent hotspots in relatively gene-rich cytogenetic bands (e.g., 13q32), implying that there is no clear consensus as to where neocentromeres are formed. Similarly, the observation that the CENPA domains of the 13q32 neocentromeres observed by Alonso et al.33 were found between active genes appears not to be a general trend, judging by the presence of active genes within the CENPA domains of three other neocentromeres (see Organization of Centromere Proteins, above). Finally, the mapped S/MAR domains [on the mardel(10) and invdup(20) neocentromeres] occupy regions filled with known protein-coding genes, at least some of which are known to be actively expressed50 (Figure 4B). From the above evidence, therefore, it seems unlikely that gene density is a deciding factor in the positioning of neocentromeres (although it might relate to the propensity of a neocentromere to become subsequently fixed in the population—see Centromere Repositioning, Karyotype Evolution, and Speciation, below).
An alternative explanation for the existence of neocentromere hotspots relates to the possible link between genome rearrangement and neocentromere formation, discussed above.10,81 This theory suggests that regions of the genome with a high content of duplications are predisposed to rearrangements, which then lead to neocentromere formation through epigenetic changes in the chromatin after DNA repair. Such a hypothesis would also explain the link between several neocentromere hotspots and the locations of ancestral centromeres (which have a high number of duplicons).10 There has not yet been a comprehensive analysis of the “older” duplicons within the genome that appear to be associated with this phenomenon,34 so assessment of whether this theory can explain all known hotspots is currently impossible. However, there is a good correlation between duplicons and sites of neocentromere formation seen at neocentromere hotspots from 15q,10 13q21,34 and 3q26,71 suggesting that such a connection between duplications and neocentromerization might be a distinct possibility.
One final point of interest regarding neocentromere hotspots is the accuracy of conventional cytogenetic mapping of the primary constriction. Two of the supposed 13q32 neocentromeres mentioned above were shown through chromatin immunoprecipitation and genomic array analysis to have CENPA domains within the next cytogenetic band, 13q33,33 suggesting that some previously reported band locations of neocentric constrictions might be inaccurate. In other words, the clustering of neocentromeres at particular cytogenetic bands might not be as extensive as the literature would suggest.
DNA Sequence Similarities
Considering the homology of repetitive satellite sequences at the normal centromeres of an organism, numerous studies have sought to find a correlation in the DNA sequences of neocentromeres, in particular in the sequence of known CENPA-binding domains.6,32–35 Currently, most analyses of these domains have failed to find significant deviations from the genome average, in terms of various centromere motifs or repetitive elements. However, one observable trend is the AT base-pair content of the CENPA binding regions: This has been shown to be consistently higher than normal in all seven neocentromeres observed, ranging from 59.9% content to 66.1%, with the genome average being 59%.6,32–34 Whether this is a major factor for determining the site of neocentromere formation is less clear, though. The three neocentromeres investigated by Alonso et al.33 were located within 6 Mb of each other, yet the AT content of their CENPA-binding domains varied between 59.9% and 65.0%. If AT content was indeed a deciding factor in determining the site of neocentromerization, it is surprising that all three neocentromeres did not form within the region of highest AT content. Similarly, a comprehensive analysis of the AT content of the seven mapped neocentromeres and the surrounding DNA suggested that there was little correlation between regions of extremely high AT content and neocentromere formation.36 Nevertheless, a threshold AT content requirement of at least the genome average is a possibility that cannot be discounted.
The only other clear significant difference in DNA content within a neocentromere was reported in the high-resolution CENPA mapping study of Chueh et al.35 Although no difference was noted over the entire CENPA binding domain of the mardel(10) neocentromere, when analyzing the separate peaks of CENPA binding within this domain a significant increase was observed in the number of retrotransposable L1 LINE elements (LRE1 [MIM 151626]) compared to the surrounding histone H3-containing regions. Analysis of the BBB neocentromere, the other neocentromere mapped at PCR-fragment resolution, also showed a higher frequency of L1 elements (and MaLR LTR elements) within the peaks of CENPA binding compared to the intervening DNA,36 and a 70bp motif associated with young L1 elements was found to be the only common feature shared between the DNA sequence of the seven mapped neocentromere domains.36 An association between L1 content and neocentromere formation is therefore a distinct possibility, although the exact basis of this association is at this stage unclear.
Epigenetic Maintenance
Once a new centromere has been formed, there remains a further interesting question of how the position and boundaries of the centromere is subsequently maintained. Without evidence of DNA sequence specificity at neocentromeres, it appears that epigenetic marks are the means through which this is achieved. CpG island methylation is an obvious candidate for this process, and a comprehensive study of CpG methylation at the mardel(10) neocentromere was recently carried out by Wong et al.67 This study looked not only at CpG islands (stretches of DNA longer than 200 bp enriched for CpG dinucleotides) but also at smaller regions (“islets”) enriched for CpG residues. The findings of this study were striking: Although islets and islands within the extended S/MAR region were generally hypermethylated at the neocentromere when compared to the progenitor chromosome 10, at the boundaries of the CENPH, HP1α, and enriched S/MAR domains, there was clear hypomethylation of islands or islets.
Such results could, of course, be coincidental—only a similar correlation seen at the boundary elements of other neocentromeres would confirm this uncategorically. Nevertheless, the results are intriguing, especially when seen in the light of a trend of hypermethylation of CpG residues located elsewhere within the neocentromere domain. Furthermore, the study showed that the hypomethylated state of these regions was preserved even after chromosomal transfer from a Chinese Hamster Ovary background to two different mouse cell lines—suggesting that the maintenance of such epigenetic marks is strong.67 However, it is currently unknown whether such marks actively demarcate the boundaries of neocentromeres or are simply created in response to the changes in chromatin caused by the formation of a neocentromere.
An alternative and perhaps additional means of epigenetic regulation of neocentromeres lies in the modification of histones through acetylation. The role of histone acetylation at the centromere appears to be complex. Histone hyperacetylation through trichostatin A (TSA) treatment has been shown to induce kinetochore formation in integrated human engineered chromosomes (HECs)83 and restore the loading of CENPA at centromeric regions when histone chaperone proteins normally associated with this loading are knocked down.84 In this context, the response of the CENPA and scaffold/matrix binding domains to trichostatin A (TSA) treatment at the mardel(10) neocentromere was surprising.85,86 Hyperacetylation was shown to cause a striking change in both domains: The CENPA domain was found to shift in position by 320 kb in response to histone hyperacetylation,85 yet at the same time, the S/MAR domain was observed to shrink to less than half of its original size.86 Both studies thus suggested a link between histone acetylation and the position of the neocentromere.
Notably, however, the shift in position of the CENPA domain observed in response to TSA treatment was found to be reversible following removal of the drug85—suggesting that other epigenetic marks such as CpG methylation discussed above might play a role in the maintenance of neocentromeres. Further studies are needed to investigate whether TSA treatment affects the hypomethylated regions found at this neocentromere and whether induced changes to the DNA methylation environment can perturb the formation of the S/MAR domain.
A further interesting observation regarding the epigenetics of neocentromeres stems from engineered neocentric minichromosomes (NC-MiCs). Several NC-MiCs have been created by telomere-associated chromosome truncation of the mardel(10) marker chromosome, the smallest of which is approximately 650 kb in size.87,88 This circular minichromosome, NC-MiC5, is thus many times smaller than the native S/MAR domain on the original neocentromere, and yet the chromosome is still capable of forming a fully functional neocentromere in cell culture. Such results would seem to indicate a pliability of centromeric chromatin that can rapidly adapt to structural changes—a hypothesis for which we have preliminary data in support (L.H.W. and K.H.A.C., unpublished data).
Centromere Repositioning, Karyotype Evolution, and Speciation
From a clinical viewpoint, neocentromere formation provides little evolutionary advantage. Although the formation of a neocentromere rescues the carrier from embryonic lethality, the chromosomal rearrangements associated with neocentromere formation are generally deleterious, resulting in either partial trisomy or tetrasomy or in a ring chromosome that is subject to aneuploidy. The ability to form a neocentromere might also aid the pathway to cancer (see Cancer, above). Why, then, does the process of neocentromere formation occur at all, and with reasonable frequency?
The answer to this intriguing question might lie with a rare type of neocentromere reported in the clinical literature. These neocentromeres are formed on an intact chromosome with the preexisting, repetitive-DNA-based centromere still present, but inactivated. In essence, the active centromere on these pseudodicentric chromosomes has been repositioned. Such neocentromeres are uncommon, with only five cases reported in the literature.71,89–92 However, considering that no obvious clinical defect is associated with the individuals carrying these repositioned centromeres, this mode of neocentromere formation might be more common than the statistics indicate.
Three examples of this pseudodicentric neocentric phenomenon were neocentromeres that formed in the heterochromatic long arm of the Y chromosome.89–91 Although one example was present with a high degree of mosaicism and instability,90 the other two examples were stably transmitted through at least three generations of males, with the alpha-satellite DNA of the preexisting centromere still present on the chromosome arms, but failing to form a constriction. Quantitative FISH analysis of the alpha-satellite remaining at one inactive centromere suggested that there might have been a partial deletion of the alphoid DNA, although the amount was only slightly outside the range of normal variation seen for the Y centromere in the population.91
Although the heterochromatic long arm of the Y chromosome might be particularly predisposed to neocentromere formation, a further two cases of centromere repositioning have involved the autosomes.71,92 In one example, on chromosome 3, the neocentromere was recorded to have been transmitted through one generation, having formed de novo in the father.71 However, with the other example, on chromosome 4, the original progenitor of the chromosome could not be determined, with the chromosome stably inherited through at least two generations without any alpha-satellite being present at the primary constriction.92 The levels of alpha-satellite DNA remaining at the old centromere in this last case were quantitated via FISH, but without the original progenitor chromosome 4 available for study, it was unclear whether the amount of satellite DNA had been reduced. Although the amount of satellite DNA was found to be low for chromosome 4, the quantity (approximately 1.3 Mb) was within the range of variation seen within the population.92
The fact that there might have been a reduction in the amount of satellite DNA at the old centromere in two cases raises an interesting possibility: Could neocentromere formation in these examples have been induced by the weakening or deactivation of the old centromere by partial deletion? However, even if all cases had involved deletions of alpha-satellite at the old centromeres, it would not necessarily mean that such deletions were the cause of neocentromere formation. In each instance where the amount of alpha-satellite remaining on the centromere was quantitated, the original progenitor of the neocentromere could not be traced, meaning that the neocentromere could potentially be many generations old.91,92 In such cases, a gradual mutation and loss of the alpha-satellite DNA at the inactive centromere would be expected.
These pseudodicentric neocentric cases are especially interesting, considering the well-documented process of centromere repositioning seen in vertebrates. Comparative studies of chromosomes in primates, other mammals, and birds have demonstrated that the positioning of centromeres changes over the course of evolution, by means unrelated to the surrounding pericentric DNA markers.34,71,81,93–96 These observations were first demonstrated for the evolution of chromosome IX in primates93 but have since been reproduced through the observations of other chromosomes in primates,81 birds,96 and other mammals.34,97 In all such cases, the order of the DNA markers surrounding the new centromeric location had remained unchanged, and the most parsimonious series of chromosomal rearrangements suggested that one centromere had been deactivated and a new centromere formed de novo at a new location.
Could such repositioning come about through a neocentric intermediate? Although the possibility cannot be excluded that repositioned centromeres are formed through the spontaneous, ectopic incorporation of centromeric satellite DNA, the examples of pseudodicentric neocentric chromosomes presented above strongly suggest that neocentromeres are indeed the means of centromere repositioning. In each of these cases, the neocentromere in question was stably transmitted through multiple generations, with the old, alphoid centromere remaining inactivated. Furthermore, there seems to be a relationship between regions known to form neocentromeres on human chromosomes and the sites of evolutionary centromere repositioning events in other organisms. On chromosomes 15, 3, and 13, centromere-repositioning events have occurred several times in regions known to favor neocentromere formation.10,34,71
Of course, such repositioned centromeres do not remain devoid of repetitive satellite DNA but must acquire it during subsequent evolution. Most intriguing in this respect are the recent studies of two rice centromeres.65,66 Although most rice chromosomes contain typical centromeres demarcated by long stretches of satellite repeats embedded within heterochromatin, the centromere on chromosome 8 (Cen8) is unusual, containing an extremely low quantity (40 kb) of satellite repeats.65 Mapping of the binding domain of CENH3 (the rice paralogue of CENPA) by ChIP and PCR demonstrated that although this block of satellite DNA lay within the centromeric chromatin, most of the 750 kb CENH3 domain was occupied by actively transcribed genes65 and was thus very similar to mapped human neocentromeres (see Transcriptional Competence within Neocentromeres, above). Cen8 might thus represent the next step in the centromere repositioning process: an example of a neocentromere that has become fixed within the species and is beginning to slowly incorporate satellite repeat sequences. A second mapped rice centromere, Cen3, possibly represents a step further in this evolutionary process. On this centromere, the satellite DNA occupied a much larger block of 450 kb, but the remainder of the 1.8 Mb region of CENH3 binding was again occupied by actively transcribed genes.66
Interestingly, it appears that the satellite repeat sequences do not gradually integrate throughout the active genes of a neocentromere, but rather expand outwards from a single location. In the case of a centromere-repositioning event within chromosome 6 in Old World monkeys, large amounts of satellite and repetitive sequences associated with the new centromere appear to have been introduced at the new centromere site without any change to the surrounding sequences at a BAC resolution level.95 The incorporation thus appears to be more in the nature of an initial insertion followed by expansion (through mechanisms such as unequal crossing over) rather than a gradual accumulation of satellite DNA over multiple regions within the neocentromere. If this is indeed the case, then the centromere must be capable of a gradual shift, from the euchromatic DNA of the initial neocentromere to the introduced satellite sequences, without overtly compromising the genetic content of the region.
There is some evidence, though, that the two gene-containing rice centromeres described above might be unusual cases in the process of centromere repositioning. Although the gene-density at both rice centromeres is lower than the surrounding regions, recent studies of chromosome 13 and the Macaque genome have suggested that centromere-repositioning events generally occur in “gene deserts”—large regions of the genome that are completely devoid of genes.34,81 Although neocentromerization clearly does not affect gene expression per se (see Transcriptional Competence within Neocentromeres, above), the eventual incorporation of heterochromatin at mature centromeres (see below) might ultimately influence the expression of genes within the centromeric region. It has thus been hypothesized that a lack of genes within sites of neocentromere formation on pseudodicentric neocentric chromosomes might be an important factor in the determination of whether the neocentromere becomes subsequently fixed in the population and incorporates satellite sequences.81
Precisely why the subsequent incorporation of satellite sequences at repositioned centromere sites occurs, however, remains a mystery. Clearly, there must be an evolutionary advantage in having repetitive DNA at centromeres, because neocentromeres are not known to have become fixed in the population of any organism studied to date without the incorporation of repetitive satellite DNA. One possibility is that repetitive satellite DNA might help to increase the loading of constitutive centromere proteins such as CENPA at centromeres. This theory has been supported by studies of the levels of CENPA present at centromeres, which has shown significantly less amounts of the protein to be present at neocentromeres (Irvine et al.,43 and see Organization of Centromere Proteins, above.) If this was the case, the presence of satellite DNA could be rapidly selected for via the phenomenon of meiotic drive.98
An alternate possibility for the incorporation of satellite DNA at neocentromeres is that repetitive DNA, being devoid of active genes, helps to promote a heterochromatic environment more favorable for sister chromatid cohesion. Centromeric heterochromatin is known to be necessary for sister-chromatid cohesion in yeast59,60 and vertebrates,61 and an increase in the amount of heterochromatin could allow the centric region to better withstand microtubule tension and aid the stability of the chromosome during mitosis and meiosis. Considering the generally high mitotic stability of balanced neocentromere marker chromosomes, this possibility might be regarded as less likely. However, significantly greater sister-chromatid separation after mechanical stress has been documented for at least one neocentromere (the pseudodicentric neocentric chromosome 4 mentioned above) indicating a possible reduction in chromatid cohesion,92 and this might be a contributing factor to the eventual fixation of satellite sequences.
Naturally, repositioning of the centromere on a chromosome provides an effective mechanism of reproductive isolation and thus evolutionary speciation. If neocentromere formation was indeed the cause of this process—something which from the above discussion appears highly likely—this would provide a simple explanation as to why mechanisms exist to drive neocentromere formation.
Conclusions
Neocentromeres are thus a fascinating example of epigenetic change within the genome. Although most commonly observed on rearranged fragments of chromosomes and occasionally in cancer cells, they appear to possess a broader role in providing a means of directing centromere repositioning and thereby speciation. By nature of their formation within nonrepetitive DNA sequences, neocentromeres have provided many insights into the processes of centromere formation and maintenance and into the structure and function of the centromere itself. Many questions remain unanswered, however, and future research is likely to focus on the mechanisms of neocentromere formation and its possible role in cancer etiology and disease development. Of particular interest is the underlying question of centromere and chromosome evolution. To what extent has neocentromere formation impacted on speciation? How is the transition between a neocentromere and a repetitive-DNA-based centromere orchestrated? The next 5 years of neocentromere research are likely to see an integration with the broader centromere field, with a recognition that neocentromeres and repetitive-DNA-based centromeres are intimately related. Ultimately, a deeper understanding of both forms of centromere is important in the deciphering of the structure and function of the primary constriction.
Web Resources
The URLs for data presented herein are as follows:
Online sSMC database, http://markerchromosomes.ag.vu/
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
Acknowledgments
We thank E. Northrop, L. Chan, and M. Pertile for stimulating discussions and critical comments on the manuscript. This work was supported by the National Health and Medical Research Council of Australia and the National Institutes of Health.
References
- 1.Obado S.O., Bot C., Nilsson D., Andersson B., Kelly J.M. Repetitive DNA is associated with centromeric domains in Trypanosoma brucei but not Trypanosoma cruzi. Genome Biol. 2007;8:R37. doi: 10.1186/gb-2007-8-3-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Willard H.F. Evolution of alpha satellite. Curr. Opin. Genet. Dev. 1991;1:509–514. doi: 10.1016/s0959-437x(05)80200-x. [DOI] [PubMed] [Google Scholar]
- 3.Choo K.H., Vissel B., Nagy A., Earle E., Kalitsis P. A survey of the genomic distribution of alpha satellite DNA on all the human chromosomes, and derivation of a new consensus sequence. Nucleic Acids Res. 1991;19:1179–1182. doi: 10.1093/nar/19.6.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voullaire L.E., Slater H.R., Petrovic V., Choo K.H. A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: Activation of a latent centromere? Am. J. Hum. Genet. 1993;52:1153–1163. [PMC free article] [PubMed] [Google Scholar]
- 5.du Sart D., Cancilla M.R., Earle E., Mao J.I., Saffery R., Tainton K.M., Kalitsis P., Martyn J., Barry A.E., Choo K.H. A functional neo-centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Nat. Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- 6.Lo A.W., Craig J.M., Saffery R., Kalitsis P., Irvine D.V., Earle E., Magliano D.J., Choo K.H. A 330 kb CENP-A binding domain and altered replication timing at a human neocentromere. EMBO J. 2001;20:2087–2096. doi: 10.1093/emboj/20.8.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rhoades M.M., Vilkomerson H. On the anaphase movement of chromosomes. Proc. Natl. Acad. Sci. USA. 1942;28:433–436. doi: 10.1073/pnas.28.10.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawe R.K., Hiatt E.N. Plant neocentromeres: Fast, focused, and driven. Chromosome Res. 2004;12:655–669. doi: 10.1023/B:CHRO.0000036607.74671.db. [DOI] [PubMed] [Google Scholar]
- 9.Depinet T.W., Zackowski J.L., Earnshaw W.C., Kaffe S., Sekhon G.S., Stallard R., Sullivan B.A., Vance G.H., Dyke D.L.V., Willard H.F. Characterization of neo-centromeres in marker chromosomes lacking detectable alpha-satellite DNA. Hum. Mol. Genet. 1997;6:1195–1204. doi: 10.1093/hmg/6.8.1195. [DOI] [PubMed] [Google Scholar]
- 10.Ventura M., Mudge J.M., Palumbo V., Burn S., Blennow E., Pierluigi M., Giorda R., Zuffardi O., Archidiacono N., Jackson M.S. Neocentromeres in 15q24–26 map to duplicons which flanked an ancestral centromere in 15q25. Genome Res. 2003;13:2059–2068. doi: 10.1101/gr.1155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voullaire L., Saffery R., Earle E., Irvine D.V., Slater H., Dale S., du Sart D., Fleming T., Choo K.H. Mosaic inv dup(8p) marker chromosome with stable neocentromere suggests neocentromerization is a post-zygotic event. Am. J. Med. Genet. 2001;102:86–94. doi: 10.1002/1096-8628(20010722)102:1<86::aid-ajmg1390>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 12.Kosztolányi G. Does “ring syndrome” exist? An analysis of 207 case reports on patients with a ring autosome. Hum. Genet. 1987;75:174–179. doi: 10.1007/BF00591082. [DOI] [PubMed] [Google Scholar]
- 13.Warburton P.C., Barwell J., Splitt M., Maxwell D., Bint S., Ogilvie C.M. Class II neocentromeres: A putative common neocentromere site in band 4q21.2. Eur. J. Hum. Genet. 2003;11:749–753. doi: 10.1038/sj.ejhg.5201047. [DOI] [PubMed] [Google Scholar]
- 14.Wandall A., Tranebjaerg L., Tommerup N. A neocentromere on human chromosome 3 without detectable alpha-satellite DNA forms morphologically normal kinetochores. Chromosoma. 1998;107:359–365. doi: 10.1007/s004120050319. [DOI] [PubMed] [Google Scholar]
- 15.Chuang L., Wakui K., Sue W.C., Su M.H., Shaffer L.G., Kuo P.L. Interstitial deletion 11(p11.12p11.2) and analphoid marker formation results in inherited Potocki-Shaffer syndrome. Am. J. Med. Genet. A. 2005;133:180–183. doi: 10.1002/ajmg.a.30362. [DOI] [PubMed] [Google Scholar]
- 16.Knegt A.C., Li S., Engelen J.J.M., Bijlsma E.K., Warburton P.E. Prenatal diagnosis of a karyotypically normal pregnancy in a mother with a supernumerary neocentric 13q21 –>13q22 chromosome and balancing reciprocal deletion. Prenat. Diagn. 2003;23:215–220. doi: 10.1002/pd.559. [DOI] [PubMed] [Google Scholar]
- 17.Dalprà L., Giardino D., Finelli P., Corti C., Valtorta C., Guerneri S., Ilardi P., Fortuna R., Coviello D., Nocera G. Cytogenetic and molecular evaluation of 241 small supernumerary marker chromosomes: Cooperative study of 19 Italian laboratories. Genet. Med. 2005;7:620–625. doi: 10.1097/01.gim.0000182876.57766.2d. [DOI] [PubMed] [Google Scholar]
- 18.Liehr T., Claussen U., Starke H. Small supernumerary marker chromosomes (sSMC) in humans. Cytogenet. Genome Res. 2004;107:55–67. doi: 10.1159/000079572. [DOI] [PubMed] [Google Scholar]
- 19.Pedeutour F., Forus A., Coindre J.M., Berner J.M., Nicolo G., Michiels J.F., Terrier P., Ranchere-Vince D., Collin F., Myklebost O. Structure of the supernumerary ring and giant rod chromosomes in adipose tissue tumors. Genes Chromosomes Cancer. 1999;24:30–41. [PubMed] [Google Scholar]
- 20.Gisselsson D., Höglund M., Mertens F., Johansson B., Cin P.D., den Berghe H.V., Earnshaw W.C., Mitelman F., Mandahl N. The structure and dynamics of ring chromosomes in human neoplastic and non-neoplastic cells. Hum. Genet. 1999;104:315–325. doi: 10.1007/s004390050960. [DOI] [PubMed] [Google Scholar]
- 21.Sirvent N., Forus A., Lescaut W., Burel F., Benzaken S., Chazal M., Bourgeon A., Vermeesch J.R., Myklebost O., Turc-Carel C. Characterization of centromere alterations in liposarcomas. Genes Chromosomes Cancer. 2000;29:117–129. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1014>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 22.Forus A., Bjerkehagen B., Sirvent N., Meza-Zepeda L.A., Coindre J.M., Berner J.M., Myklebost O., Pedeutour F. A well-differentiated liposarcoma with a new type of chromosome 12-derived markers. Cancer Genet. Cytogenet. 2001;131:13–18. doi: 10.1016/s0165-4608(01)00516-7. [DOI] [PubMed] [Google Scholar]
- 23.Italiano A., Attias R., Aurias A., Pérot G., Burel-Vandenbos F., Otto J., Venissac N., Pedeutour F. Molecular cytogenetic characterization of a metastatic lung sarcomatoid carcinoma: 9p23 neocentromere and 9p23-p24 amplification including JAK2 and JMJD2C. Cancer Genet. Cytogenet. 2006;167:122–130. doi: 10.1016/j.cancergencyto.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 24.Abeliovich D., Yehuda O., Ben-Neriah S., Kapelushnik Y., Ben-Yehuda D. dup(10q) lacking alpha-satellite DNA in bone marrow cells of a patient with acute myeloid leukemia. Cancer Genet. Cytogenet. 1996;89:1–6. doi: 10.1016/0165-4608(95)00300-2. [DOI] [PubMed] [Google Scholar]
- 25.Tomonaga T., Matsushita K., Yamaguchi S., Oohashi T., Shimada H., Ochiai T., Yoda K., Nomura F. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003;63:3511–3516. [PubMed] [Google Scholar]
- 26.Tomonaga T., Matsushita K., Ishibashi M., Nezu M., Shimada H., Ochiai T., Yoda K., Nomura F. Centromere protein H is up-regulated in primary human colorectal cancer and its overexpression induces aneuploidy. Cancer Res. 2005;65:4683–4689. doi: 10.1158/0008-5472.CAN-04-3613. [DOI] [PubMed] [Google Scholar]
- 27.Heun P., Erhardt S., Blower M.D., Weiss S., Skora A.D., Karpen G.H. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev. Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saffery R., Irvine D.V., Griffiths B., Kalitsis P., Wordeman L., Choo K.H. Human centromeres and neocentromeres show identical distribution patterns of >20 functionally important kinetochore-associated proteins. Hum. Mol. Genet. 2000;9:175–185. doi: 10.1093/hmg/9.2.175. [DOI] [PubMed] [Google Scholar]
- 29.Craig J.M., Earle E., Canham P., Wong L.H., Anderson M., Choo K.H.A. Analysis of mammalian proteins involved in chromatin modification reveals new metaphase centromeric proteins and distinct chromosomal distribution patterns. Hum. Mol. Genet. 2003;12:3109–3121. doi: 10.1093/hmg/ddg330. [DOI] [PubMed] [Google Scholar]
- 30.Palmer D.K., O'Day K., Wener M.H., Andrews B.S., Margolis R.L. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullivan K.F., Hechenberger M., Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J. Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lo A.W., Magliano D.J., Sibson M.C., Kalitsis P., Craig J.M., Choo K.H. A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromere DNA. Genome Res. 2001;11:448–457. doi: 10.1101/gr.167601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alonso A., Mahmood R., Li S., Cheung F., Yoda K., Warburton P.E. Genomic microarray analysis reveals distinct locations for the CENP-A binding domains in three human chromosome 13q32 neocentromeres. Hum. Mol. Genet. 2003;12:2711–2721. doi: 10.1093/hmg/ddg282. [DOI] [PubMed] [Google Scholar]
- 34.Cardone M.F., Alonso A., Pazienza M., Ventura M., Montemurro G., Carbone L., de Jong P.J., Stanyon R., D'Addabbo P., Archidiacono N. Independent centromere formation in a capricious, gene-free domain of chromosome 13q21 in Old World monkeys and pigs. Genome Biol. 2006;7:R91. doi: 10.1186/gb-2006-7-10-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chueh A.C., Wong L.H., Wong N., Choo K.H.A. Variable and hierarchical size distribution of L1-retroelement-enriched CENP-A clusters within a functional human neocentromere. Hum. Mol. Genet. 2005;14:85–93. doi: 10.1093/hmg/ddi008. [DOI] [PubMed] [Google Scholar]
- 36.Alonso A., Fritz B., Hasson D., Abrusan G., Cheung F., Yoda K., Radlwimmer B., Ladurner A., Warburton P. Co-localization of CENP-C and CENP-H to discontinuous domains of CENP-A chromatin at human neocentromeres. Genome Biol. 2007;8:R148. doi: 10.1186/gb-2007-8-7-r148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blower M.D., Sullivan B.A., Karpen G.H. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zinkowski R.P., Meyne J., Brinkley B.R. The centromere-kinetochore complex: A repeat subunit model. J. Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Filipski J., Leblanc J., Youdale T., Sikorska M., Walker P.R. Periodicity of DNA folding in higher order chromatin structures. EMBO J. 1990;9:1319–1327. doi: 10.1002/j.1460-2075.1990.tb08241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paulson J.R., Laemmli U.K. The structure of histone-depleted metaphase chromosomes. Cell. 1977;12:817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- 41.Laemmli U.K., Cheng S.M., Adolph K.W., Paulson J.R., Brown J.A., Baumbach W.R. Metaphase chromosome structure: The role of nonhistone proteins. Cold Spring Harb. Symp. Quant. Biol. 1978;42:351–360. doi: 10.1101/sqb.1978.042.01.036. [DOI] [PubMed] [Google Scholar]
- 42.Jackson D.A., Dickinson P., Cook P.R. The size of chromatin loops in HeLa cells. EMBO J. 1990;9:567–571. doi: 10.1002/j.1460-2075.1990.tb08144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Irvine D.V., Amor D.J., Perry J., Sirvent N., Pedeutour F., Choo K.H.A., Saffery R. Chromosome size and origin as determinants of the level of CENP-A incorporation into human centromeres. Chromosome Res. 2004;12:805–815. doi: 10.1007/s10577-005-5377-4. [DOI] [PubMed] [Google Scholar]
- 44.Ando S., Yang H., Nozaki N., Okazaki T., Yoda K. CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol. Cell. Biol. 2002;22:2229–2241. doi: 10.1128/MCB.22.7.2229-2241.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masumoto H., Ikeno M., Nakano M., Okazaki T., Grimes B., Cooke H., Suzuki N. Assay of centromere function using a human artificial chromosome. Chromosoma. 1998;107:406–416. doi: 10.1007/s004120050324. [DOI] [PubMed] [Google Scholar]
- 46.Ohzeki J., Nakano M., Okada T., Masumoto H. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J. Cell Biol. 2002;159:765–775. doi: 10.1083/jcb.200207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Basu J., Stromberg G., Compitello G., Willard H.F., Bokkelen G.V. Rapid creation of BAC-based human artificial chromosome vectors by transposition with synthetic alpha-satellite arrays. Nucleic Acids Res. 2005;33:587–596. doi: 10.1093/nar/gki207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okamoto Y., Nakano M., Ohzeki J.I., Larionov V., Masumoto H. A minimal CENP-A core is required for nucleation and maintenance of a functional human centromere. EMBO J. 2007;26:1279–1291. doi: 10.1038/sj.emboj.7601584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foltz D.R., Jansen L.E.T., Black B.E., Bailey A.O., Yates J.R., Cleveland D.W. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 50.Saffery R., Sumer H., Hassan S., Wong L.H., Craig J.M., Todokoro K., Anderson M., Stafford A., Choo K.H.A. Transcription within a functional human centromere. Mol. Cell. 2003;12:509–516. doi: 10.1016/s1097-2765(03)00279-x. [DOI] [PubMed] [Google Scholar]
- 51.Adolphs K.W., Cheng S.M., Paulson J.R., Laemmli U.K. Isolation of a protein scaffold from mitotic HeLa cell chromosomes. Proc. Natl. Acad. Sci. USA. 1977;74:4937–4941. doi: 10.1073/pnas.74.11.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tavormina P.A., Côme M.G., Hudson J.R., Mo Y.Y., Beck W.T., Gorbsky G.J. Rapid exchange of mammalian topoisomerase II alpha at kinetochores and chromosome arms in mitosis. J. Cell Biol. 2002;158:23–29. doi: 10.1083/jcb.200202053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Christensen M.O., Larsen M.K., Barthelmes H.U., Hock R., Andersen C.L., Kjeldsen E., Knudsen B.R., Westergaard O., Boege F., Mielke C. Dynamics of human DNA topoisomerases IIalpha and IIbeta in living cells. J. Cell Biol. 2002;157:31–44. doi: 10.1083/jcb.200112023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maeshima K., Laemmli U.K. A two-step scaffolding model for mitotic chromosome assembly. Dev. Cell. 2003;4:467–480. doi: 10.1016/s1534-5807(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 55.Coelho P.A., Queiroz-Machado J., Sunkel C.E. Condensin-dependent localisation of topoisomerase II to an axial chromosomal structure is required for sister chromatid resolution during mitosis. J. Cell Sci. 2003;116:4763–4776. doi: 10.1242/jcs.00799. [DOI] [PubMed] [Google Scholar]
- 56.Kireeva N., Lakonishok M., Kireev I., Hirano T., Belmont A.S. Visualization of early chromosome condensation: A hierarchical folding, axial glue model of chromosome structure. J. Cell Biol. 2004;166:775–785. doi: 10.1083/jcb.200406049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strissel P.L., Espinosa R., Rowley J.D., Swift H. Scaffold attachment regions in centromere-associated DNA. Chromosoma. 1996;105:122–133. doi: 10.1007/BF02509522. [DOI] [PubMed] [Google Scholar]
- 58.Sumer H., Craig J.M., Sibson M., Choo K.H.A. A rapid method of genomic array analysis of scaffold/matrix attachment regions (S/MARs) identifies a 2.5-Mb region of enhanced scaffold/matrix attachment at a human neocentromere. Genome Res. 2003;13:1737–1743. doi: 10.1101/gr.1095903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bernard P., Maure J.F., Partridge J.F., Genier S., Javerzat J.P., Allshire R.C. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 60.Nonaka N., Kitajima T., Yokobayashi S., Xiao G., Yamamoto M., Grewal S.I.S., Watanabe Y. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- 61.Fukagawa T., Nogami M., Yoshikawa M., Ikeno M., Okazaki T., Takami Y., Nakayama T., Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- 62.Black B.E., Foltz D.R., Chakravarthy S., Luger K., Woods V.L., Cleveland D.W. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 63.Black B.E., Jansen L.E.T., Maddox P.S., Foltz D.R., Desai A.B., Shah J.V., Cleveland D.W. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol. Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 64.Lam A.L., Boivin C.D., Bonney C.F., Rudd M.K., Sullivan B.A. Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc. Natl. Acad. Sci. USA. 2006;103:4186–4191. doi: 10.1073/pnas.0507947103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nagaki K., Cheng Z., Ouyang S., Talbert P.B., Kim M., Jones K.M., Henikoff S., Buell C.R., Jiang J. Sequencing of a rice centromere uncovers active genes. Nat. Genet. 2004;36:138–145. doi: 10.1038/ng1289. [DOI] [PubMed] [Google Scholar]
- 66.Yan H., Ito H., Nobuta K., Ouyang S., Jin W., Tian S., Lu C., Venu R.C., Wang G.L., Green P.J. Genomic and genetic characterization of rice Cen3 reveals extensive transcription and evolutionary implications of a complex centromere. Plant Cell. 2006;18:2123–2133. doi: 10.1105/tpc.106.043794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wong N.C., Wong L.H., Quach J.M., Canham P., Craig J.M., Song J.Z., Clark S.J., Choo K.H.A. Permissive transcriptional activity at the centromere through pockets of DNA hypomethylation. PLoS Genet. 2006;2:e17. doi: 10.1371/journal.pgen.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Williams B.C., Murphy T.D., Goldberg M.L., Karpen G.H. Neocentromere activity of structurally acentric mini-chromosomes in Drosophila. Nat. Genet. 1998;18:30–37. doi: 10.1038/ng0198-30. [DOI] [PubMed] [Google Scholar]
- 69.Maggert K.A., Karpen G.H. The activation of a neocentromere in Drosophila requires proximity to an endogenous centromere. Genetics. 2001;158:1615–1628. doi: 10.1093/genetics/158.4.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nasuda S., Hudakova S., Schubert I., Houben A., Endo T.R. Stable barley chromosomes without centromeric repeats. Proc. Natl. Acad. Sci. USA. 2005;102:9842–9847. doi: 10.1073/pnas.0504235102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ventura M., Weigl S., Carbone L., Cardone M.F., Misceo D., Teti M., D'Addabbo P., Wandall A., Björck E., de Jong P.J. Recurrent sites for new centromere seeding. Genome Res. 2004;14:1696–1703. doi: 10.1101/gr.2608804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Van Hooser A.A., Ouspenski I.I., Gregson H.C., Starr D.A., Yen T.J., Goldberg M.L., Yokomori K., Earnshaw W.C., Sullivan K.F., Brinkley B.R. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 2001;114:3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- 73.Collins K.A., Furuyama S., Biggins S. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 2004;14:1968–1972. doi: 10.1016/j.cub.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 74.Moreno-Moreno O., Torras-Llort M., Azorín F. Proteolysis restricts localization of CID, the centromere-specific histone H3 variant of Drosophila, to centromeres. Nucleic Acids Res. 2006;34:6247–6255. doi: 10.1093/nar/gkl902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Amor D.J., Choo K.H.A. Neocentromeres: Role in human disease, evolution, and centromere study. Am. J. Hum. Genet. 2002;71:695–714. doi: 10.1086/342730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sullivan B.A., Willard H.F. Stable dicentric X chromosomes with two functional centromeres. Nat. Genet. 1998;20:227–228. doi: 10.1038/3024. [DOI] [PubMed] [Google Scholar]
- 77.Platero J.S., Ahmad K., Henikoff S. A distal heterochromatic block displays centromeric activity when detached from a natural centromere. Mol. Cell. 1999;4:995–1004. doi: 10.1016/s1097-2765(00)80228-2. [DOI] [PubMed] [Google Scholar]
- 78.Platero J.S., Csink A.K., Quintanilla A., Henikoff S. Changes in chromosomal localization of heterochromatin-binding proteins during the cell cycle in Drosophila. J. Cell Biol. 1998;140:1297–1306. doi: 10.1083/jcb.140.6.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jansen L.E.T., Black B.E., Foltz D.R., Cleveland D.W. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schuh M., Lehner C.F., Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr. Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- 81.Ventura M., Antonacci F., Cardone M.F., Stanyon R., D'Addabbo P., Cellamare A., Sprague L.J., Eichler E.E., Archidiacono N., Rocchi M. Evolutionary formation of new centromeres in macaque. Science. 2007;316:243–246. doi: 10.1126/science.1140615. [DOI] [PubMed] [Google Scholar]
- 82.Warburton P.E. Chromosomal dynamics of human neocentromere formation. Chromosome Res. 2004;12:617–626. doi: 10.1023/B:CHRO.0000036585.44138.4b. [DOI] [PubMed] [Google Scholar]
- 83.Nakano M., Okamoto Y., Ohzeki J., Masumoto H. Epigenetic assembly of centromeric chromatin at ectopic alpha-satellite sites on human chromosomes. J. Cell Sci. 2003;116:4021–4034. doi: 10.1242/jcs.00697. [DOI] [PubMed] [Google Scholar]
- 84.Fujita Y., Hayashi T., Kiyomitsu T., Toyoda Y., Kokubu A., Obuse C., Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev. Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 85.Craig J.M., Wong L.H., Lo A.W.I., Earle E., Choo K.H.A. Centromeric chromatin pliability and memory at a human neocentromere. EMBO J. 2003;22:2495–2504. doi: 10.1093/emboj/cdg232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sumer H., Saffery R., Wong N., Craig J.M., Choo K.H.A. Effects of scaffold/matrix alteration on centromeric function and gene expression. J. Biol. Chem. 2004;279:37631–37639. doi: 10.1074/jbc.M401051200. [DOI] [PubMed] [Google Scholar]
- 87.Saffery R., Wong L.H., Irvine D.V., Bateman M.A., Griffiths B., Cutts S.M., Cancilla M.R., Cendron A.C., Stafford A.J., Choo K.H. Construction of neocentromere-based human minichromosomes by telomere-associated chromosomal truncation. Proc. Natl. Acad. Sci. USA. 2001;98:5705–5710. doi: 10.1073/pnas.091468498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wong L.H., Saffery R., Choo K.H.A. Construction of neocentromere-based human minichromosomes for gene delivery and centromere studies. Gene Ther. 2002;9:724–726. doi: 10.1038/sj.gt.3301756. [DOI] [PubMed] [Google Scholar]
- 89.Rivera H., Vassquez A.I., Ayala-Madrigal M.L., Ramirez-Dueñas M.L., Davalos I.P. Alphoidless centromere of a familial unstable inverted Y chromosome. Ann. Genet. 1996;39:236–239. [PubMed] [Google Scholar]
- 90.Bukvic N., Susca F., Gentile M., Tangari E., Ianniruberto A., Guanti G. An unusual dicentric Y chromosome with a functional centromere with no detectable alpha-satellite. Hum. Genet. 1996;97:453–456. doi: 10.1007/BF02267065. [DOI] [PubMed] [Google Scholar]
- 91.Tyler-Smith C., Gimelli G., Giglio S., Floridia G., Pandya A., Terzoli G., Warburton P.E., Earnshaw W.C., Zuffardi O. Transmission of a fully functional human neocentromere through three generations. Am. J. Hum. Genet. 1999;64:1440–1444. doi: 10.1086/302380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Amor D.J., Bentley K., Ryan J., Perry J., Wong L., Slater H., Choo K.H.A. Human centromere repositioning “in progress”. Proc. Natl. Acad. Sci. USA. 2004;101:6542–6547. doi: 10.1073/pnas.0308637101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Montefalcone G., Tempesta S., Rocchi M., Archidiacono N. Centromere repositioning. Genome Res. 1999;9:1184–1188. doi: 10.1101/gr.9.12.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ventura M., Archidiacono N., Rocchi M. Centromere emergence in evolution. Genome Res. 2001;11:595–599. doi: 10.1101/gr.152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Eder V., Ventura M., Ianigro M., Teti M., Rocchi M., Archidiacono N. Chromosome 6 phylogeny in primates and centromere repositioning. Mol. Biol. Evol. 2003;20:1506–1512. doi: 10.1093/molbev/msg165. [DOI] [PubMed] [Google Scholar]
- 96.Kasai F., Garcia C., Arruga M.V., Ferguson-Smith M.A. Chromosome homology between chicken (Gallus gallus domesticus) and the red-legged partridge (Alectoris rufa); evidence of the occurrence of a neocentromere during evolution. Cytogenet. Genome Res. 2003;102:326–330. doi: 10.1159/000075770. [DOI] [PubMed] [Google Scholar]
- 97.Szpirer C., Rivière M., VanVooren P., Moisan M.P., Haller O., Szpirer J. Chromosome evolution of MMU16 and RNO11: Conserved synteny associated with gene order rearrangements explicable by intrachromosomal recombinations and neocentromere emergence. Cytogenet. Genome Res. 2005;108:322–327. doi: 10.1159/000081526. [DOI] [PubMed] [Google Scholar]
- 98.Henikoff S., Ahmad K., Malik H.S. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 99.Warburton P.E., Dolled M., Mahmood R., Alonso A., Li S., Naritomi K., Tohma T., Nagai T., Hasegawa T., Ohashi H. Molecular cytogenetic analysis of eight inversion duplications of human chromosome 13q that each contain a neocentromere. Am. J. Hum. Genet. 2000;66:1794–1806. doi: 10.1086/302924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Morrissette J.D., Celle L., Owens N.L., Shields C.L., Zackai E.H., Spinner N.B. Boy with bilateral retinoblastoma due to an unusual ring chromosome 13 with activation of a latent centromere. Am. J. Med. Genet. 2001;99:21–28. doi: 10.1002/1096-8628(20010215)99:1<21::aid-ajmg1122>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 101.Li S., Malafiej P., Levy B., Mahmood R., Field M., Hughes T., Lockhart L.H., Wu Z., Huang M., Hirschhorn K. Chromosome 13q neocentromeres: molecular cytogenetic characterization of three additional cases and clinical spectrum. Am. J. Med. Genet. 2002;110:258–267. doi: 10.1002/ajmg.10454. [DOI] [PubMed] [Google Scholar]
- 102.Barwell J., Mazzaschi R., Bint S., Ogilvie C.M., Elmslie F. A new neocentromere locus on chromosome 13 resulting in mosaic tetrasomy for distal 13q and an asymmetric phenotype. Am. J. Med. Genet. A. 2004;130:295–298. doi: 10.1002/ajmg.a.30208. [DOI] [PubMed] [Google Scholar]
- 103.Amor D.J., Voullaire L., Bentley K., Savarirayan R., Choo K.H.A. Mosaic monosomy of a neocentric ring chromosome maps brachyphalangy and growth hormone deficiency to 13q31.1–13q32.3. Am. J. Med. Genet. A. 2005;133:151–157. doi: 10.1002/ajmg.a.30527. [DOI] [PubMed] [Google Scholar]
- 104.Tonnies H., Gerlach A., Heineking B., Starke H., Neitzel H., Neumann L.M. Molecular cytogenetic identification and characterization of a de novo supernumerary neocentromeric derivative chromosome 13. Cytogenet. Genome Res. 2006;114:325–329. doi: 10.1159/000094221. [DOI] [PubMed] [Google Scholar]
- 105.Hubbard T., Barker D., Birney E., Cameron G., Chen Y., Clark L., Cox T., Cuff J., Curwen V., Down T. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Slater H.R., Nouri S., Earle E., Lo A.W., Hale L.G., Choo K.H. Neocentromere formation in a stable ring 1p32-p36.1 chromosome. J. Med. Genet. 1999;36:914–918. [PMC free article] [PubMed] [Google Scholar]
- 107.Constantinou M., Zajac E., Plowas I., Kaluzewski B. Two new cases of neocentric marker chromosomes (NMCs). Molecular cytogenetic and clinical characterization. Chromosome Res. 2005;13(Suppl):66. [Google Scholar]
- 108.Higgins R.R., Wright E., Baldinger S., Tschider E., Ahmad J., Schwartz S., Curtis C.A. Neocentromere in a ring-shaped chromosome 1. Am. J. Hum. Genet. 2000;67(Suppl):A775. [Google Scholar]
- 109.Kucerova M., Polivkova Z., Dluholucky S., Kvasnicova M. Terminal deletion with stable acentric fragment of 1q in a child with congenital malformations. Am. J. Hum. Genet. 1983;35:91–95. [PMC free article] [PubMed] [Google Scholar]
- 110.Spiegel M., Hickmann G., Senger G., Kozlowski P., Bartsch O. Two new cases of analphoid marker chromosomes. Am. J. Med. Genet. A. 2003;116:284–289. doi: 10.1002/ajmg.a.10916. [DOI] [PubMed] [Google Scholar]
- 111.Petit P., Fryns J.P. Interstitial deletion 2p accompanied by marker chromosome formation of the deleted segment resulting in a stable acentric marker chromosome. Genet. Couns. 1997;8:341–343. [PubMed] [Google Scholar]
- 112.Pietrzak J., Mrasek K., Obersztyn E., Stankiewicz P., Kosyakova N., Weise A., Cheung S.W., Cai W.W., von Eggeling F., Mazurczak T. Molecular cytogenetic characterization of eight small supernumerary marker chromosomes originating from chromosomes 2, 4, 8, 18, and 21 in three patients. J. Appl. Genet. 2007;48:167–175. doi: 10.1007/BF03194675. [DOI] [PubMed] [Google Scholar]
- 113.Maraschio P., Tupler R., Rossi E., Barbierato L., Uccellatore F., Rocchi M., Zuffardi O., Fraccaro M. A novel mechanism for the origin of supernumerary marker chromosomes. Hum. Genet. 1996;97:382–386. doi: 10.1007/BF02185778. [DOI] [PubMed] [Google Scholar]
- 114.Gimelli G., Zuffardi O., Giglio S., Zeng C., He D. CENP-G in neocentromeres and inactive centromeres. Chromosoma. 2000;109:328–333. doi: 10.1007/s004120000082. [DOI] [PubMed] [Google Scholar]
- 115.Batanian J.R., Bernreuter K., Koslosky L., Frater J.L. Coexistence of neocentromeric marker 3q and trisomy 3 in two different tissues in a 3-year-old boy with peripheral T-cell lymphoma: Support for a gene dosage effect hypothesis. Cancer Genet. Cytogenet. 2006;170:152–157. doi: 10.1016/j.cancergencyto.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 116.Teshima I., Bawle E.V., Weksberg R., Shuman C., Dyke D.L.V., Schwartz S. Analphoid 3qter markers. Am. J. Med. Genet. 2000;94:113–119. doi: 10.1002/1096-8628(20000911)94:2<113::aid-ajmg3>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 117.Cockwell A.E., Gibbons B., Moore I.E., Crolla J.A. An analphoid supernumerary marker chromosome derived from chromosome 3 ascertained in a fetus with multiple malformations. J. Med. Genet. 2000;37:807–810. doi: 10.1136/jmg.37.10.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yu J., Qi Z., Thompson K., Modaff P., Wells W., Meisner L., Pauli R. Characterization of a rare neocentric marker chromosome using chromosome microdissection. Am. J. Hum. Genet. 2004;74(Suppl.):A982. [Google Scholar]
- 119.Sullivan C.M., Mountford S., Emmerson J.M., Ellis R.J., Turmbull C., Waters K.S. A mosaic karyotype with an additional inv dup(3)(qter->q26.2:q26.2->qter), containing a neocentromere, detected in a skin biopsy from a girl with skeletal abnormalities, abnormal skin pigmentation and developmental delay. J. Med. Genet. 2005;42(Suppl.):71. [Google Scholar]
- 120.Papenhausen P., Gadi I., Tepperberg J., Mowrey P., Singh-Kahlon P., Wisniewski L., Goodwin D. Trisomy 3q secondary to a terminal deletion/generation of a mirrorimage analphoid marker in a neonate. Am. J. Hum. Genet. 2003;73(Suppl.):A841. [Google Scholar]
- 121.Portnoi M.F., Boutchnei S., Bouscarat F., Morlier G., Nizard S., Dersarkissian H., Crickx B., Nouchy M., Taillemite J.L., Belaich S. Skin pigmentary anomalies and mosaicism for an acentric marker chromosome originating from 3q. J. Med. Genet. 1999;36:246–250. [PMC free article] [PubMed] [Google Scholar]
- 122.Barbi G., Spaich C., Adolph S., Kehrer-Sawatzki H. Analphoid de novo marker chromosome inv dup(3)(q28qter) with neocentromere in a dysmorphic and developmentally retarded girl. J. Med. Genet. 2003;40:e27. doi: 10.1136/jmg.40.3.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gimelli G., Giorda R., Beri S., Gimelli S., Zuffardi O. A large analphoid invdup(3)(q22.3qter) marker chromosome characterized by array-CGH in a child with malformations, mental retardation, ambiguous genitalia and Blaschko's lines. Eur. J. Med. Genet. 2007;50:264–273. doi: 10.1016/j.ejmg.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 124.Grimbacher B., Dutra A.S., Holland S.M., Fischer R.E., Pao M., Gallin J.I., Puck J.M. Analphoid marker chromosome in a patient with hyper-IgE syndrome, autism, and mild mental retardation. Genet. Med. 1999;1:213–218. doi: 10.1097/00125817-199907000-00008. [DOI] [PubMed] [Google Scholar]
- 125.Fritz B., Dietze I., Wandall A., Aslan M., Schmidt A., Kattner E., Schwerdtfeger R., Friedrich U. A supernumerary marker chromosome with a neocentromere derived from 5p14–>pter. J. Med. Genet. 2001;38:559–565. doi: 10.1136/jmg.38.8.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Qin N., Bartley J., Wang J., Jogenson J., Warburton P.E. A neocentromere derived from a supernumerary marker deleted from the long arm of chromosome 6. Am. J. Hum. Genet. 2005;76(Suppl.):A829. doi: 10.1159/000109633. [DOI] [PubMed] [Google Scholar]
- 127.Sala E., Crosti F., Villa N., Oldrini A., Lalatta F., Gandolfi P., Gautiero E., Solano R., Dalpra L. First report of a supernumerary marker chromosome 6. Chromosome Res. 2005;13(Suppl.):131. [Google Scholar]
- 128.de Pater J.M., Kroes H.Y., Verschuren M., van Oppen A.C.C., Albrechts J.C.M., Engelen J.J.M. Mosaic trisomy (8)(p22 –> pter) in a fetus caused by a supernumerary marker chromosome without alphoid sequences. Prenat. Diagn. 2005;25:151–155. doi: 10.1002/pd.1097. [DOI] [PubMed] [Google Scholar]
- 129.Herry A., Morel F., Bris M.J.L., Bellec V., Lallaoui H., Parent P., Braekeleer M.D. Molecular cytogenetic characterization of two small chromosome 8 derived supernumerary mosaic markers. Am. J. Med. Genet. A. 2004;128:33–38. doi: 10.1002/ajmg.a.30077. [DOI] [PubMed] [Google Scholar]
- 130.Ohashi H., Wakui K., Ogawa K., Okano T., Niikawa N., Fukushima Y. A stable acentric marker chromosome: Possible existence of an intercalary ancient centromere at distal 8p. Am. J. Hum. Genet. 1994;55:1202–1208. [PMC free article] [PubMed] [Google Scholar]
- 131.Neumann T., Exeler R., Wittwer B., Muller-Navia J., Schrors E., Kennerknecht I., Horst J. A small supernumerary acentric marker chromosome 8 in a 23 year old slightly dysmorphic patient without mental retardation. Cytogenet. Cell Genet. 1999;85:158. [Google Scholar]
- 132.Velinov M., Gu H., Genovese M., Duncan C., Warburton P., Brooks S.S., Jenkins E.C. Characterization of an analphoid, neocentromere-positive inv dup 8p marker chromosome using multiplex whole chromosome and sub-telomere FISH analyses. Ann. Genet. 2004;47:199–205. doi: 10.1016/j.anngen.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 133.Sulcova V., Reddy K.S., Schwartz S., Noble J., Lin H. An analphoid marker chromosome shown to be an inverted duplication 8q23qter with a neocentromere. Am. J. Hum. Genet. 1999;65(Suppl.):A2026. [Google Scholar]
- 134.Reddy K.S., Sulcova V., Schwartz S., Noble J.E., Phillips J., Brasel J.A., Huff K., Lin H.J. Mosaic tetrasomy 8q: inverted duplication of 8q23.3qter in an analphoid marker. Am. J. Med. Genet. 2000;92:69–76. [PubMed] [Google Scholar]
- 135.Satinover D.L., Vance G.H., Dyke D.L.V., Schwartz S. Cytogenetic analysis and construction of a BAC contig across a common neocentromeric region from 9p. Chromosoma. 2001;110:275–283. doi: 10.1007/s004120100143. [DOI] [PubMed] [Google Scholar]
- 136.Vance G.H., Curtis C.A., Heerema N.A., Schwartz S., Palmer C.G. An apparently acentric marker chromosome originating from 9p with a functional centromere without detectable alpha and beta satellite sequences. Am. J. Med. Genet. 1997;71:436–442. doi: 10.1002/(sici)1096-8628(19970905)71:4<436::aid-ajmg13>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 137.Levy B., Papenhausen P., Tepperberg J., Dunn T., Fallet S., Magid M., Kardon N., Hirschhorn K., Warburton P. Prenatal molecular cytogenetic diagnosis of partial tetrasomy 10p due to neocentromere formation in an inversion duplication analphoid marker chromosome. Cytogenet. Cell Genet. 2000;91:165–170. doi: 10.1159/000056839. [DOI] [PubMed] [Google Scholar]
- 138.Dufke A., Walczak C., Liehr T., Starke H., Trifonov V., Rubtsov N., Schöning M., Enders H., Eggermann T. Partial tetrasomy 12pter-12p12.3 in a girl with Pallister-Killian syndrome: Extraordinary finding of an analphoid, inverted duplicated marker. Eur. J. Hum. Genet. 2001;9:572–576. doi: 10.1038/sj.ejhg.5200673. [DOI] [PubMed] [Google Scholar]
- 139.Vermeesch J.R., Melotte C., Salden I., Riegel M., Trifnov V., Polityko A., Rumyantseva N., Naumchik I., Starke H., Matthijs G. Tetrasomy 12pter-12p13.31 in a girl with partial Pallister-Killian syndrome phenotype. Eur. J. Med. Genet. 2005;48:319–327. doi: 10.1016/j.ejmg.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 140.Rivera H., Vasquez A.I., García-Cruz D., Crolla J.A. Neocentromere at 13q32 in one of two stable markers derived from a 13q21 break. Am. J. Med. Genet. 1999;85:385–388. doi: 10.1002/(sici)1096-8628(19990806)85:4<385::aid-ajmg15>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 141.Magnani I., Sacchi N., Darfler M., Nisson P.E., Tornaghi R., Fuhrman-Conti A.M. Identification of the chromosome 14 origin of a C-negative marker associated with a 14q32 deletion by chromosome painting. Clin. Genet. 1993;43:180–185. doi: 10.1111/j.1399-0004.1993.tb04444.x. [DOI] [PubMed] [Google Scholar]
- 142.Sacchi N., Magnani I., Fuhrman-Conti A.M., Monard S.P., Darfler M. A stable marker chromosome with a cryptic centromere: evidence for centromeric sequences associated with an inverted duplication. Cytogenet. Cell Genet. 1996;73:123–129. doi: 10.1159/000134322. [DOI] [PubMed] [Google Scholar]
- 143.Li M.M., Howard-Peebles P.N., Killos L.D., Fallon L., Listgarten E., Stanley W.S. Characterization and clinical implications of marker chromosomes identified at prenatal diagnosis. Prenat. Diagn. 2000;20:138–143. [PubMed] [Google Scholar]
- 144.Blennow E., Telenius H., de Vos D., Larsson C., Henriksson P., Johansson O., Carter N.P., Nordenskjöld M. Tetrasomy 15q: Two marker chromosomes with no detectable alpha-satellite DNA. Am. J. Hum. Genet. 1994;54:877–883. [PMC free article] [PubMed] [Google Scholar]
- 145.Tonnies H., Pietrzak J., Bocian E., Macdermont K., Kuechler A., Belitz B., Trautmann U., Schmidt A., Schulze B., Rodriguez L. New immortalized cell lines of patients with small supernumerary marker chromosome (sSMC): Towards the establishment of a cell bank. J. Histochem. Cytochem. 2007;55:651–660. doi: 10.1369/jhc.6A7161.2007. [DOI] [PubMed] [Google Scholar]
- 146.Huang B., Ning Y., Lamb A.N., Sandlin C.J., Jamehdor M., Ried T., Bartley J. Identification of an unusual marker chromosome by spectral karyotyping. Am. J. Med. Genet. 1998;80:368–372. [PubMed] [Google Scholar]
- 147.Van den Enden A., Verschraegen-Spae M.R., Van Roy N., Decaluwe W., De Praeter C., Speleman F. Mosaic tetrasomy 15q25–>qter in a newborn infant with multiple anomalies. Am. J. Med. Genet. 1996;63:482–485. doi: 10.1002/(SICI)1096-8628(19960614)63:3<482::AID-AJMG13>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 148.Huang X.L., de Michelena M.I., Mark H.F.L., Harston R., Benke P.J., Price S.J., Milunsky A. Characterization of an analphoid supernumerary marker chromosome derived from 15q25–>qter using high-resolution CGH and multiplex FISH analyses. Clin. Genet. 2005;68:513–519. doi: 10.1111/j.1399-0004.2005.00523.x. [DOI] [PubMed] [Google Scholar]
- 149.Hu J., McPherson E., Surti U., Hasegawa S.L., Gunawardena S., Gollin S.M. Tetrasomy 15q25.3 –> qter resulting from an analphoid supernumerary marker chromosome in a patient with multiple anomalies and bilateral Wilms tumors. Am. J. Med. Genet. 2002;113:82–88. doi: 10.1002/ajmg.10708. [DOI] [PubMed] [Google Scholar]
- 150.Chen C.P., Lin C.C., Li Y.C., Chern S.R., Lee C.C., Chen W.L., Lee M.S., Wang W., Tzen C.Y. Clinical, cytogenetic, and molecular analyses of prenatally diagnosed mosaic tetrasomy for distal chromosome 15q and review of the literature. Prenat. Diagn. 2004;24:767–773. doi: 10.1002/pd.977. [DOI] [PubMed] [Google Scholar]
- 151.Rowe A.G., Abrams L., Qu Y., Chen E., Cotter P.D. Tetrasomy 15q25–>qter: Cytogenetic and molecular characterization of an analphoid supernumerary marker chromosome. Am. J. Med. Genet. 2000;93:393–398. [PubMed] [Google Scholar]
- 152.Mahjoubi F., Peters G.B., Malafiej P., Shalhoub C., Turner A., Daniel A., Hill R.J. An analphoid marker chromosome inv dup(15)(q26.1qter), detected during prenatal diagnosis and characterized via chromosome microdissection. Cytogenet. Genome Res. 2005;109:485–490. doi: 10.1159/000084207. [DOI] [PubMed] [Google Scholar]
- 153.Tabet A.C., Gosset P., Elghezal H., Fontaine S., Martinovic J., Razavi F.E., Romana S., Vekemans M., Morichon-Delvallez N. Prenatal diagnosis and characterization of an analphoid marker chromosome 16. Prenat. Diagn. 2004;24:733–736. doi: 10.1002/pd.804. [DOI] [PubMed] [Google Scholar]
- 154.Ravnan J., Ouellette K., Fabre A., Crenshaw D., Guillory S., Siewert R., McCoy S., Kothari J. A stable acentric marker chromosome formed by interstitial deletion of 17q and subsequent inverted duplication of the deleted segment resulting in partial trisomy 17q22 to 17q23 diagnosed in dysmorphic newborn. Am. J. Hum. Genet. 1999;65(Suppl.):A356. [Google Scholar]
- 155.Rauch A., Pfeiffer R.A., Trautmann U., Liehr T., Rott H.D., Ulmer R. A study of ten small supernumerary (marker) chromosomes identified by fluorescence in situ hybridization (FISH) Clin. Genet. 1992;42:84–90. doi: 10.1111/j.1399-0004.1992.tb03145.x. [DOI] [PubMed] [Google Scholar]
- 156.Voullaire L., Saffery R., Davies J., Earle E., Kalitsis P., Slater H., Irvine D.V., Choo K.H. Trisomy 20p resulting from inverted duplication and neocentromere formation. Am. J. Med. Genet. 1999;85:403–408. [PubMed] [Google Scholar]
- 157.Barbi G., Kennerknecht I., Wöhr G., Avramopoulos D., Karadima G., Petersen M.B. Mirror-symmetric duplicated chromosome 21q with minor proximal deletion, and with neocentromere in a child without the classical Down syndrome phenotype. Am. J. Med. Genet. 2000;91:116–122. doi: 10.1002/(sici)1096-8628(20000313)91:2<116::aid-ajmg7>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 158.Kaiser-Rogers K.A., Davenport M.L., Powell C.M., Rao K.W. A recombinant X chromosome with an atypical centromere observed in a child with Turner syndrome. Am. J. Hum. Genet. 1995;57(Suppl.):A658. [Google Scholar]
- 159.Yu S., Barbouth D., Benke P.J., Warburton P.E., Fan Y.S. Characterization of a neocentric supernumerary marker chromosome originating from the Xp distal region by FISH, CENP-C staining, and array CGH. Cytogenet. Genome Res. 2007;116:141–145. doi: 10.1159/000097434. [DOI] [PubMed] [Google Scholar]
- 160.Conde C., Chheng S., Wu J., Santini M., Kashork C., Ware S., Scaglia F., Shaffer L.G. A novel analphoid marker of the Y chromosome. Am J. Hum. Genet. 2001;69(Suppl.):A765. [Google Scholar]
- 161.Floridia G., Gimelli G., Zuffardi O., Earnshaw W.C., Warburton P.E., Tyler-Smith C. A neocentromere in the DAZ region of the human Y chromosome. Chromosoma. 2000;109:318–327. doi: 10.1007/s004120000081. [DOI] [PubMed] [Google Scholar]
- 162.Warburton P.E., Cooke C.A., Bourassa S., Vafa O., Sullivan B.A., Stetten G., Gimelli G., Warburton D., Tyler-Smith C., Sullivan K.F. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- 163.Assumpção J.G., Berkofsky-Fessler W., Campos N.V., Maciel-Guerra A.T., Li S., Melaragno M.I., de Mello M.P., Warburton P.E. Identification of a neocentromere in a rearranged Y chromosome with no detectable DYZ3 centromeric sequence. Am. J. Med. Genet. 2002;113:263–267. doi: 10.1002/ajmg.10701. [DOI] [PubMed] [Google Scholar]


