Abstract
Through extended survey of mitochondrial DNA (mtDNA) diversity in the Nganasan, Yukaghir, Chuvantsi, Chukchi, Siberian Eskimos, and Commander Aleuts, we filled important gaps in previously unidentified internal sequence variation within haplogroups A, C, and D, three of five (A–D and X) canonical mtDNA lineages that defined Pleistocenic extension from the Old to the New World. Overall, 515 mtDNA samples were analyzed via high-resolution SNP analysis and then complete sequencing of the 84 mtDNAs. A comparison of the data thus obtained with published complete sequences has resulted in the most parsimonious phylogenetic structure of mtDNA evolution in Siberia-Beringia. Our data suggest that although the latest inhabitants of Beringia are well genetically reflected in the Chukchi-, Eskimo-Aleut-, and Na-Dene-speaking Indians, the direct ancestors of the Paleosiberian-speaking Yukaghir are primarily drawn from the southern belt of Siberia when environmental conditions changed, permitting recolonization the high arctic since early Postglacial. This study further confirms that (1) Alaska seems to be the ancestral homeland of haplogroup A2 originating in situ approximately 16.0 thousand years ago (kya), (2) an additional founding lineage for Native American D, termed here D10, arose approximately 17.0 kya in what is now the Russian Far East and eventually spread northward along the North Pacific Rim. The maintenance of two refugial sources, in the Altai-Sayan and mid-lower Amur, during the last glacial maximum appears to be at odds with the interpretation of limited founding mtDNA lineages populating the Americas as a single migration.
Introduction
During the Ice Age (43.0–12.0 thousand years ago [kya]), the whole Siberian subcontinent was vastly enlarged in the northeast due to lower levels of the Bering and Chukchi Sea, creating the Beringian refugium in the ice-free part connecting Siberia and Alaska, permitting human migrations into the New World.1,2 Recent discovery of an undoubted association of a set of specific artifacts with extinct rhinoceros and mammoth at the lower Yana River Site, dating to 27.0 kya (radiocarbon years), indicates that the harsh glacial environment didn't prevent modern human occupation of Siberian Arctic before the Last Glacial Maximum (approximately 18.0 kya).3 Other Paleolithic sites in Siberia lie far south of the Arctic Circle, implying that the early populations of extreme northeastern Siberia either became extinct or retreated to the south unless environmental conditions changed and permitted repeopling of the Siberian Arctic.4 The presence of Beringia would have important implications for resolving initial human settling of northwestern edge of the New World.5–7 The last inhabitants of former Beringia, the Yukaghir, Chukchi, Eskimo-Aleuts, and Na-Dene Indians, are likely the survivors of rapid environmental changes that took place in the late Pleistocene to early Holocene.8–10 Hence, these populations deserve special attention focused on the issues, such as the number of New World founders, where in Siberia-Asia and Beringia progenitors of the First Americans arose, when and how they spread into the deglaciated habitats of Alaska and Northwest Coast, and for how long they persisted there before migrating south of what is now the United States-Canadian border.11–13
Studies of maternally inherited mitochondrial DNA (mtDNA) variation on different sides of the Bering Sea, based on phylogeographic and molecular clock principle, have led to a number of important insights into the genetic history of Beringia and peopling of the New World by placing timescales on evolutionary events that would otherwise be difficult to date.14–20 However, most of existing mtDNA data were obtained by examination of a small part of mitochondrial genome, its control region (CR) only. Although very helpful in tracing population affinities, the mtDNA data thus obtained lack resolving power to discern closely related haplotypes within the lineages (A, B, C, D, and X) classified as founding Native American haplogroups. Therefore, they may produce equivocal phylogenies making phylogenetic inferences questionable.21 In the meantime, the human colonization process of the New World, as well as potential population sources in Siberia-Asia and the route they took in Northeast Asia and Americas, remains the focus of considerable genetic research.22,23 Most recently, Tamm et al.24 and Derenko et al.25 have contributed significantly to the evolutionary history of the Eurasian and Native American mtDNA haplogroups, though with little work on populations indigenous to Arctic Siberia or Beringia itself.
In the present study, we employed the detailed population, molecular, and phylogeographic resolution of haplogroup mtDNA lineages dominating in extreme northeastern Eurasia. Accordingly, we considerably expanded our previous survey of mtDNA diversity in the Nganasan, Yukaghir, Chuvantsi, Chukchi, Siberian Eskimos, and Commander Aleuts via high-resolution SNP analysis and then complete sequencing. Newly obtained sequences were integrated with those previously published, and intricate phylogenies were constructed to obtain the sequence-divergence estimates for trans-Beringian branches of haplogroups A, C, and D and define their pre- or post-Last Glacial Maximum (LGM) population dispersions. As a result, more detailed description of the evolutionary history of our species in this part of the globe since the time when Alaska was the end of Siberia has become possible.
Material and Methods
Populations and Samples
Blood samples were collected from well-defined territorial groups and/or population subdivisions of aboriginal Arctic Siberians with appropriate informed consent during multiple expeditions conducted by R.I.S., E.B.S., and N.V.V. in 2001–2007. The individuals who participated in this study were interviewed and had their family histories verified by senior members of the community for accuracy of the compiled genealogies prior to blood being drawn only from unrelated subjects who lacked nonnative maternal ancestors. The sample areas are shown in Figure 1, and a brief description of each population follows.
Figure 1.
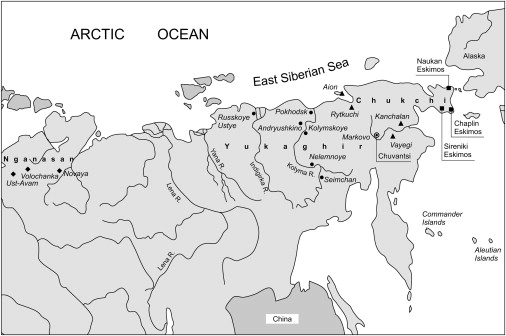
Map of Siberia and Adjacent Part of North Pacific, Showing mtDNA Sampling Locations
Black squares represent locations of the Nganasan, dots the Yukaghir, triangles the Chukchi, squares the Eskimos, and the white circle the Chuvantsi.
Nganasan
According to archeological records, since approximately 9.0 kya, the huge territory of boreal forest that extended from the lower Yenisei River in the west to the Anadyr River basin in the east has been continuously occupied by sparse reindeer hunting groups.26 When the first Russians appeared in the Kheta-Khatanga-Anabara River region in 1618, they encountered the Yukaghir-speaking Tavghi of over 600 individuals, who sustained on reindeer hunting. By that time, the Tavghi represented all that remained of the westernmost Yukaghir tribes. Having been decimated by small pox in 1630–1631, the Tavghi retreated to the boreal forest-tundra refuge of the Taimyr peninsula, where they subsequently fused with odd and scanty bands of the Samoyed-speaking Entsi and Tungusic-speaking Evenki. Shortly, two interrelated tribes called the Avam and Vadei Samoyed, or the Nganasan, have been established.27,28 In traditional times, the Nganasan numbered 700–800 individuals who spoke their own language belonging to the Uralic linguistic family.29 Because the Nganasan lived in isolation, in remote parts of the Taimyr peninsula, they retained the main features of traditional kinship and population structure, as well as the genetic heritage, until 1960s.30–32 Recent decades saw dramatic changes in their way of life; the Nganasan were almost totally dissolved in Turkic-speaking Dolgan, themselves derived from the Yakuts.
The current study includes 39 Nganasan, 22 individuals of whom come from the villages of Ust-Avam and Volochanka,33 whereas 17 are new samples drawn from the members of the Vadei tribe currently residing in the factory of Novaya on the lower Kheta River.
Yukaghir
When the vanguard of Russian Cossaks, the tribute collectors, appeared in the Yana (1635–1638), Indigirka and Alaseya (1642), and Kolyma and Anadyr (1643–1647) River regions, they discovered the Yukagir, typical elk-and-reindeer-hunting bands with stone and bone weapons who used canoes and rafts for traveling in summer and snowshoes and dog sleds in winter.34,35 The Yukaghir territory has the severest climate of all Siberia. In it was found the coldest spots on the earth's surface, which are located close to the Verkhoyansk range. The Stanovoi and Verkhoynsk Ranges—where Yana, Indigirka, and Kolyma, the main Yukaghir Rivers, arise—formed an apparent barrier isolating the Yukaghir from southern herding groups, the Evenki or Even and Yakut.28,35–37 The modern mountain ranges were not an overwhelming barrier to these Altaic-speakers who have predominated in the region after the Yukaghir population dwindled since the period of Russian contact.
Prior to contact, the Yukaghir population size was estimated to be approximately 4800, and the total population was subdivided into at least 12 tribes scattered across their vast geographic range.28 At that time, the Yukaghir would meet criteria characteristics for primary genetic isolates classified by James V. Neel38 as “a tribal population of presumably very ancient origin which, since it emerged as a distinct entity, has had relatively little biological exchange with other similar groups.” With the passage of time, small-pox epidemics augmented by the changes in wild reindeer ecology led to the extinction of some of the Yukagirs tribes, the decimation of others, and, hence, to the reduction of their traditional territory. Their tribal integrity and kinship structure had deteriorated, and the interbreeding remnants of a few tribes were living in the midst of alien groups in their ancient territory.
In 1897, when the first All-Russian Census was undertaken, the Yukaghir numbered 544 individuals,39 with approximately 130 inhabiting the upper Kolyma region and 410 residing in the lower-Kolyma-Alaseia region.35,40 The upper-Kolyma Yukaghir spoke a forest dialect of the Yukaghir language and sustained on elk hunting and fishing, whereas the lower-Kolyma Yukaghir retained the tundra dialect of the Yukaghir language, and their economy was based largely on seasonal reindeer hunting supplemented by fishing. The relationship of the Yukaghir language with those of the surrounding populations is unclear. According to Jochelson,35 the Yukaghir language is a Paleosiberian language isolate, though Kreinovitch41 assumed that the Yukaghir language was related with the easternmost branch of the Uralic language family.
The present-day Yukaghir have been almost totally assimilated by the Even, Chukchi, Yakut, or Russians. However, quite a few families can trace their Yukaghir ancestry through the maternal side42 (this study). Accordingly, we collected 18 blood samples from the upper-Kolyma Yukaghir in the villages of Nelemnoye and Seimchan, whereas 82 samples come from the lower-Kolyma and Indigirka region in the villages of Kolymskoye, Cherski, Andryushkino, Pokhodsk, and Russkoye Ustye. The majority of the samples were collected from individuals who reported their Yukaghir maternal ancestry, but quite a few blood donors were uncertain about their Yukaghir or Even ancestral continuity. Efforts were made to avoid taking blood from individuals who had the Yakut on their maternal side.
Of 82 tundra Yukaghir samples collected, 36 were drawn from the Old Russian Settlers in the villages of Pokhodsk (n = 20) and Russkoye Ustye (n = 16) located in the Kolyma and Indigirka delta, respectively. Historical documents and their family histories indicate that many could be descendants of the first Cossaks who settled in the lower Indigirka-Alaseya-Kolyma River region in 1640–1650. Mating with native women taken from local Yukaghir tribes marked the formation of an isolated group of fishermen, hunters, and dog breeders of several hundred persons.34,43,44
Chuvantsi
Historically, the Chuvantsi represented the Yukaghir tribe to the east of the Kolyma watershed.28,35,45 In the upper-Anadyr region, the census records of 1897 noted 81 Yukaghir and 262 Chuvantsi. Of these, 43 Yukaghir and 147 Chuvantsi lived alongside with Russians (Cossacks, local traders, and merchants) in a few small settlements. Accordingly, we collected blood samples from 32 elder individuals in the village of Markovo (Chukotkan Autonomous Region) whose pedigrees suggested the Chuvantsi-Yukaghir continuity in their maternal ancestry.
Chukchi
Of 182 Chukchi samples included in this study, 66 mtDNAs are from easternmost Chukotka16 that were revised and extended through the course of this study. In addition, 54 new samples were recently collected, of which 40 were drawn from the Chukchi in the villages of Aion, Yanranai, and Rytkuchi located at the northernmost edge of Chukotka, whereas 14 samples come from the Chukchi currently residing in Cherski and Kolymskoye (Nizhnekolymskiy district, Yakut Republic). The remaining 62 samples represent two Chukchi subdivisions—Vayegi and Kanchalan—located in the middle-lower Anadyr River region. These were randomly chosen from a much larger collection of old samples46 not investigated for mtDNA variation previously.
Siberian Eskimos
The present report is based on 126 mtDNA Eskimo samples, 39 of which represent Naukan tribe, whereas 37 and 50 come from Sireniki and Chaplin populations, respectively. Previously published data with 79 Eskimo mtDNAs samples16 are revised and extended, as well as supplemented by previously collected but nonsurveyed samples. Of the 39 Naukan Eskimos included in the present study, 33 are new samples collected in the village of Lavrentiya (Chukotskiy district, Chukotkan Autonomous Region) in May 2002. In historical time, the Naukan Eskimos inhabited the mountain terrace on the northeastern coast of the Chukchi peninsula. They spoke an isolated dialect of Siberian Yupik and occasionally intermarried with the nearby Eskimo tribe (Imaklik) of Little Diomide.47
Commander Aleut
Of 36 Commander Aleut individuals surveyed or revised through this study, mtDNA data from 30 individuals were collected in the village of Nikolskoye on the Bering Island.48 Six additional samples were drawn in 2007 from the Aleuts who were born in the village of Preobrazhenskoye on Copper Island (closed in the 1960s) and currently residing in Petropavlovsk-Kamchatski.
mtDNA Analysis
Genomic DNAs were extracted from buffy coats with standard procedures. The first step consisted of mtDNA variation surveyed by digestion with a battery of restriction endonucleases, sequencing hypervariable segment I (HVS-I) of the control region and diagnostic SNPs in the coding region. Subhaplogroup structure was extended through complete sequencing of the selected mtDNAs exhibiting both the identical and distinctive CR sequence motifs. In this way, we have selected the samples for complete sequencing so as to have them represent the widest possible range of intrinsic diversity of haplogroup A, C, and D in Siberia-Beringia. The procedure of complete sequencing entailed polymerase chain reaction (PCR) amplification of eight overlapping mtDNA fragments that were sequenced in both forward and reversed directions by use of BigDye terminator chemistry (PE Applied Biosystems) and an ABI Prism 3100 DNA Analyzer. Trace files were analyzed with the Sequencher (v.4.0.5 GeneCode Corporation) software. Mutations were scored relative to the revised Cambridge Reference Sequence.49 We used facilities of the Sequencing Center at the Institute of Molecular Biology, Russian Academy of Sciences (RAS), Moscow, Cytology and Genetics Institute, Siberian Branch, Russian Academy of Sciences (SBRAS), Novosibirsk, and those of the Center for Molecular and Mitochondrial Medicine and Genetics, University of California, Irvine, CA.
Phylogenetic and Statistical Analyses
Network Analysis
mtDNA complete-sequence data, revealing shared lineages or sublineages within and among Siberian-Beringian populations, were assembled into phylogenetic trees. The focus was primarily placed on the intrinsic diversity of A, C, and D haplogroups because of their prevalence across northern perimeter of former Beringia. The mtDNA phylogenetic nomenclature initiated by Torroni et al.50 and updated by Forster et al.,14 Saillard et al.,18 Derbeneva et al.,48 Starikovskaya et al.,51 Volodko et al.,52 Helgason et al.,20 is used. The mtDNA data obtained through this study allowed us to reach the level of resolution sufficient to further define, redefine, or revise the haplogroups and, hence, delineate population and regional stratification.
Estimates of Coalescence Time
The coalescence dates were estimated with the ρ statistics.14 Standard error was calculated according to Saillard et al.18 We applied the phylogenetically based rates by using a calibration of one substitution per 5138 years for the coding region53 under unrealistic but necessary assumption that the effect of selection does not unduly bias the phylogeny.
Relationship-Matrix Analysis
To define the genetic relationships between populations, we used the relationship (R) matrix method of Harpending and Jenkins.54
Results
mtDNA Diversity
The mtDNAs of 515 individuals from nine Siberian-Beringian populations were first characterized by high-resolution RFLP/HVS-I screening and surveyed for additional diagnostic SNPs in the coding region (Table 1). The majority of the mtDNA types were found to fall into different sublineages of A, C, and D haplogroups, and only a few remaining mtDNAs were identified as U4, G1, and Z1. For example, west-Eurasian haplogroup U4 harbored by the Nganasan has not been found in the main body of the Yukaghir, making it less probable that U4 was ever present in the Yukaghir-speaking Tavghi, the main ancestral stock of the Nganasan. When Avam Nganasan mtDNA sample attributable to U4 was subjected to complete sequencing, an unreported haplotype emerged, as it has harbored a unique set of mutations in the coding region: 629, 2405+C, 3637A, 5567, 10692, 11326, 11518, and 13105. The identity of this sublineage, which we identified as U4c, is confirmed by sharing of 626 HaeIII+ and 16189 variant also revealed in the Mansi.55 This finding indicates that U4c is a part of the ancestral mtDNA pool of eastern Uralic-speaking populations, and it should be clustered to previously nonencountered sublineage of the haplogroup U4.56 In the other direction, the haplogroup G1 mtDNAs are encountered in the tundra Yukaghir, Chuvantsi, and Chukchi (Table 2). Most likely this is an implication of northward expansion of the bearers of G1 from the Sea of Okhotsk-Kamchatka region where this lineage is prevalent in the Koryak, Itelmen, and Even.17 A similar trend in the gene flow across the linguistic and geographic boundaries is relevant to the Z1 mtDNAs, revealed at low frequencies in the Nganasan and Yukaghir.
Table 1.
mtDNA Diversity in Siberian-Beringian Populations
|
Haplogroup |
RFLP(s) |
HVS-I (−16000) |
Nganasan |
Yukaghir |
Chukchi |
Siberian Eskimos |
Aleut (Commander Islands) |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lower Kolyma-Indigirka | Upper Kolyma | Upper Anadyr (Chuvantsi) | Sireniki | Chaplin | Naukan | ||||||
| A2a | 663e (3330) (−/−) | 111 223 290 319 362 | − | − | − | − | 2 | − | − | 1 | − |
| A2a | 663e (3330) (−/−) | 111 192 223 290 319 362 | − | − | − | 1 | 50 | 14 | 22 | 5 | − |
| A2a | 663e (3330) (−/−) | 111 192 223 290 319 362 519 | − | − | − | − | − | − | − | 1 | − |
| A2a | 663e (3330) −3337k (−/−) | 111 192 223 290 319 362 | − | − | − | − | − | 2 | 13 | − | − |
| A2a | 663e (3330) (−/−) | 111 192 223 290 311 319 362 | − | − | − | 4 | 3 | − | − | 6 | − |
| A2a | 663e (3330) (−/−) | 111 192 223 290 311 319 | − | − | − | − | 2 | − | − | − | − |
| A2a | 663e (3330) (−/−) | 111 192 223 261 290 319 362 | − | − | − | − | 6 | − | − | − | − |
| A2a | 663e (3330) (−/−) | 111 176 192 223 290 319 362 | − | − | − | 1 | 5 | − | 1 | − | − |
| A2b | 663e (−/−) (11365) | 111 223 290 319 362 | − | − | − | − | 2 | − | − | − | − |
| A2b | 663e (−/−) (11365) | 111 223 265 290 319 362 | − | − | − | 1 | 25 | 8 | 9 | 10 | − |
| A2b | 663e (−/−) (11365) | 111 176 223 265 290 319 362 | − | − | − | 1 | 1 | − | − | 6 | − |
| A2b | 663e (−/−) (11365) | 111 129 223 265 290 319 362 | − | − | − | − | 1 | 2 | − | − | − |
| A2b | 663e (8281d9) (−/−) (11365) | 111 212 223 265 290 319 362 | − | − | − | − | 2 | − | − | − | − |
| A2b | 663e (−/−) (11365) | 111 223 265 290 319 325 362 | − | − | − | − | 1 | − | − | − | − |
| U4a | 4643k (8818) (−/−) 11329a 12308 g | 134 356 519 | 5 | − | − | − | − | − | − | − | − |
| U4c | 626e (2405+C) 4643k (−/−) 11329a 12308 g | 189 356 519 | 3 | − | − | − | − | − | − | − | − |
| C2a | (3816) (+/+) −13259o | 223 298 327 519 | − | 26 | 2 | 6 | − | − | − | − | − |
| C2a | (3816) (+/+) −13259o | 223 291 298 327 519 | − | 2 | 3 | − | − | − | − | − | − |
| C2a | (3816) (+/+) −13259o | 223 291 298 327 399 519 | − | 3 | − | − | − | − | − | − | − |
| C2a | (3816) (+/+) −13259o | 223 291 298 311 327 399 519 | − | − | 4 | − | − | − | − | − | − |
| C2a | (3816) (+/+) −13259o | 189 223 298 327 519 | 1 | − | − | − | − | − | − | − | |
| C2a | (3816) (+/+) −13259o | 093 223 298 327 519 | 6 | − | − | − | − | − | − | − | − |
| C2a | (3816) (+/+) −13259o | 093 223 298 317 327 519 | 1 | − | − | − | − | − | − | − | − |
| C2a | (3816) (+/+) −13259o | 223 259+A 298 311 327 519 | 1 | − | − | − | − | − | − | − | − |
| C2a | (3816) (+/+) −13259o | 124 223 298 318T 327 519 | − | − | − | − | 8 | − | − | − | − |
| C2a | (3816) (+/+) −13259o | 124 223 298 311 318T 327 519 | − | − | − | − | 4 | − | − | − | − |
| C2a | (3816) (+/+) −13259o | 223 298 318T 327 519 | − | − | − | − | 1 | − | − | − | − |
| C2b1 | −1715c (+/+) (12672) −13259o | 129 223 298 327 519 | − | − | 1 | − | − | − | − | − | − |
| C2b1 | −1715c (+/+) (12672) −13259o | 093 129 223 298 327 519 | 3 | 5 | − | 1 | − | − | − | − | − |
| C2b1 | −1715c (+/+) (12672) −13259o | 093 129 223 327 519 | − | − | − | 3 | − | − | − | − | − |
| C2b1 | −1715c (8281d9) (+/+) (12672) −13259o | 093 129 223 298 327 519 | − | 2 | − | − | − | − | − | − | − |
| C2b2 | (+/+) (12672) −13259o | 171 223 298 327 344 357 519 | − | 3 | 2 | − | − | − | − | − | − |
| C2b2 | 3397k (+/+) (12672) −13259o | 145 171 223 298 327 344 357 519 | 1 | − | − | − | − | − | − | − | − |
| C3 | (+/+) −13259o | 223 288 298 519 | − | − | − | − | 1 | − | − | − | − |
| C3 | (+/+) −13259o | 223 261 288 298 519 | − | − | − | − | 2 | − | − | − | − |
| C3 | (+/+) −13259o | 093 189 223 261 288 298 519 | − | 2 | 1 | − | 3 | − | − | − | − |
| C3 | (+/+) −13259o | 093 189 223 261 288 298 309 519 | − | 1 | − | − | 3 | − | − | − | − |
| C3 | (+/+) −13259o | 189 223 261 288 298 390 519 | − | 3 | − | − | 3 | − | − | − | − |
| C3 | (+/+) −13259o | 189 223 261 288 298 299 519 | − | − | − | − | 1 | − | − | − | − |
| C3 | −1413l (+/+) −13259o | 093 223 288 298 327 390 519 | − | 5 | − | − | − | − | − | − | − |
| C3 | −1413l (+/+) −13259o | 093 145 223 288 298 327 390 519 | − | 1 | − | − | − | − | − | − | − |
| C3 | −1715c 3397k (+/+) −13259o | 148 223 288 298 327 519 | 4 | − | − | − | − | − | − | − | − |
| C3 | −1715c 3397k (8289i9) (+/+) −13259o | 148 223 288 298 327 519 | 1 | − | − | − | − | − | − | − | − |
| C3 | 1718e 3397k (+/+) −13259o 15847a | 148 223 288 298 327 519 | 2 | − | − | − | − | − | − | − | − |
| C3 | 1718e (+/+) −13259o 15847a | 148 223 288 298 327 519 | 1 | − | − | − | − | − | − | − | − |
| Z1a | (6752) (10325) (+/+) (15261) | 129 185 223 224 260 298 519 | 1 | 1 | 1 | 2 | − | − | − | − | − |
| D2∗ | −5176a (+/+) (11215) | 129 189 223 362 | − | 1 | − | − | − | − | − | − | − |
| D2a1a | −3315e −5176a (+/+) (11215) | 129 223 271 362 | − | − | − | − | − | − | − | − | 3 |
| D2a1a | −3315e (4991) −5176a (+/+) (11215) | 129 223 271 362 | − | − | − | − | 11 | 2 | − | − | − |
| D2a1a | −3315e (4991) −5176a (+/+) (11215) | 111 129 223 271 362 | − | − | − | − | 1 | 4 | 4 | − | − |
| D2a1a | −3315e −5176a (+/+) (11215) | 129 223 271 362 366 519 | − | − | − | − | 1 | − | − | − | |
| D2a1a | −3315e −5176a (+/+) (11215) | 111 129 223 271 362 366 | − | − | − | 1 | 2 | 3 | 1 | − | − |
| D2a1a | −3315e −5176a (+/+) (11215) | 111 129 223 271 362 366 519 | − | − | − | − | − | 1 | − | − | − |
| D2a1a | −3315e −5176a (+/+) (11215) | 111 129 223 271 294 362 366 | − | − | − | − | − | 1 | − | − | − |
| D2a1a | −3315e −5176a (+/+) (11215) | 092 129 271 223 362 | − | − | − | − | − | − | − | − | 2 |
| D2a1a1 | −3315e −5176a (8910A) (+/+) (11215) | 129 223 271 362 | − | − | − | − | − | − | − | − | 23 |
| D2a1a1 | −3315e −5176a (8910A) (+/+) (11215) | 129 223 271 311 362 | − | − | − | − | − | − | − | − | 4 |
| D2a1a1 | −3315e −5176a (8910A) (+/+) (11215) | 129 223 271 362 519 | − | − | − | − | − | − | − | − | 4 |
| D3a1 | −951j −5176a −10180l (+/+) 15437e | 223 319 362 | 7 | 2 | − | − | 2 | − | − | − | − |
| D3a2 | 3397k −5176a −10180l (+/+) 14923c 15437e | 223 319 320 362 | − | 1 | − | − | − | − | − | − | − |
| D3a2a | −5176a −10180l (+/+) 13717a 14923c 15437e | 093 173 223 319 362 | − | − | − | − | 6 | − | − | 9 | − |
| D3a2a | −5176a −10180l (+/+) 13717a 14923c 15437e | 093 150 173 223 319 362 | − | − | − | − | − | − | − | 1 | − |
| D3a2a | −5176a −10180l (+/+) 13717a 14923c 15437e | 093 173 223 234 255 319 362 | − | − | − | − | 1 | − | − | − | − |
| D3a2a | −5176a −10180l (+/+) 13717a 14923c 15437e | 093 172 173 223 255 319 362 | − | − | − | 2 | − | − | − | − | − |
| D4a | −5176a (+/+) 10646k | 093 223 232 290 362 471 | 1 | − | − | − | − | − | − | − | − |
| D5a1 | −5176a (−/−) −12026h (12705) | 092 172 189 223 362 | − | 1 | − | − | − | − | − | − | − |
| D5a1 | −5176a (−/−) −12026h (12705) | 092 172 189 223 266 362 | − | 1 | 3 | − | − | − | − | − | − |
| D6 | −5176a (+/+) (11696) | 223 362 519 | 2 | − | − | − | − | − | − | − | − |
| D6 | −5176a (7445C) (+/+) (11696) | 223 319 362 | − | − | 1 | − | − | − | − | − | − |
| D7 | −5176a (+/+) (10427) | 223 274 362 368 | − | 1 | − | − | − | − | − | − | − |
| D7 | −5176a (+/+) (10427) | 145 223 311 362 368 | − | 1 | − | − | − | − | − | − | − |
| D8 | −1715e −5176a (8762) (+/+) (12651C) | 042 093 214 223 362 | − | 2 | − | − | − | − | − | − | − |
| D9 | 4830n −5176a (+/+) | 223 294 362 | − | 5 | − | − | − | − | − | − | − |
| G1 | 4830n 8198a (+/+) | 017 093 129 189 223 519 | − | − | − | − | 1 | − | − | − | − |
| G1 | 4830n 8198a (+/+) | 017 129 223 311 519 | − | − | − | − | 2 | − | − | − | − |
| G1 | 4830n 8198a (+/+) | 017 129 223 519 | − | 2 | − | 6 | 19 | − | − | − | − |
| G1 | 4830n 8198a (+/+) | 017 051 093 129 223 519 | − | − | − | − | 1 | − | − | − | − |
| G1 | 4830n 8198a (+/+) | 017 093 129 223 519 | − | 10 | − | 1 | 5 | − | − | − | − |
| G1 | 4830n 8198a (+/+) | 017 093 207 223 311 519 | − | − | − | − | 1 | − | − | − | − |
| G1 | 4830n 8198a (+/+) | 017 093 129 172 223 265 519 | − | − | − | 1 | 2 | − | − | − | − |
| G1 | 4830n 8198a (+/+) | 017 093 129 153 223 319 519 | − | − | − | 1 | 1 | − | − | − | − |
| Total: 515 | 39 | 82 | 18 | 32 | 182 | 37 | 50 | 39 | 36 |
RFLP sites are numbered from the first nucleotide of the enzyme recognition sequence. “−”indicates the absence of restriction site. The restriction enzymes are given with the following single-letter codes: a, AluI; c, DdeI; e, HaeIII; g, HinfI; h, HpaI; j, MboI; k, RsaI; l, TaqI; n, HaeII; and o, HincII. The presence or absence of the associated 10394 DdeI/10397 AluI sites is denoted with slash brackets (+/+), (−/−), or (+/−). 8281d9 indicates 9 bp COII/tRNALys deletion; 8289i9 indicates 9 bp COII/tRNALys insertion. Additional mutations in the coding region (SNPs) verified through sequencing are shown in brackets. Mutations are transitions unless the base change is specified explicitly. Founding RFLP/HVS-I haplotypes are shown in boldface. Only those nucleotide positions between 16013 and 16520 that differ from the revised Cambridge Reference Requence49 are shown.
Table 2.
Haplogroup Composition and Frequencies in Siberian-Beringian Populations
| Lineage | Nganasan (39) |
Yukaghir |
Eskimos |
Aleut |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lower Kolyma-Indigirka (82) | Upper Kolyma (18) | Upper Anadyr (Chuvantsi) (32) | Chukchi (182) | Sireniki (37) | Chaplin (50) | Naukan (39) | Canadian∗ (96) | Greenland∗ (385) | Commander Islands (36) | Aleutian Islands∗∗ (163) | ||
| A2a | − | − | − | 18.8 | 33.7 | 43.2 | 72.0 | 33.3 | 16.7 | 45.2 | − | 34.4 |
| A2b | − | − | − | 6.3 | 13.9 | 27.0 | 18.0 | 41.0 | 70.8 | 50.9 | − | − |
| U4a | 12.8 | − | − | − | − | − | − | − | − | − | − | − |
| U4c | 7.7 | − | − | − | − | − | − | − | − | − | − | − |
| C2a | 20.5 | 41.5 | 50.0 | 18.8 | 6.9 | − | − | − | − | − | − | − |
| C2b | 10.3 | 12.2 | 11.1 | 12.5 | − | − | − | − | − | − | − | − |
| C3 | 20.5 | 12.2 | 11.1 | − | 11.9 | − | − | − | − | − | − | − |
| Z1a | 2.6 | 2.4 | 5.6 | 6.3 | − | − | − | − | − | − | − | − |
| D2∗ | − | 1.2 | − | − | − | − | − | − | − | − | − | − |
| D2a1a | − | − | − | 3.1 | 2.0 | 29.7 | 10.0 | − | − | − | 100.0 | 65.6 |
| D3a1 | 17.9 | 2.4 | − | − | 2.0 | − | − | − | − | − | − | − |
| D3a2 | − | 1.2 | − | − | − | − | − | − | − | − | − | − |
| D3a2a | − | − | − | 6.3 | 3.0 | − | − | 25.6 | 12.5 | 4.0 | − | − |
| D4a | 2.6 | − | − | − | − | − | − | − | − | − | − | − |
| D5a1 | − | 2.4 | 16.6 | − | − | − | − | − | − | − | − | − |
| D6 | 5.1 | − | 5.6 | − | − | − | − | − | − | − | − | − |
| D7 | − | 2.4 | − | − | − | − | − | − | − | − | − | − |
| D8 | − | 2.4 | − | − | − | − | − | − | − | − | − | − |
| D9 | − | 4.9 | − | − | − | − | − | − | − | − | − | − |
| G1 | − | 14.6 | − | 28.1 | 26.7 | − | − | − | − | − | − | − |
A, C, and D mtDNAs: Distribution and Network Analysis
Haplogroup A
A total of 14 distinct A2 haplotypes were revealed in the Chukchi and Siberian Eskimos (Table 1). These were subdivided into two phylogenetically verified subclusters, A2a and A2b, depending on the presence or absence of the characteristic coding region/HVS-I variants: 3330–16192 or 11365–16265, respectively (Figure 2). The varying proportions of A2a and A2b types, with the highest frequency of A2b in Canadian Eskimos (70.8%) and the lowest in the Chuvantsi (6.3%), presented in Table 2, reflect the expansion history of the Eskimos as a whole entity and place the Bering Strait at the center of their population dispersal20 (this study). With the exception of the Chuvantsi, all Yukaghir subdivisions lack the haplogroup A2 mtDNAs. Presence of the A2 (A2a+A2b) mtDNAs at a frequency of 25.1% in the Chuvantsi would reflect the present rather than the past situation at the eastern edge of the Yukaghir distribution range. By the middle of the 19th century, Chuvantsi were partly exterminated, as well as partly assimilated by rapidly expanding Chukchi.28,35,39
Figure 2.
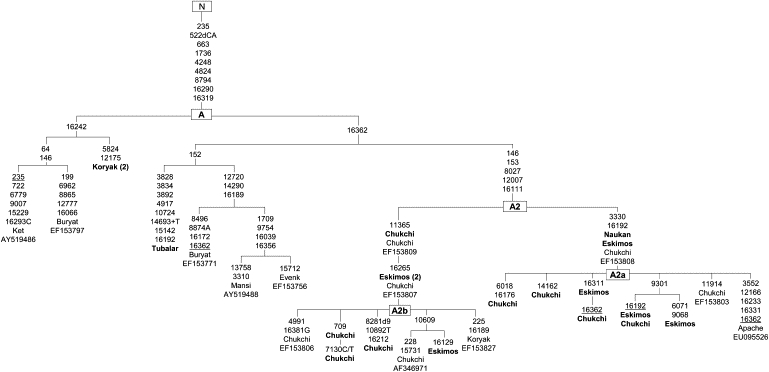
The Phylogenetic Tree of Haplogroup A Complete Sequences Revealed in Siberia-Beringia
Mutation positions, relative to the revised Cambridge Reference Requence,49 are transitions unless the base change is specified. Deletions are indicated by a “d” preceding the deleted nucleotides. Insertions are indicated by a “+” preceding the inserted nucleotide. Reversal mutations are underlined, whereas heteroplasmy is indicated as C/T, A/G, or vice versa. Point mutations at 16182 and 16183 are excluded because of their dependence on the presence of C-T transition at 16189; the length variation in the poly-C stretch at nts 309–315 and point mutation at 16519 was omitted because of their hypervariability. In bold are new sequences generated through the course of this study. When two or more identical samples belong to the same group, their number is given in brackets.
The network analysis embraces 31 haplogroup A sequences. Of these, 18 are new, (eight Chukchi, seven Eskimos, one Tubalar, and two Koryak) and 13 are gleaned from our previously published studies and GenBank (Figure 2). These are one Ket, one Koryak, one Evenk, two Buryat, one Mansi, seven Chukchi, and one Apache. In Beringia, the A2 includes two star-like subclusters (A2a and A2b), but A2a is the only sublineage shared by the Chukchi, Siberian, and North American Eskimos, Aleuts, and Na-Dene populations. Notably, the A2a root type (3330–16192) revealed in one Naukan Eskimos appears to be ancestral not only to the subset of the Chukchi-Eskimo mtDNAs but also Na-Dene (Apache) A2 mtDNA subjected to complete sequencing by Tamm et al.24 On the other hand, the Amerindian A2 mtDNA coding regions available from the GenBank share no mutations with the Chukchi-Eskimo and Na-Dene lineages except for the A2 root.
Haplogroup C
An updated haplogroup C network, including 50 entire sequences, of which 27 are new, is given in Figure 3. An entire tree reveals that haplogroup C splits into four distinct lineages (C1–C4) branching off from the C root. The relationships within two major subsets, the C2 and C3, previously outlined by Starikovskaya et al.,51 are confirmed and extended. Specifically, three distinct lineages sharing the C2 root have been identified: The C2a and C2b are common in Siberia51 (this study), whereas the C2c (originally identified as C4c) is represented by a sole sequence from Ijka-speaking Amerindian tribe from Colombia.24 Within haplogroup C3 phylogeny, a few distinct lineages sharing C3 root in populations of different linguistic affiliation have been identified in Siberia but none in the Americas. Finally, through sequencing rare Okhotsk Evenk mtDNA belonging to haplogroup C, we have identified previously unrecognized subhaplogroup categorized here as C4. It harbored three transitions (5821, 6338, and 7853) shared with two “Asian” haplogroup C sequences from GenBank.
Figure 3.
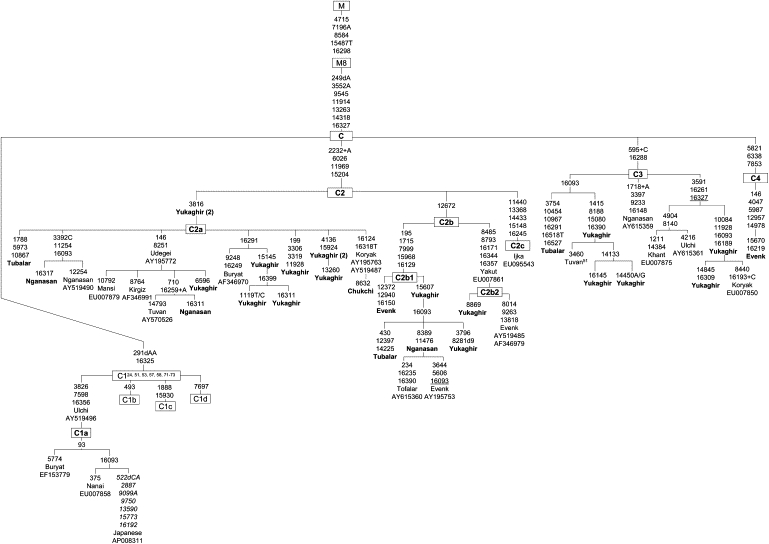
The Phylogeny of Haplogroup C Complete Sequences
For additional information, see the Figure 2 legend. For expediency, we redefine the assignment of C4c used by Tamm et al.,24 now C2c.
Haplogroup D
The phylogeny of 92 haplogroup D complete sequences, of which 39 are new, shows that the wide range of distinct lineages revealed in the Nganasan, Yukaghir, Chukchi, and Eskimo-Aleuts is only a subset of the entire haplogroup D variation subdivided into two distinct clusters (Figures 4 and 5). The main cluster is distinguished by three coding-region mutations (3010, 8414, and 14668), and it encompasses a wide range of independent basal branches (D1–D4 and D6–D11) identifiable by subhaplogroup-specific mutations. The other descendant cluster is D5 rooted in eastern Asia, where it is most frequent and diverse.
Figure 4.
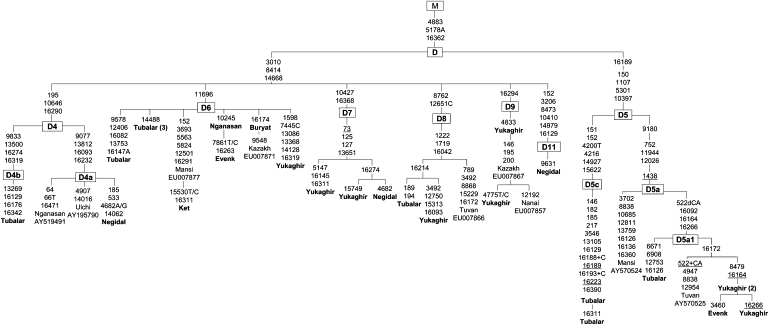
The Phylogeny of Haplogroup D Complete Sequences
The phylogeny of haplogroup D complete sequences found in Arctic Siberians in comparison to their South Siberian counterparts.
Figure 5.
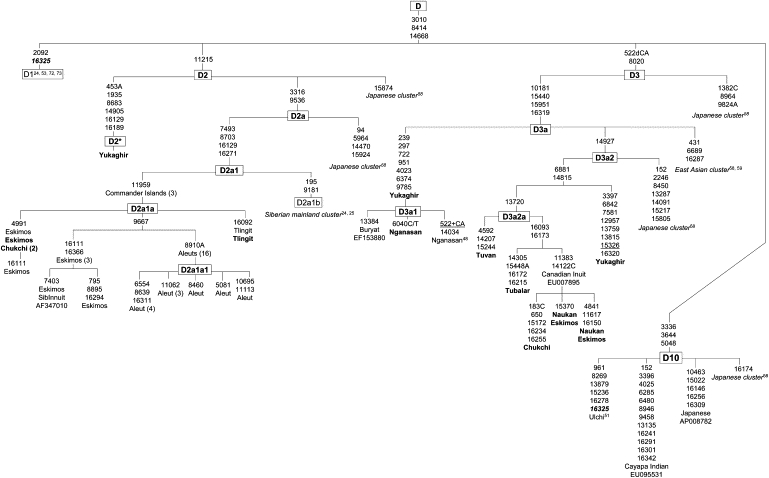
The Phylogeny of Haplogroup D Complete Sequences, Continued
For expediency, we redefine the assignment of D4h3 (now D10) used by Tamm et al.24 and Derenko et al.25
A few of the Yukaghir-Nganasan mtDNAs cluster to the lineage defined here as D6. This lineage is conspicuous for its nodal mutation at 11696 and embraces different haplotypes from Siberia, Japan, and northern China48,51,57–59 (this study). We also sequenced one rare haplogroup D mtDNA exhibited by the Negidal and compared it with a subset of similar Japanese mtDNAs.58 The comparison showed that they all share the coding region variants 3206, 8473, 10410, and 14979, suggesting that this lineage, which we categorized as D11, originated in the Lower Amur-Sea of Okhotsk region, and it most likely represents the traces of an additional source of the genes for peopling of the Japan archipelago. Lineage D11, as well as the D4–D9 mtDNAs, has not been observed in contemporary Beringia or the Americas and appear to have contributed solely to recolonization of continental Siberia.
The genealogy of the haplogroup D2 shows that the founding sequence marked by the coding region transition at 11215 gives rise to two star-like branches and previously unreported D2∗ sequence variant (Figure 5). Most prominent is the node that gave rise to D2a containing two mutations, 3316 and 9536. Next is the D2a1 cluster distinguished by the presence of 7493-8703-16129-16271. It splits into two distinct lineages, the D2a1a defined by the transition at 11959 and D2a1b distinguished by the motif 195–9181, with the former being restricted to Beringian populations48,51 (this study), whereas the latter occurred in the ethnically heterogenous Altaic-speakers of Siberian interior, though in very low frequencies.24,25
Of 36 Commander mtDNA samples, three non-Aleut, probably of the Tlingit origin, harbored the D2a1a root type (sequence VIII in Derbeneva et al.48), thus delineating a founding haplotype for all Central Beringian haplogroup D2 mtDNAs. It is apparent that the Aleut 8910A lineage is only a small portion of larger radiation of D2a1a, which gave rise to the Na-Dene, Aleut, and Chukchi-Eskimo lineages.
New haplogroup D3 sequences from Siberia (one Tubalar, one Tuvan, one Nganasan, two Yukaghir, one Chukchi, and two Naukan Eskimos) have refined the ancestral D3 type distinguished by the coding region transition at 8020. One of the two subsets of D3, designated here as D3a, is distinguished by motif 10181-15440-15951-16319, categorized previously as D3.51 It encompasses all Siberian D3 sequences and a large fraction of their Chinese and Japanese counterparts screened by Kong et al.59 and Tanaka et al.58 One of the sublineages represented by four sequences (one Chukchi, two Naukan Eskimos, and one Canadian Inuit), distinguished by the coding region transition at 11383 and transvertion at 14122C within the D3a2a phylogeny, appears to be the distinctive feature of the North American Eskimos but it is absent from the Sireniki and Chaplin Eskimo tribes. Its presence in the Naukan Eskimos and Chukchi is not surprising when the traditional gene flow across the Bering Strait is taken into account; in our Naukan sample, almost every fourth individual was born or derived from Little Diomede Island, located in the narrowest portion of the Bering Strait.
Time Estimates
The coalescence time and variance computed from the root of the A2, C1, C2, C3, D2, and D3 lineages and of their younger nodes are given in Table 3. We focused on these lineages because they are close or restricted to the Bering Strait and because the remainder members of the A, C, and D haplogroups are absent or rare, suggesting a significant discontinuity in the northeast Asian gene pool. When the Beringian and American A2 mtDNAs are grouped together, the coalescence of A2 mtDNAs, based on 44 coding region sequences, dates to 16.2 ± 2.1 kya. The coalescence time estimate of the C2a subcluster is 7.9 ± 2.2 kya, whereaes the ages of the entire C2 and C3 clusters are significantly older (18.9 ± 4.6 and 18.0 ± 4.6 kya, respectively).
Table 3.
Age Estimates
| Lineage | Number of Coding Regionsa | ρ ± δb | T ± ΔT (kya)c |
|---|---|---|---|
| A2 | 44 | 3.10 ± 0.41 | 16.2 ± 2.1 |
| A2a | 11 | 0.91 ± 0.36 | 4.7 ± 1.9 |
| A2b | 12 | 0.50 ± 0.26 | 2.6 ± 1.4 |
| C1 | 24 | 4.67 ± 0.75 | 24.0 ± 3.8 |
| C2 | 34 | 3.68 ± 0.90 | 18.9 ± 4.6 |
| C2a | 22 | 1.55 ± 0.42 | 7.9 ± 2.2 |
| C2b | 11 | 4.91 ± 1.41 | 25.2 ± 7.3 |
| C3 | 12 | 3.50 ± 0.90 | 18.0 ± 4.6 |
| D1 | 20 | 3.35 ± 0.55 | 17.2 ± 2.8 |
| D2 | 80 | 5.06 ± 1.50 | 26.0 ± 7.7 |
| D2a | 56 | 4.27 ± 1.57 | 21.9 ± 8.1 |
| D2a1 | 48 | 2.33 ± 1.13 | 12.0 ± 5.8 |
| D2a1a | 42 | 1.33 ± 0.79 | 6.9 ± 4.1 |
| D2a1b | 6 | 1.33 ± 0.88 | 6.9 ± 4.5 |
| D3a | 30 | 6.00 ± 1.17 | 30.8 ± 6.0 |
| D3a2 | 12 | 6.25 ± 1.52 | 32.1 ± 7.8 |
| D3a2a | 6 | 2.17 ± 0.83 | 11.1 ± 4.3 |
| D10 | 9 | 3.33 ± 0.77 | 17.1 ± 4.0 |
The sequences from this study plus the coding-region sequences from the studies by Ingman et al.,70 Maca-Meyer et al.,71 Derbeneva et al.,48 Herrnstadt et al.,72 Kong et al.,59 Mishmar et al.,53 Tanaka et al.,58 Starikovskaya et al.,51 Kivisild et al.,73 Tamm et al.,24 Derenko et al.,25 and Ingman and Gyllensten.57
Coalescence time was calculated by consideration of one base substitution between nucleotides 577–16023 equal to 5138 years.53
The ages for the D2a (21.9 ± 8.1 kya) and D2a1 (12.0 ± 5.8 kya) falls within the range of late Pleistocene to early Holocene, but the split between the Chukchi-Eskimo-Aleut-Na-Dene D2a1a and Evenki-Buryat-Yakut-Mongolian D2a1b seems to occurr later, at 6.9 ± 4.1 kya. Although the coalescence time for the Chukotkan-Alaskan D2a1a is based on the average divergence of 42 sequences encompassing two Chukchi, seven Sireniki Eskimos, two Chaplin Eskimos, 26 Aleut, and five non-Aleut from the Commanders, its antiquity should be considered as rough estimate because the D2a1a root gave rise to a few subbranches with quite a few descendants that used to generate high standard error.18,60
The estimated age of the D3a cluster is 30.8 ± 6.0 kya, in consistency with its pattern of geographic distribution. In turn, the age of the D3a2a subcluster, calculated on the basis of only six entire sequences (one Tuvan, one Tubular, one Chukchi, two Naukan Eskimos, and one Canadian Inuit), is 11.1 ± 4.3 kya, suggesting a separate Upper Paleolithic dispersal initiated northward from the Altai-Sayan region.
R Matrix Analysis
Figure 6 plots the populations included in the R matrix analysis on the basis of the subhaplogroup frequencies given in Table 3. The resemblances and distinctions of samples representing 12 Circumpolar groups are well reflected on the map, notwithstanding the first two eigenvectors extract only 63% of the total variation. The first eigenvector clearly separates Yukaghir subdivisions, the Nganasan included, from the Eskimo-Aleut language group. Not surprisingly, the Chuvantsi, who possess a mixture of Chukotkan and Yukaghir mtDNA lineages, are placed closer to the Chukchi rather than Yukaghir. The second axis differentiates the Commanders from Aleutian Islands. The Aleuts from Commanders are virtually missing haplogroup A2a mtDNAs. This is not a surprise, when the founder and bottleneck events in their history that eventually led to the impoverishment of the Commander Aleuts gene pool are taken into account48 (this study). This conclusion is also supported by parallel study of autosomal HLA class II genes variation.61
Figure 6.
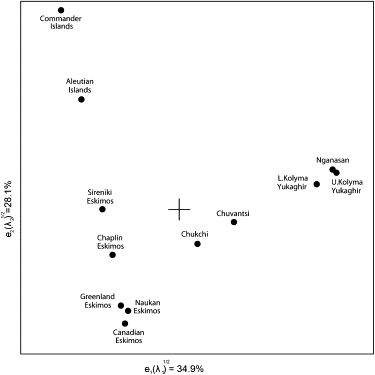
Phylogenetic Relationships between 12 Populations, Determined with the Frequencies of the Observed Subhaplogroups.
Haplogroup composition and frequencies in Canadian and Greenland Eskimos and in Aleutian Islands Aleut mtDNAs are adopted from Helgason et al.20 and Rubicz et al.,19 respectively.
Of three Siberian Eskimo tribes (Chaplin, Sireniki, and Naukan), only Naukan Eskimos cluster to two, geographically separated populations, the Inupik-speaking Canadian and Greenland Eskimos, primarily because they share subhaplogroup D3a2a mtDNAs. With respect to the Aleuts, they differed from the Chaplin and Sireniki Eskimos in that they have haplogroup D2 mtDNAs at a higher frequency.
Discussion
Origins of Beringian-Specific Lineages
One of the significant findings of this study is the genetic discontinuity between the Paleosiberian-speaking Yukaghir and adjoining Chukchi, who are linguistically related to the Koryaks.62 Unlike the Chukchi and Siberian Eskimos of Chukotka, who harbored A2a, A2b, and D2, the core of genetic makeup of the Yukaghir consisted of a unique combination of the C and D sublineages that apparently were continental Siberian. This implies that the A2a, A2b, and D2a1a lineages differentiate the Chukotkan-Alaskan area not only from the rest of the Americas, but also from the rest of Siberia-Asia.
The difference in the age of A2 (16.2 ± 2.1 kya), in comparison with A2a (4.7 ± 1.9 kya) and A2b (2.6 ± 1.4), suggests that during thousands of years the bearers of haplogroup A2 root evolved in isolation, which would have occurred in the deglaciated enclaves of the southern Alaska and Northwest Coast. Hence, Alaska appears to be a good candidate for the ancestral homeland of the lineage A2, its in situ differentiation and subsequent expansion. The D2 cluster dating to 26.0 ± 7.7 kya is essentially older than that of A2. The coalescence dates and spatial distribution of the D2 subhaplogroups across Siberia-Beringia25,48,51 (this study) suggest that the haplogroup D2 root emerged in the mid-Amur region prior to or during LGM, but only much later were its particular derivatives involved in the southern Beringian coastal expansion. Thus, the geographic specificity of the lineages confined to Chukotka and Alaska (A2a and D2a1a) is the main argument in favor of the refugial hypothesis, which assumes the origin of the founding population of Eskimo-Aleut and Na-Dene Indians in southern Alaska at the terminal Pleistocene to early Holocene.10
Haplogroup D3 is also diverse; several subsets of this haplogroup that share a common root, but diverged early with very different subsequent evolutionary histories, have been found. A wide geographic distribution of the D3a lineage and its time depth (30.8 ± 6.0 kya) may be attributed to those early Siberians who underwent pronounced differentiation in the Altai-Sayan region, followed by far-reaching dispersals and subsequent isolation between ancestral and descendant groups. This conjecture is also supported by the overlap of the Japanese, Yukaghir, Tubalar, Tuvan, Chukchi, Naukan, and Canadian Eskimo (Inuit) mtDNA nodal sequences within the D3a2 phylogeny. An age of the 11383–14122C subcluster within D3a2a that contributed to formation of the Chukchi-Eskimos is 6.4 ± 2.9 kya, and it is compatible with archeological records placing the onset of colonization of the North American Arctic in early-mid Holocene.63
Origin of the Yukaghirs and Colonization of Arctic Siberia
Unlike adjacent Chukotkan populations, the genetic prehistory of the Yukaghir remains almost unknown. The early analysis of mtDNA variation in the 27 Yukaghir mtDNA samples64 showed that they largely exhibited two haplogroups, C and D, recognizable at partial RFLP analysis, which was insufficient to reliably quantify the variation that had accumulated within each of the Yukaghir C and D lineages. Likewise, the mtDNA data of Pakendorf et al.,65 obtained through screening the control region of 31 Yukaghir mtDNA samples, has contributed little to the Yukaghir origin and their affinities with surrounding populations.
Molecular dissection of the haplogroups C and D into clusters conducted in the course of this study reveals a wide range of distinct subhaplogroups. Some of them are rare being represented by single previously unreported sequences. Together, the Yukaghir, Nganasan, and other rare sequences belonging to A, C, and D haplogroups observed in Siberian populations may be helpful in discrimination between the primary and secondary colonization events. For example, the ancestral C2a sequence was revealed in two individuals from the Lower Kolyma. This finding, along with the high frequency (43.0%) of the C2a lineage in the Yukaghir, would define original Yukaghir territory as the geographic origin of the C2a range expansion dating to early-middle Holocene. We have also shown that although the majority of mtDNA diversity in the remnants of the Yukaghir tribes, the Nganasan included, are accounted for by the lineages C2a, C2b, C3, and D4–D9, around 6% of their samples harbored the D3a, the derivatives of which have ultimately reached Northern North America.20,66 Such a genetic structure could represent a recent amalgamation of northern and southern populations. Alternatively, and more likely, the core of the Yukaghir genetic makeup would reflect ancient distribution of the C and D lineages associated with the Selemdzha culture (hunting-gathering-fishing populations) of blade industries originated in the mid-Amur region approximately 25.0 kya.67
Pioneers reached the Siberian Arctic by approximately 27.0 kya,3,68 and the stage was set for colonization the New World. Recent identification of C4c by Tamm et al.24 rooted separately to Siberian C2 ancestral sequence (Figure 3) introduces the possibility that the founding haplotype was among the initial colonizers of Siberian Arctic who later emerged among Native Americans. For the bearers of the C4c (called for expediency C2c), the now-submerged coastal shelves of the East Siberian and Chukchi Sea seem to have been the only practical route from Siberia to the Americas before the LGM.
The principle weakness of this hypothesis is the failure thus far to demonstrate the presence of representatives of the C2c lineage in Siberia or Beringia along presumed route of migrations. Sample sizes are still relatively small, and it is difficult to detect C2c in the remnants of the hunting-gathering groups, readily explained by their extinction through the action of genetic drift and/or selection in cold climate. Moreover, the populations of this type have undergone marked nonrandom postcontact decimation. As a consequence, the extinction of C2c in small isolated groups such as the Yukaghir would lead to reduction of original genetic diversity yielding coalescence estimates at a minimum time depths.
Concerning the C1 lineage, network analysis encompassing four sequences, one of the Ulchi,51 one Nanai57 from the Lower Amur, one of the Japanese,58 and one of the Buryat,25 appears to confirm that the founding haplotype for Native American C1 originated in the Amur River region as was previously proposed by Starikovskaya et al.51
A Number of Founding Lineages and Peopling of the Americas
Recently, it has become evident that the list of founding lineages that define the migrations from Eurasia to the Americas (A–D and X) is incomplete. Thus, an additional, independent, subhaplogroup of haplogroup D, postulated by Rickards et al.69 by analysis of the Cayapa HVS-I sequences from Ecuador, has been recently confirmed through identification of ancient mtDNA type in approximately 10.3-kya-old human remains from Alaska.22 Comparison of similar haplogroup D complete sequences from the Cayapa, Ulchi, and Japanese shows that they share three coding region transitions, 3336, 3644, and 5048, thus delineating additional haplogroup D cluster, which was attributed to D4h3 by Tamm et al.24 but here designated D10 (Figure 5). The divergent time for this lineage is 17.1 ± 4.0 kya (Table 3), and it is approximately 7000 years older than the age of the ancient haplogroup D mtDNA sample from Alaska, which evidently is related to the D10 cluster.
So far, there is no clearly identifiable progenitor of the Native American D1 in contemporary populations of Siberia and Beringia, and it is also absent from Asia. Contrary to our presumptive phylogeny of haplogroup D1,51 it remains unclear whether the D1, dating to 17.2 ± 2.8 kya (Table 3), has evolved in situ within North America or derived from a much earlier migration from Siberia-Asia. Though genealogical relationships among major lineages of the exceptionally diverse haplogroup D have become more pronounced, still some intermediate haplotypes may be missing, and this suggests long evolutionary history for Native American D1. Indirect support for this interpretation comes from the antiquity of Siberian-derived C1 aged 24.0 ± 3.8 kya.
Most of the models of the New World colonization, in which one to two major expansions from Siberia-Asia gave rise to Amerindian populations, generally indicate that colonization was after the LGM. However, based on the mtDNA data available at this phase of development, we propose that the first migrants to the New World were derived from the Altai-Sayan and/or the mid-lower-Amur region approximately 25.0–30.0 kya. With the global climate change that brought the Ice Age to an end (approximately 11.8 kya), the second wave of migrants comes from populations inhabiting regions of the Amur-Mongolia-Manchuria. The maintenance of more than one refugial sources, in the Altai-Sayan and mid-lower Amur, during the last glacial maximum appear to be at odds with the interpretation of limited founding mtDNA lineages populating the Americas as a single migration.
Accession Numbers
The 84 mtDNA sequences have been deposited in the GenBank with the accession numbers EU482303–EU482386. Simultaneously, these were placed on laboratory website (www.bionet.nsc.ru/labs/mtgenome/database.html).
Acknowledgments
This paper is an essential part of the collaborative molecular and evolutionary studies on the gene pool of indigenous Siberian and Native American populations initiated and put into life by Dr. James V. Neel in early 1990s, and we dedicate this paper to his memory. We are indebted to native people of Siberia, including Old Russian Settlers from Pokhodsk and Russkoye Ustye (Yakutia), for their help and hospitality. Special thanks to Sergei A. Frolov (District Hospital, Pevek, Chukotka) and Vladimir N. Dobrinin (President of the Association of Northern Peoples of the Aleut region, Kamchatka, Russian Federation). The authors have greatly benefited from conversations with Anatoliy P. Derevianko and Andrei V. Tabarev (Institute of Archeology and Ethnography, SBRAS, Novosibirsk). We would also like to thank Maria A. Lvova and Olga A. Derbeneva for their technical assistance provided at the initial phase of this project. We are indebted to two anonymous reviewers for their helpful comments that allowed us to improve the manuscript considerably. The help of Egene F. Voronov and Andre M. Sukernik in editing the final draft of the manuscript is gratefully acknowledged. This work received support from the Russian Academy of Science (Grant-Programs on Molecular Biology and Evolution), U.S. Civilian Research and Development Foundation (CRDF, #RB1-2501-NO-03), Wenner-Gren Foundation for Anthropological Research (WG Int. Res. Grant #65), and Russian Foundation for Basic Research (RFBR, #06-04-048182).
References
- 1.Goebel T. The “Microblade Adaptation” and Recolonization of Siberia during the Late Upper Pleistocene. Archeological Papers of the American Anthropological Association. 2002;12:117–131. [Google Scholar]
- 2.Goebel T., Waters M., Dikova M. The archaeology of Ushki Lake, Kamchatka, and the Pleistocene peopling of the Americas. Science. 2003;301:501–505. doi: 10.1126/science.1086555. [DOI] [PubMed] [Google Scholar]
- 3.Pitulko V., Nikolsky P., Girya E., Basilyan A., Tumskoy V., Koulakov S., Astakhov S., Pavlova E., Anisimov M. The Yana RHS site: Humans in the Artic before the Last Glacial Maximum. Science. 2004;303:52–56. doi: 10.1126/science.1085219. [DOI] [PubMed] [Google Scholar]
- 4.Vasil'ev S., Kuzmin Y., Orlova L., Dementyev V. Radiocarbon-based chronology of the Paleolithic in Siberia and its relevance to the peopling of the New World. Radiocarbon. 2002;44:503–530. [Google Scholar]
- 5.Hoffecker J., Powers W., Goebel T. The Colonization of Beringia and the peopling of the New World. Science. 1993;259:46–53. doi: 10.1126/science.259.5091.46. [DOI] [PubMed] [Google Scholar]
- 6.Kuntz M., Reanier R. Paleoindians in Beringia: Evidence from arctic Alaska. Science. 1994;263:660–662. doi: 10.1126/science.263.5147.660. [DOI] [PubMed] [Google Scholar]
- 7.Derevianko A.P. Selemdzha and the Eastern Siberian, Far Eastern, and Beringian Records. In: Derevianko A., editor. The Paleolithic of Siberia: New Discoveries and Interpretations. University of Illinois Press; Urbana, Illinois: 1998. pp. 285–286. [Google Scholar]
- 8.Laughlin W.S. Holt, Rinehart, and Winston; New York: 1980. Survivors of the Bering Land Bridge. [Google Scholar]
- 9.Rogers R.A. Languague, human subspecification, and ice age barriers in Nothern Siberia. Canadian Journal of Anthropology. 1986;5:11–22. [Google Scholar]
- 10.Rogers R., Rogers L., Hoffmann R., Martin L. Native American biological diversity and the biogeographic influence of Ice Age refugia. J. Biogeogr. 1991;18:623–630. [Google Scholar]
- 11.Neel J., Biggar R., Sukernik R. Virologic and genetic studies relate Amerind origins to the indigenous people of the Mongolia-Manchuria-Southeastern Siberia region. Proc. Natl. Acad. Sci. USA. 1994;91:10737–10741. doi: 10.1073/pnas.91.22.10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalton R. The coast road. Nature. 2003;422:10–12. doi: 10.1038/422010a. [DOI] [PubMed] [Google Scholar]
- 13.Hey J. On the number of New World founders: A population genetic portrait of the peopling of the Americas. PLoS Biol. 2005;3:e193. doi: 10.1371/journal.pbio.0030193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forster P., Harding R., Torroni A., Bandelt H. Origin and evolution of Native American mtDNA variation: a reappraisal. Am. J. Hum. Genet. 1996;59:935–945. [PMC free article] [PubMed] [Google Scholar]
- 15.Bonnato S., Salzano F. Diversity and age of the four major mtDNA haplogroups, and their implications for the peopling of the New World. Am. J. Hum. Genet. 1997;61:1413–1423. doi: 10.1086/301629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Starikovskaya E., Sukernik R., Schurr T., Kogelnik A., Wallace D. Mitochondrial DNA diversity in Chukchi and Siberian Eskimos: Implications for the genetic history of ancient Beringia and the peopling of the New World. Am. J. Hum. Genet. 1998;63:1473–1491. doi: 10.1086/302087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schurr T., Sukernik R., Starikovskaya Y., Wallace D. Mitochondrial DNA variation in Koryaks and Itel'men: Population replacement in the Okhotsk Sea-Bering Sea region during the Neolithic. Am. J. Phys. Anthropol. 1999;108:1–39. doi: 10.1002/(SICI)1096-8644(199901)108:1<1::AID-AJPA1>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 18.Saillard J., Forster P., Lynnerup N., Bandelt H., Norby S. mtDNA variation among Greenland Eskimos: The edge of the Beringian expansion. Am. J. Hum. Genet. 2000;67:718–726. doi: 10.1086/303038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubicz R., Schurr T., Babb P., Crawford M. Mitochondrial DNA variation and the origins of the Aleuts. Hum. Biol. 2003;75:809–835. doi: 10.1353/hub.2004.0009. [DOI] [PubMed] [Google Scholar]
- 20.Helgason A., Palsson G., Pedersen H., Angulalik E., Gunnarsdottir E., Yngvadottir B., Stefansson K. mtDNA variation in Inuit populations of Greenland and Canada: Migration history and population structure. Am. J. Phys. Anthropol. 2006;130:123–134. doi: 10.1002/ajpa.20313. [DOI] [PubMed] [Google Scholar]
- 21.Hewitt G.M. The structure of biodiversity – insights from molecular phylogeography. Front. Zool. 2004;I:4. doi: 10.1186/1742-9994-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kemp B., Malhi R., McDonough J., Bolnick D., Eshleman J., Rickards O., Martinez-Labarga C., Johnson J., Lorenz J., Dixon E. Genetic analysis of early holocene skeletal remains from Alaska and its implications for the settlement of the Americas. Am. J. Phys. Anthropol. 2007;132:605–621. doi: 10.1002/ajpa.20543. [DOI] [PubMed] [Google Scholar]
- 23.Wang S., Lewis C., Jakobsson M., Ramachandran S., Ray N., Bedoya G., Rojas W., Parra M., Molina J., Gallo C. Genetic variation and population structure in native Americans. PLoS Genet. 2007;3:e185. doi: 10.1371/journal.pgen.0030185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamm E., Kivisild T., Reidla M., Metspalu M., Smith D., Mulligan C., Bravi C., Rickards O., Martinez-Labarga M., Khusnutdinova E. Beringian standstill and spread of Native American founders. PLoS ONE. 2007;2:e829. doi: 10.1371/journal.pone.0000829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Derenko M., Malyarchuk B., Grzybowski T., Denisova G., Dambueva I., Perkova M., Dorzhu C., Luzina F., Lee H., Vanecek T. Phylogeographic analysis of mitochondrial DNA in Northern Asian Populations. Am. J. Hum. Genet. 2007;81:1025–1041. doi: 10.1086/522933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khlobystin L.P. Dmitriy Bulanin; St. Petersburg, Russia: 1998. Ancient History of Taimyr and the Formation of the North Eurasian Cultures. [Google Scholar]
- 27.Dolgikh B. Origin of the Nganasan. Academy of Sciences USSR. 1952;18:5. [Google Scholar]
- 28.Dolgikh B. Tribal composition of native Siberian people in the 17th century. Proceedings of the Institute of Ethnography of the Academy of Sciences USSR. 1960;55:3. [Google Scholar]
- 29.Simchenko Y.B. Nauka; Moscow: 1976. The Culture of Reindeer Hunters of North Eurasia. [Google Scholar]
- 30.Goltsova T., Sukernik R. Genetic structure of an isolated group of the indigenous population of northern Siberia, the Nganasans (Tavginians) of Taimir. Genetika. 1979;15:734–744. [PubMed] [Google Scholar]
- 31.Karaphet T., Sukernik R., Osipova L., Simchenko Y. Blood groups, serum proteins, and red cell enzymes in the Nganasans (Tavghi)-reindeer hunters from Taimir Peninsula. Am. J. Phys. Anthropol. 1981;56:139–145. doi: 10.1002/ajpa.1330560205. [DOI] [PubMed] [Google Scholar]
- 32.Osipova L.P., Sukernik R. Immunoglobulin allotypes in aboriginal populations of the Taimir Peninsula. J. Immunogenet. 1983;10:11–16. doi: 10.1111/j.1744-313x.1983.tb01011.x. [DOI] [PubMed] [Google Scholar]
- 33.Derbeneva O., Starikovskaia E., Volod'ko N., Wallace D., Sukernik R. Mitochondrial DNA variation in Kets and Nganasans and the early peoples of Northern Eurasia. Genetika. 2002;38:1554–1560. [PubMed] [Google Scholar]
- 34.Wrangel F.P. St. Petersburg, Russia; 1841. Puteshestvie po severnim beregam Sibiri i po ledovitomu moriu, soverishennoye v 1820–1824 godakh. [Google Scholar]
- 35.Jochelson W. The Yukaghir and the Yukaghirized Tungus. Publications of the The Jesup North Pacific Expedition. 1910;9 part 1. [Google Scholar]
- 36.Okladnikov A.P. Academiya nauk SSSR; Moscow, Leningrad, Russia: 1955. Yakutiya do prisoedineniya k russkomu gosudarstvu. [Google Scholar]
- 37.Okladnikov A.P. Nauka; Novosibirsk, Russia: 1975. Yukaghiry (Istoriko-etnograficheskiy ocherk) [Google Scholar]
- 38.Neel J.V. The distinction between primary and secondary isolates. In: Roberts D., Fujiki N., Torizuka K., editors. Symposium 33. Isolation, Migration and Health. Cambridge University Press; 1992. pp. 17–23. [Google Scholar]
- 39.Bogoras W. The Chukchee. Publications of the Jesup North Pacific Expedition. 1910;8 part 1. [Google Scholar]
- 40.Spiridonov N.I. Severoved; Yakutsk, Russia: 1935. Oduly (Yukaghiry) Kolymskogo okruga. [Google Scholar]
- 41.Kreinovitch E. O nekotoryh jukagirsko-ural'skih jazykovyh paralleljah. Sovetskoe finno-ugrovedenie. 1978;4:241–249. [Google Scholar]
- 42.Tugolukov V.A. Nauka; Moscow: 1979. Who Are You Yukaghir? [Google Scholar]
- 43.Bogoras W. Russkie na reke Kolyme. Zhizn. 1899;6:103–125. [Google Scholar]
- 44.Chikachov A.G. Nauka; Novosibirsk, Russia: 1990. Russkie na Indigirke: Istoriko-etnograficheskij ocherk. [Google Scholar]
- 45.Dyachkoff G. Tiopgraphiya Sibirskogo Flotskogo Ekipazha; Vladivostok, Russia: 1893. Anadyrski Krai. [Google Scholar]
- 46.Sukernik R., Lemza S., Karaphet T., Osipova L. Reindeer Chukchi and Siberian Eskimos: Studies on blood groups, serum proteins, and red cell enzymes with regard to genetic heterogeneity. Am. J. Phys. Anthropol. 1981;55:121–128. doi: 10.1002/ajpa.1330550116. [DOI] [PubMed] [Google Scholar]
- 47.Menovshshikov G.A. Nauka; Leningrad, Russia: 1980. Language of the Bering Strait Eskimos. [Google Scholar]
- 48.Derbeneva O., Sukernik R., Volodko N., Hosseini S., Lott M., Wallace D. Analysis of mitochondrial DNA diversity in the Aleuts of the Commander Islands and its implications for the genetic history of Beringia. Am. J. Hum. Genet. 2002;71:415–421. doi: 10.1086/341720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andrews R., Kubacka I., Chinnery P., Lightowlers R., Turnbull D., Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 50.Torroni A., Schurr T., Cabell M., Brown M., Neel J., Larsen M., Smith D., Vullo M., Wallace D. Asian affinities and continental radiation of the four founding Native American mtDNAs. Am. J. Hum. Genet. 1993;53:563–590. [PMC free article] [PubMed] [Google Scholar]
- 51.Starikovskaya E., Sukernik R., Derbeneva O., Volodko N., Ruiz-Pesini E., Torroni A., Brown M., Lott M., Hosseini S., Huoponen K., Wallace D. Mitochondrial DNA diversity in indigenous populations of the southern extent of Siberia, and the origins of Native American haplogroups. Ann. Hum. Genet. 2005;69:67–89. doi: 10.1046/j.1529-8817.2003.00127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Volodko, N., Sukernik, R., Starikovskaya, E., Lvova, M., and Wallace, D. (2006). Mitochondrial DNA lineages in the Yukaghir, Chukchi and Siberian Eskimos, and resettlement of Arctic Siberia after the Last Glacial Maximum (LGM). Abstract. 56th Annual Meeting of ASHG, New Orleans, USA. Oct. 9–13.
- 53.Mishmar D., Ruiz-Pesini E., Golik P., Macaulay V., Clark A., Hosseini S., Brandon M., Easley K., Chen E., Brown M. Natural selection shaped regional mtDNA variation in humans. Proc. Natl. Acad. Sci. USA. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harpending H., Jenkins T. Genetic distance among Southern African populations. In: Crawford M., Workman P., editors. Methods and Theories of Anthropological Genetics. University of New Mexico Press; Albuquerque, New Mexico: 1973. pp. 177–199. [Google Scholar]
- 55.Derbeneva O., Starikovskaya E., Wallace D., Sukernik R. Traces of early Eurasians in the Mansi of northwest Siberia revealed by mitochondrial DNA analysis. Am. J. Hum. Genet. 2002;70:1009–1014. doi: 10.1086/339524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Achilli A., Rengo C., Battaglia V., Pala M., Olivieri A., Fornarino S., Magri C., Scozzari R., Babudri N., Santachiara-Benerecetti A. Saami and Berbers–an unexpected mitochondrial DNA link. Am. J. Hum. Genet. 2005;5:883–886. doi: 10.1086/430073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ingman M., Gyllensten U. Rate variation between mitochondrial domains and adaptive evolution in humans. Hum. Mol. Genet. 2007;16:2281–2287. doi: 10.1093/hmg/ddm180. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka M., Cabrera V., Conzales A., Larruga J., Takeyasu T., Fuku N., Guo L.-J., Hirose R., Fujita Y., Kurata M. Mitochondrial genome variation in Eastern Asia and the Peopling of Japan. Genet. Res. 2004;14:1832–1850. doi: 10.1101/gr.2286304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kong Q.-P., Yao Y.-G., Sun C., Bandelt H.-J., Zhu C.-L., Zhang Y.-P. Phylogeny of east Asian mitochondrial DNA lineages inferred from complete sequences. Am. J. Hum. Genet. 2003;73:671–676. doi: 10.1086/377718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Forster P. Ice Ages and the mitochondrial DNA chronology of human dispersals: A review. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004;359:255–264. doi: 10.1098/rstb.2003.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Volodko N., Derbeneva O., Uinuk–ool T., Sukernik R. Genetic history of Aleuts of the Commander islands as revealed by the analysis of the HLA class II gene variability. Genetika. 2003;12:1710–1718. [PubMed] [Google Scholar]
- 62.Krauss M.E. Many tongues-ancient tales. Peoples of the Amur and maritime regions. In: Fitzhugh W., Crowell A., editors. Crossroads of Continents: Cultures of Siberia and Alaska. Smithsonian Institution Press; Washington, D.C.: 1988. pp. 145–150. [Google Scholar]
- 63.West F.H. University of Chicago Press; Chicago: 1996. American Beginnings, the Prehistory and Paleoecology of Beringia. [Google Scholar]
- 64.Torroni A., Sukernik R., Schurr T., Starikovskaya Y., Cabell M., Crawford M., Comuzzie A., Wallace D. Analysis of mitochondrial DNA variation of indigenous Siberians confirms their genetic diversity and reveals distinctive affinity with Native Americans. Am. J. Hum. Genet. 1993;53:591–608. [PMC free article] [PubMed] [Google Scholar]
- 65.Pakendorf B., Novgorodov I., Osakovskij V., Danilova A., Protod'jakonov A., Stoneking M. Investigating the effects of prehistoric migrations in Siberia: Genetic variation and the origins of Yakuts. Hum. Genet. 2006;120:334–353. doi: 10.1007/s00439-006-0213-2. [DOI] [PubMed] [Google Scholar]
- 66.Hayes M., O'Rourke D. Genetic signatures of migrations and population replacements during human colonization of the North American Arctic. Am. J. Hum. Genet. 2001;69(Suppl.):179. [Google Scholar]
- 67.Derevianko A., Volkov P., Khondjon L. Institute of Archeology and Ethnography, Russian Academy of Sciences; Novosibirsk, Russia: 1998. Selemdzha Late Paleolithic Culture. [Google Scholar]
- 68.Finlayson C., Carrion J. Rapid ecological turnover and its impact on Neanderthal and other human populations. Trends Ecol. Evol. 2007;22:213–222. doi: 10.1016/j.tree.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 69.Rickards O., Martinez-Labarga C., Lum J., De Stefano G., Cann R. mtDNA history of the Cayapa Amerinds of Ecuador: Detection of additional founding lineages for the Native American populations. Am. J. Hum. Genet. 1999;65:519–530. doi: 10.1086/302513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ingman M., Kaessmann H., Paabo S., Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- 71.Maca-Meyer N., Gonzalez A., Larruga J., Flores C., Cabrera V. Major genomic mitochondrial lineages delineate early human expansions. BMC Genet. 2001;2:13. doi: 10.1186/1471-2156-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Herrnstadt C., Elson J., Fahy E., Preston G., Turnbull D., Anderson C., Ghosh S., Olefsky J., Beal M., Davis R., Howell N. Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am. J. Hum. Genet. 2002;70:1152–1171. doi: 10.1086/339933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kivisild T., Shen P., Wall D.P., Do B., Sung R., Davis K., Passarino G., Underhill P.A., Scharfe C., Torroni A. The role of selection in the evolution of human mitochondrial genomes. Genetics. 2006;172:373–387. doi: 10.1534/genetics.105.043901. [DOI] [PMC free article] [PubMed] [Google Scholar]


