Abstract
Germline mutations in the gene encoding phosphatase and tensin homolog deleted on chromosome ten (PTEN [MIM 601728]) are associated with a number of clinically distinct heritable cancer syndromes, including both Cowden syndrome (CS) and Bannayan-Riley-Ruvalcaba syndrome (BRRS). Seemingly identical pathogenic PTEN mutations have been observed in patients with CS and BRRS, as well as in patients with incomplete features of CS, referred to as CS-like (CSL) patients. These observations indicate that additional, unidentified, genetic and epigenetic factors act as phenotypic modifiers in these disorders. These genetic factors could also contribute to disease in patients with CS, CSL, or BRRS without identifiable PTEN mutations. Two potential modifiers are miR-19a and miR-21, which are previously identified PTEN-targeting miRNAs. We investigated the role of these miRNAs by characterizing their relative expression levels in PTEN-mutation-positive and PTEN-mutation-negative patients with CS, CSL, or BRRS. Interestingly, we observed differential expression of miR-19a and miR-21 in our PTEN-mutation-positive patients. Both were found to be significantly overexpressed within this group (p < 0.01) and were inversely correlated with germline PTEN protein levels. Similarly, the relative expression of miR-19a and miR-21 was differentially expressed in a series of PTEN-mutation-negative patients with CS or CSL with variable clinical phenotypes and decreased full-length PTEN protein expression. Among PTEN-mutation-positive patients with CS, both miRNAs were significantly overexpressed (p = 0.006–0.013). Taken together, our study results suggest that differential expression of PTEN-targeting miR-19a and miR-21 modulates the PTEN protein levels and the CS and CSL phenotypes, irrespective of the patient's mutation status, and support their roles as genetic modifiers in CS and CSL.
Introduction
Cowden syndrome (CS [MIM 158350]) is an underdiagnosed autosomal-dominant disorder characterized by multiple hamartomatous lesions and a vast phenotypic spectrum that includes predisposition to malignancies of the breast, thyroid, and endometrium.1 The majority of patients with CS (85%) have been found to harbor pathogenic germline mutations in the gene encoding phosphatase and tensin homolog deleted on chromosome ten (PTEN [MIM 601728]), a tumor-suppressor gene located on 10q23.2 The inactivation of PTEN protein, an antagonist of the phosphatidylinositol-triphosphate kinase (PI3K)-signaling pathway, in CS results in the constitutive activation of Akt.3–8 As a consequence, hyperactive Akt-mediated signaling of several cascades results in increased and uncontrolled cellular survival and proliferation.
Germline mutations in PTEN also cause a subset of Bannayan-Riley-Ruvalcaba syndrome (BRRS [MIM 153480]), a related hamartomatous tumor syndrome that shares, in addition to its genetic etiology, a few of the clinical manifestations seen in CS.3 Interestingly, identical pathogenic PTEN mutations have been observed among patients with these seemingly disparate disorders.9 Furthermore, these same identical mutations have also been observed in two additional hamartomatous tumor syndromes, Proteus syndrome (PS [MIM 176920]) and Proteus-like syndrome (PLS), as well as in a subset of patients referred to as CS-like (CSL) patients who do not meet the full diagnostic criteria for CS.1,3 Whereas germline PTEN mutations have been identified in the majority of patients diagnosed with CS and BRRS, for approximately 15% and 35%, respectively, as well as for 90% of patients with CSL, the pathogenic mutations have yet to be identified.2,9 Interestingly, decreased PTEN protein expression has been observed in a large number of these PTEN-mutation-negative patients (Waite and Eng, unpublished observation). The lack of detectable germline PTEN mutations in subsets of patients with CS, CSL, or BRRS with altered PTEN protein expression, as well as the imprecise genotype-phenotype correlation associated with these syndromes, suggests the presence of alternate mechanisms that contribute to PTEN dysfunction and to the development and progression of the disease.
MicroRNAs (miRNAs), a novel class of negative gene regulators, have recently been shown to regulate the expression of several tumor suppressors and oncogenes and contribute to carcinogenesis.10–14 To date, two miRNAs, miR-19a and miR-21, have been reported to specifically target and to downregulate PTEN.15,16 Furthermore, the overexpression of each of these miRNAs has been correlated with decreased PTEN levels in human cancer.17,18
Because PTEN dysfunction is common in patients with CS, CSL, and BRRS, irrespective of their mutation status, we hypothesized that aberrant miR-19a and/or miR-21 expression could modulate PTEN protein levels and, thereby, modulate phenotypic expression in patients with these syndromes. To investigate this hypothesis, we chose to characterize the relative expression levels of both miR-19a and miR-21 in PTEN-mutation-positive and PTEN-mutation-negative patients with CS, CSL, and BRRS relative to healthy control subjects.
Subjects and Methods
Study Subjects
A total of 178 unrelated subjects were included in the present study, including 20 control subjects and 158 patients with CS, CSL, or BRRS (Table 1). Mutation analysis of the entire PTEN coding sequence and its exon-intron boundaries and promoter region was performed for all patients with CS, CSL, or BRRS who were included in the present study.2 Additionally, all patient samples were also screened for PTEN protein expression by Western blot analysis. Briefly, patient protein lysates were isolated from immortalized lymphoblastoid cell lines (LBCLs) by the Genomic Medicine Biorepository and protein concentrations were determined by use of the BCA assay, with BSA as a standard. Thirty micrograms of protein was applied to nitrocellulose with a dotblot apparatus (Bio-Rad, Hercules, CA, USA). Equal protein loading was confirmed by staining of the nitrocellulose blots with Ponceau stain (Bio-Rad). Nitrocellulose blots were then subjected to Western blot analysis with α-PTEN (monoclonal antibody clone 6H2.1, Cascade Biosciences, Portland, OR, USA) at 1:1000 dilution and then incubated with appropriate secondary antibody as previously described.7 Proteins were visualized via enhanced chemiluminescence. Full-length PTEN protein levels were compared to controls and scored by two individuals as either normal, decreased (<50% loss, compared to controls), half (∼50% of controls), or faint (>50% loss, compared to controls). PTEN-mutation-negative patients with decreased, half, or faint full-length PTEN protein levels were also scanned for large deletions and rearrangements, and those with the latter were excluded. Among the patients with CS, CSL, or BRRS, 28/158 of the individuals selected for inclusion were previously found to harbor pathogenic germline mutations (i.e., mutation-positive patients). Mutation analysis in the remaining 130/158 patients revealed no detectable germline PTEN mutations (i.e., mutation-negative patients). For interrogation of miR-19a and miR-21 expression in patients with CS, CSL, or BRRS without germline PTEN mutations but with deregulated PTEN protein levels, all 130 mutation-negative patients selected for inclusion in the current study were previously found to have decreased full-length PTEN protein expression relative to normal controls (data not shown). All subjects were enrolled by referral from centers throughout the United States, Canada, and Europe after granting informed consent in accordance with procedures approved by the human-subjects-protection committees of each respective institution.
Table 1.
Summary of Control and Patient Clinical Features
| Controls (n = 20)a | PTEN-Mutation-Positive Patients (n = 28)a | PTEN-Mutation-Negative Patients (n = 130)a | |
|---|---|---|---|
| Gender | |||
| Male | 7 (35.0) | 12 (42.9) | 10 (7.7) |
| Female | 13 (65.0) | 16 (57.1) | 120 (92.3) |
| Phenotype | |||
| CS | – | 17 (60.7)b | 63 (48.5) |
| CSL | – | 5 (17.9) | 62 (47.7) |
| BRRS | – | 5 (17.9) | 5 (3.8) |
| Mutation | |||
| R130X | – | 13 (46.4) | – |
| R233X | – | 5 (17.9) | – |
| R335X | – | 10 (35.7) | – |
| Clinical Features | |||
| Macrocephaly | – | 25 (89.3) | 53 (40.8) |
| Pathognomonic | – | 16 (57.1) | 28 (21.5) |
| Breast CA | – | 7 (43.8)c | 80 (66.7)c |
| Thyroid CA | – | 2 (7.1) | 32 (24.6) |
| Endometrial CA | – | 2 (12.5)c | 14 (11.7)c |
Pathognomonic features include the mucocutaneous lesions associated with CS (trichilemmomas, acral keratoses, and papillomatous papules).
Values are expressed as n (%).
One PTEN-mutation-positive sample had features consistent with both CS and BRRS.
Frequency among female patients.
Among the cohort of PTEN-mutation-positive patients included in our analysis were 13/28 patients with R130X mutations (c.388 C/T), 5/28 patients with R233X mutations (c.697 C/T), and 10/28 patients with R335X mutations (c.1003 C/T) (Figure 1 and Table 1). Of those patients with R130X mutations, 9/13 had classic CS, 1/13 exhibited a CSL phenotype, and 2/13 had classic BRRS. Additionally, 1/13 patients with an R130X mutation had phenotypic features consistent with both CS and BRRS, and this patient thus was considered as a CS/BRRS overlap patient. Among the patients with an R233X mutation, 1/5 met classic CS criteria, 3/5 were considered as having CSL, and 1/5 had classic BRRS. Lastly, of patients with R335X mutations, 7/10 met classic CS criteria, 1/10 had CSL, and 2/10 had BRRS.
Figure 1.
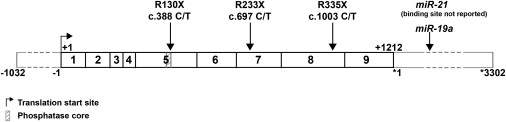
Schematic Diagram of PTEN's mRNA Sequence
PTEN's full-length mRNA sequence (NM_000314.4), including its 1032 nucleotide (nt) 5′ UTR, 1212 nt coding sequence, and 3302 nt 3′ UTR, is shown along with the miR-19a binding site (position ∗1208 to ∗1228). miR-21's binding site has not been reported. Additionally, the positions for the three truncating mutations R130X, R233X, and R335X are shown. PTEN's translation start site and the region encoding PTEN's phosphatase core are also annotated.
Among the cohort of PTEN-mutation-negative patients with decreased full-length PTEN expression, 63/130 had classic CS, 62/130 had CSL, and 5/130 were diagnosed with BRRS. All mutation-positive and mutation-negative patients with CS included in the present study were classified in accordance with criteria established by the International Cowden Consortium and incorporated into the practice guidelines of the National Comprehensive Cancer Network.1
Cell Lines and Culture
LBCLs from all healthy control samples and those from patients with CS, CSL, or BRRS were generated by the Genomic Medicine Biorepository. LBCLs were cultured in RPMI-1640 media supplemented with 20% FBS and 100 units/ml each of Penicillin and Streptomycin and maintained at 37°C with 5% CO2.
Isolation of Total RNA and cDNA Synthesis
Total RNA was isolated from control and patient LBCLs with the miRVana miRNA Isolation Kit (Ambion, Austin, TX, USA) according to the manufacturer's protocol and, subsequently, subjected to DNase treatment with DNA-free (Ambion). For analysis of PTEN expression levels, 1 μg of DNase-treated RNA was converted to cDNA with Superscript II Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) as specified by the manufacturer.
In order to assay mature miR-19a and miR-21 expression in our control and patient samples, stem-loop reverse transcription (RT) was performed with TaqMan MicroRNA Assays (Applied Biosystems, Foster City, CA). For each miRNA, individual RT reactions were carried out with 10 ng of DNase-treated RNA and either miR-19a-, miR-21-, or U6 small nuclear RNA (RNU6)-specific TaqMan MicroRNA RT primers (Applied Biosystems). Twenty-microliter RT reactions were performed with the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer's protocol.
Real-Time Quantitative PCR
Relative expression of the full-length PTEN transcript was assayed for all samples with 12.5 μl SYBR Green PCR Master Mix (Applied Biosystems), 10 mM forward primer (5′-TCCACAAACAGAACAAGATG-3′), 10 mM reverse primer (5′-CTGGTCCTGGTATGAAGAAT-3′), and 20 ng of template cDNA. Thermal cycling conditions comprised 50°C for 2 min, 95°C for 10 min, and 40 cycles at 95°C for 15 s followed by 60°C for 1 min with an ABI 7500 Sequence Detection System (Applied Biosystems). Real-time PCR reactions were also performed for GAPDH, a normalization control (F: 5′-GGGCTGCTTTTAACTCTGGTAA-3′ and R: 5′-ATGGGTGGAATCATATTGGAAC-3′).
Mature miR-19a and miR-21 expression was assayed with a 20 μl reaction containing 10.0 μl TaqMan 2X Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems), 1.0 μl 20X TaqMan MicroRNA assay (Applied Biosystems), and 1.3 μl of RT product. Real-time PCR conditions were performed as follows: 50°C for 2 min, 95°C for 10 min, and 40 cycles at 95°C for 15 s followed by 60°C for 1 min with an ABI 7500 Sequence Detection System (Applied Biosystems). Additionally, RNU6 expression was assayed for normalization.
All reactions were performed in triplicate, and relative gene-expression determinations were made with the comparative delta-delta CT method (2−ΔΔCT) as described by Livak et al.19
Statistical Analysis
After expression analysis was performed, statistical comparisons of relative levels of PTEN, miR-19a, and miR-21 expression between all patient groups and control samples were performed with Welch's t test. All results were considered significant at the p < 0.05 level. In addition, we performed classification and regression tree (CART) analysis as implemented in the R software package.
Results
Differential Expression of miR-19a and miR-21 in Three Groups of Patients with CS, CSL, or BRRS with Germline PTEN Mutation R130X, R233X, or R335X
In the present study, we sought to investigate whether miRNAs previously shown to target and regulate PTEN protein expression are differentially regulated in patients with CS, CSL, and BRRS relative to healthy controls and, more specifically, whether these miRNAs are modifiers of these phenotypes. To address this question, we initially chose to assess the relative expression of miR-19a and miR-21 in a series of 28 PTEN-mutation-positive patients sharing identical truncating mutations located in each of PTEN's three most frequently mutated exons (Figure 1) and in 20 normal controls.3 Included among these patients were 13 carriers of R130X mutations, five carriers of R233X mutations, and ten carriers of R335X mutations. Previous studies have reported a broad clinical spectrum in patients even within each of these three mutation groups.9,20 Similar observations were apparent in this collection, given that nine patients with CS, one with CSL, one with CS and BRRS, and two with BRRS were among those with R130X mutations; one patient with CS, three with CSL, and one with BRRS were among those with R233X mutations; and seven patients with CS, one with CSL, and two with BRRS were among those with R335X mutations (Table S1, available online).
In addition to the variable clinical spectrum observed among patients with identical mutations, levels of PTEN mRNA expression were also found to be variable. Among all 28 PTEN-mutation-positive patients, expression of PTEN transcript levels were significantly reduced relative to controls (p = 0.015) (Table 2). However, when we divided this group on the basis of the three different germline PTEN mutations found in these patients, significantly decreased PTEN transcript expression was only observed in patients with the R130X and R233X mutations (p < 0.001) (Table 2) and not in carriers of the R335X mutation (p = 0.287). Similarly, we observed differential expression of miR-19a and miR-21 among our mutation-positive samples. Overall, the average relative expression of both miR-19a and miR-21 was higher in mutation-positive patients compared to controls (2−ΔΔCT = 1.54 versus 1.08, p = 0.003; and 2−ΔΔCT = 1.49 versus 1.15, p = 0.006, respectively) (Figure 2A and Table 2). For mutation-positive patients with R130X and R233X mutations, miR-19a was found to be overexpressed as compared to that of controls (p = 0.037 and p = 0.015, respectively), whereas miR-21 was overexpressed only in carriers of the R130X mutation (p = 0.044). miR-19a and miR-21 were not differentially expressed in carriers of the R335X mutation relative to our control population. Interestingly, 90% of patients with this mutation had decreased full-length PTEN protein levels, compared to only 54% of those with R130X mutations. Moreover, although miR-19a and miR-21 were not differentially expressed in carriers of the R335X mutation, the majority of these patients (70%) exhibit classic CS features, compared to only 56% of patients with R130X and/or R233X mutations.
Table 2.
Summary of Comparisons of Relative Levels of PTEN Transcript, miR-19a, and miR-21 Expression among Control Samples and PTEN-Mutation-Positive and PTEN-Mutation-Negative Patient and Patient Subgroup Samples
| PTEN | miR-19a | miR-21 | |
|---|---|---|---|
| PTEN+ | 0.015 | 0.003 | 0.006 |
| R130X | < 0.001 | 0.037 | 0.044 |
| R233X | < 0.001 | 0.015 | 0.184 |
| R335X | 0.287 | 0.137 | 0.147 |
| PTEN+ dec. versus PTEN+ norm. | 0.336 | 0.009 | 0.003 |
| R130X/R233X dec. versus R130X/R233X norm. | 0.234 | 0.020 | 0.008 |
| PTEN+ CS | 0.069 | 0.006 | 0.013 |
| PTEN+ CSL | 0.055 | 0.034 | 0.085 |
| PTEN+ BRRS | 0.209 | 0.932 | 0.772 |
| PTEN− | 0.969 | < 0.001 | 0.141 |
| PTEN− CS | 0.900 | < 0.001 | 0.835 |
| PTEN− CSL | 0.544 | < 0.001 | 0.018 |
| PTEN− BRRS | 0.148 | 0.492 | 0.215 |
| PTEN+ versus PTEN− | 0.001 | 0.977 | < 0.001 |
| PTEN+ CS versus PTEN− CS | 0.020 | 0.878 | 0.015 |
| PTEN+ CSL versus PTEN− CSL | 0.079 | 0.368 | 0.029 |
| PTEN+ BRRS versus PTEN− BRRS | 0.067 | 0.496 | 0.326 |
p values were determined from t tests comparing the relative expression of PTEN, miR-19a, and miR-21 between controls and patients or patient subgroups, except where indicated. Significant results are indicated as underlined.
“PTEN+” denotes PTEN-mutation-positive patients.
“PTEN−” denotes PTEN-mutation-negative patients.
“PTEN+ dec.” denotes PTEN+ patients with decreased PTEN protein expression. PTEN protein status “calls” are illustrated in Figure S1.
“PTEN+ norm.” denotes PTEN+ patients with normal PTEN protein expression. PTEN protein status “calls” are illustrated in Figure S1.
“R130X/R233X dec.” denotes patients with R130X and/or R233X mutations with decreased PTEN protein expression.
“R130X/R233X norm.” denotes patients with R130X and/or R233X mutations with normal PTEN protein expression.
Figure 2.
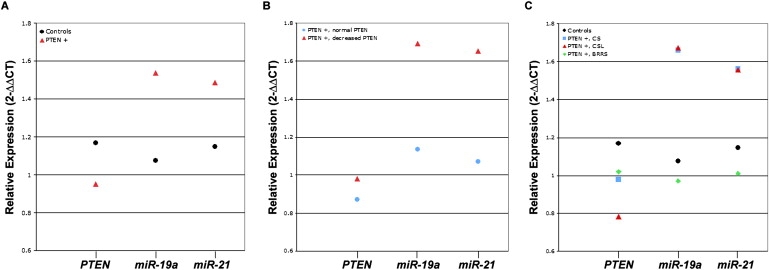
Relative Expression Values for PTEN Transcript, miR-19a, and miR-21 in PTEN-Mutation-Positive Patients
Relative expression values for PTEN transcript and each miRNA are shown for (A) controls and all mutation-positive patients, (B) mutation-positive patients who express normal PTEN protein and those who express decreased PTEN protein, and (C) controls and mutation-positive patients subdivided on the basis of their clinical phenotypes.
All relative expression values are expressed as n-fold change (2−ΔΔCT).
PTEN+ = PTEN-mutation-positive patients.
miR-19a and miR-21 Overexpression are Associated with Decreased Full-Length PTEN Protein Levels and Clinical Phenotype in PTEN-Mutation-Positive Patients with CS, CSL, or BRRS
Our data show that PTEN transcript and PTEN protein levels, as determined by Western blot analysis, do not correspond well among many of the mutation-positive patients included in this analysis, suggesting that other biological processes are likely involved in the regulation of these products (Table S1). On the basis of this observation, we then chose to examine the relationship between the relative expression of each miRNA and PTEN protein levels in these patients. More specifically, we examined levels of miR-19a and miR-21 expression in mutation-positive patients with normal full-length PTEN protein expression compared to those with decreased full-length PTEN protein levels. Interestingly, both miRNAs were significantly overexpressed in PTEN-mutation-positive patients with decreased full-length PTEN protein levels as compared to those with normal full-length PTEN protein levels (miR-19a: 2−ΔΔCT = 1.69 versus 1.13, p = 0.009; miR-21: 2−ΔΔCT = 1.65 versus 1.07, p = 0.003, respectively) (Figure 2B and Table 2). Moreover, patients with R130X and/or R233X mutations with decreased full-length PTEN protein expressed approximately 54% higher levels of miR-19a (p = 0.020) and 60% higher levels of miR-21 (p = 0.008) relative to patients with R130X and/or R223X mutations with normal PTEN protein levels (Table 2).
Next, we subdivided our PTEN-mutation-positive group on the basis of their clinical diagnoses (CS, CSL, or BRRS) and compared their relative levels of miR-19a, miR-21, and PTEN expression to those of the control group. Mutation-positive patients with CS displayed relative overexpression of both miR-19a and miR-21 compared to controls (p = 0.006 and p = 0.013, respectively) (Figure 2C and Table 2). A modest association was observed between the mutation-positive group of patients with CSL and miR-19a expression status (p = 0.034), whereas miR-21 overexpression in this same group did not reach statistical significance when compared to controls (p = 0.085), which was due in part to the small sample size in this latter subgroup. No differences in PTEN transcript expression were observed in either phenotypic subgroup. Lastly, PTEN transcript, miR-19a, and miR-21 expression did not differ between the mutation-positive patients with BRRS and the controls (p > 0.20).
Additionally, PTEN transcript, miR-19a, and miR-21 expression did not differ between PTEN-mutation-positive patients with CS and PTEN-mutation-positive patients with CSL (p > 0.200). Both miR-19a and miR-21 levels were lower in patients with BRRS as compared to levels in patients with CS (miR-19a: 2−ΔΔCT = 1.07 versus 1.63 and miR-21: 2−ΔΔCT = 1.25 versus 1.53) and in patients with CSL (miR-19a: 2−ΔΔCT = 1.07 versus 1.67 and miR-21: 2−ΔΔCT = 1.25 versus 1.56), although statistical significance was only reached in comparisons of miR-19a expression (p < 0.05).
Differential Expression of miR-19a and miR-21 in PTEN-Mutation-Negative Patients with CS, CSL, or BRRS
Differential expression of miR-19a and miR-21 in PTEN-mutation-positive patients with CS, CSL, or BRRS with variable PTEN protein expression suggests that these miRNAs play a role in modulation of both the PTEN protein expression and the disease phenotype in patients with these syndromes. On the basis of these results, we further hypothesized that these miRNAs could play a similar role in patients with CS, CSL, or BRRS who lack detectable PTEN mutations. To investigate this, we chose, therefore, to assess the relative expression of miR-19a and miR-21 in 130 selected patients with CS, CSL, or BRRS with both undetectable PTEN mutations and decreased full-length PTEN protein expression.
In contrast to the PTEN-mutation-positive patients, decreased expression of PTEN transcript levels was not observed in mutation-negative patients (p = 0.969). Interestingly, similar to our mutation-positive patients, mutation-negative patients displayed relative overexpression of miR-19a as compared to our control population (p < 0.001) (Figure 3 and Table 2). However, contrary to what was observed in mutation-positive patients, no difference in miR-21 expression was observed in this group (p = 0.141).
Figure 3.
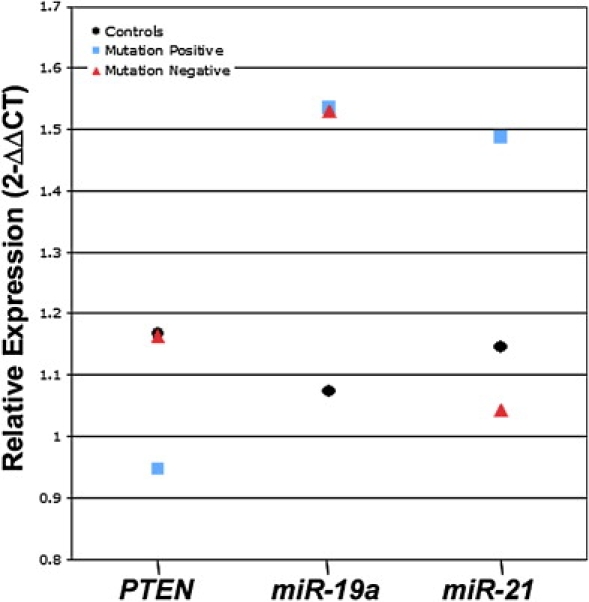
Relative Expression Values for PTEN Transcript, miR-19a, and miR-21 among PTEN-Mutation-Positive Patients, PTEN-Mutation-Negative Patients, and Controls
All relative expression values are expressed as n-fold change (2−ΔΔCT).
Following this analysis, PTEN-mutation-negative patients were subsequently subdivided on the basis of their clinical diagnoses (CS, CSL, or BBRS). We then carried out analyses of genotype-phenotype association based on these groupings. Similar to the mutation-positive cohort, miR-19a overexpression was observed both in mutation-negative patients with CS and in mutation-negative patients with CSL (p < 0.001 in both groups) (Table 2). In contrast, miR-21 was underexpressed in mutation-negative patients with CSL compared to controls (p = 0.018), whereas no difference was observed in patients with CS from this same group (p = 0.835). We did not detect any difference in miR-19a and miR-21 expression among our mutation-negative patients with BRRS (p > 0.215).
miR-21 is Differentially Expressed between PTEN-Mutation-Positive and PTEN-Mutation-Negative Patients
Because miR-19a is overexpressed in patients with CS or CSL irrespective of PTEN mutation status, whereas overexpressed miR-21 occurs only in PTEN-mutation-positive patients, we then chose to compare the relative expression of each miRNA between patients with and without PTEN mutations. miR-21 was significantly overexpressed in PTEN-mutation-positive patients relative to patients without PTEN mutations (2−ΔΔCT = 1.48 versus 1.05, p < 0.001) (Figure 3 and Table 2), whereas miR-19a expression did not differ between the two groups (p = 0.977). PTEN transcript levels were significantly lower in the mutation-positive patient group (p = 0.001). When subdivided on the basis of their clinical diagnoses, miR-21 overexpression was observed both in groups of mutation-positive patients with CS and in groups of mutation-positive patients with CSL relative to mutation-negative patients with these same phenotypes (p = 0.015 and p = 0.029, respectively), whereas miR-19a expression did not differ among these subgroups (p > 0.50).
miR-21 Expression Could Contribute to Phenotypic Features Associated with CS
To investigate whether differential expression of miR-19a and miR-21 was associated with the cancers commonly seen in CS, as well as with other clinical features of the syndrome, we compared their relative expression between patient groups with and without each of the key phenotypic features of CS. These comparisons failed to yield any significant associations with breast cancer, thyroid cancer, or macrocephaly between miRNA or PTEN expression among all patient samples and patient subgroups (p > 0.05), probably due to small sample sizes in subgroup analyses. We did observe a trend of overexpression of miR-21 in patients diagnosed with endometrial cancer as compared to those not diagnosed with this cancer, irrespective of mutation status (2−ΔΔCT = 1.39 versus 1.06 for PTEN-mutation-positive patients and 2−ΔΔCT = 1.37 versus 1.01 for PTEN-mutation-negative patients; p = 0.12-0.15). Despite our small sample size, we did find that miR-21 was significantly overexpressed among patients with CS and one or more pathognomonic feature (adult Lhermitte-Duclos disease, trichilemmomas, acral keratoses, and papillomatous papules) relative to those without any of these features (p = 0.02).
To examine the predictive value each miRNA might contribute to the CS phenotypes observed in PTEN-mutation-negative patients, we performed CART analysis in an attempt to identify subgroups of patients at higher risk of developing four major clinical features associated with this syndrome (breast cancer, endometrial cancer, thyroid cancer, and macrocephaly) by using relative expression values for both miR-19a and miR-21. Additional CART analysis was performed for the CS and CSL phenotypes. Approximately 76% of mutation-negative patient samples with relative miR-19a expression > 2.26 had developed breast cancer. Similarly, 70% of those with high miR-21 expression (2−ΔΔCT > 1.81) also developed breast cancer. Although the total number of mutation-negative patients included in our study who developed endometrial cancer was small (N = 16), none of these patients were among the subgroups identified with relatively low miR-19a and miR-21 expression (2−ΔΔCT < 0.87 and < 0.64, respectively).
Macrocephaly was observed in 64% of PTEN-mutation-negative patients with miR-19a expression > 2.60, and this feature was observed in 71% of patients with miR-21 expression > 1.94. Analysis of the CS and CSL phenotypes revealed that 72% of mutation-negative patients with high miR-19a expression (2−ΔΔCT > 2.32) had classic CS, whereas 67% of those with low miR-19a expression (2−ΔΔCT < 0.75) had CSL. Similarly, 79% of patients with high miR-21 expression (2−ΔΔCT > 1.56) had CS. CART analysis was not able to identify any unique clusters of patients with thyroid cancer among the mutation-negative patients with the use of either miRNA as a predictor.
Each phenotype was also assessed with the use of the relative expression of both miRNAs jointly as phenotypic predictors. Although expression of neither miRNA was predictive of thyroid cancer, as was the case when each was considered separately, high miR-21 expression proved to be the single strongest predictor for endometrial cancer, macrocephaly, and the CS or CSL phenotype given that the joint analysis with both miR-19a and miR-21 together identified the same clusters as did miR-21 considered independently.
Discussion
Alternate mechanisms of PTEN dysfunction are becoming increasingly germane in the PTEN hamartoma tumor syndromes, particularly in CS and BRRS.21–24 Our investigation of two PTEN-targeting miRNAs, miR-19a and miR-21, suggests that their differential expression modulates PTEN protein levels and the CS or CSL phenotype in patients with these syndromes, irrespective of their PTEN mutation status.
Variable PTEN protein levels were inversely correlated with miR-19a and miR-21 expression levels in PTEN-mutation-positive patients and, more specifically, only in patients with R130X and/or R233X mutations. In patients with the R335X mutation, in which the relationship among miRNA expression, PTEN protein levels, and clinical diagnosis was absent, seven of ten had classic CS phenotypic features. This observation suggests that the R335X PTEN genotype strongly influences phenotype, and indeed, this is corroborated by the miR-19a- and/or miR-21-independent overall decreased full-length PTEN protein levels in this group of patients with the R335X mutation. In contrast, we believe that differential expression of miR-19a and miR-21, directly correlated with PTEN protein levels in R130X and/or R233X mutation carriers, could help modulate phenotype beyond the germline mutations.
Similarly, both miRNAs were differentially expressed in a series of PTEN-mutation-negative patients with CS and CSL with variable clinical phenotypes and decreased expression of full-length PTEN protein. Importantly, decreased expression of PTEN transcript levels was not observed in these mutation-negative patients, suggesting that their decreased PTEN protein expression is probably due to dysregulation at the protein level. Taken together, our data in both mutation-positive and mutation-negative patients demonstrate that miR-19a and miR-21 can modulate PTEN protein levels. Because PTEN's sufficient activity is dependent on its protein levels, perturbation of its expression can enhance disease progression and facilitate tumorigenesis.25 Therefore, our data suggest essential roles for miR-19a and miR-21 in the modulation of the diverse clinical and molecular phenotypes observed in CS.
The identification of clear genotype-phenotype correlations has, for the most part, proven elusive in CS and prompted speculation that other loci contribute to the variable clinical spectrum observed in this syndrome.9,20 Although there is a lack of genetic heterogeneity in CS, recent studies have suggested that other genetic factors, such as modifier loci, contribute to disease susceptibility and the variable phenotypes observed in patients with this disorder.1,9,26,27 Interestingly, a study by Freeman et al. demonstrated that phenotypic differences, including the onset and incidence of tumor formation, differed among mice with identical PTEN mutations yet with different genetic backgrounds.26 These differences correspond well with the vast clinical spectrum common in patients with CS and BRRS, suggesting that genetic modifiers, which probably include other genes, miRNAs, and proteins (for example PICT-1), account for the observed differences in human patients.28 Our data strongly suggest that miR-19a and miR-21, at least in part, contribute to this phenotypic variability.
Interestingly, in accordance with this, neither miR-19a nor miR-21 were differentially expressed in patients with the BRRS phenotype. Although we interpret this with caution because of the small number of patients with BRRS included in our study, this observation suggests that although these disorders are allelic, their underlying modifiers differ. If this can be replicated in a larger series of patients with BRRS, then it is tempting to speculate that miR-19a and/or miR-21 modulation of PTEN leads to CS or CSL, whereas the absence of this modulatory influence, probably together with other mechanisms, results in the BRRS phenotype.
Although the results of our investigation of miR-19a and miR-21 in patients with CS and CSL are highly suggestive of their role in these diseases, the precise mechanism by which these miRNAs are deregulated in these patients is currently unknown. Both miRNAs localize to chromosomal regions in which loss of heterozygosity in sporadic cancer has been described.27,29–31 However, these loci have not been genetically linked to CS, CSL, or BRRS. Furthermore, although genetic polymorphisms at miRNA-binding sites have recently been shown to alter miRNA:target gene interactions, no genetic alterations have been identified in PTEN's 3′ untranslated region (UTR) near miR-19a's reported binding site (miR-21's precise binding site is unknown).1,3,32 Although these alterations are likely to result in the loss of miRNA function, it is interesting to speculate whether genetic variations in PTEN's 3′UTR further contribute to the variable phenotypic spectrum of CS and CSL by altering the binding of other potential miRNAs. To begin to address this question, we recently screened PTEN's 3′UTR for genetic alterations in a subset of PTEN-mutation-negative patients with decreased full-length PTEN protein expression. However, we failed to identify any sequence differences in these individuals (Zbuk and Eng, unpublished data). Given these data, we hypothesize that the differential expression of miR-19a and miR-21 that we observed in our patients with CS or CSL probably results from the dysregulation of their expression, perhaps through the oncogenic activation of miR-19a's and miR-21's primary miRNA or through the aberrant processing of their mature miRNA. Although the mechanisms that contribute to the oncogenic activation of these miRNAs are currently unknown, our data suggest that, both in the presence and absence of specific PTEN mutations, this dysregulation modulates disease phenotype. On the basis of our findings, additional investigation for better understanding of these potential mechanisms in patients with CS and CSL is warranted.
Our study is the first patient-oriented study to examine miR-19a and miR-21 expression in CS, CSL, and BRRS. Along with other, yet to be identified, genetic modifiers, these miRNAs might contribute to disease susceptibility and phenotypic modulation and could serve as potential biomarkers of phenotypic variation associated with each syndrome. It is our hope that these findings will improve our understanding of the pathogenesis of CS, CSL, and BRRS in patients, both in those with defined germline PTEN mutations and in those for which traditional mutational-scanning methodologies have been unable to uncover a genetic etiology. We believe that an improved understanding of the role of these, and other, modifier loci in CS, CSL, and BRRS will enable more accurate clinical and molecular phenotyping and lead to advances in both the diagnostic and the preventive care afforded to patients with these and related syndromes as well as with sporadic neoplasias in which the PTEN pathway is germane.
Supplemental Data
One supplemental figure and one supplemental table are available with this article online at http://www.ajhg.org/.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.gov/Omim/
National Comprehensive Cancer Network, www.nccn.org
Genomic Medicine Biorepository, www.lerner.ccf.org/gmi/gmb/methods.php
R software package, http://cran.r-project.org
Acknowledgments
MGP would like to thank Kevin Zbuk, Meng Xu, and Jeanie Na for helpful discussions. The authors thank Tammy Sadler for expert technical assistance during the preparation of Figure S1. This work has been partially supported by the American Cancer Society, Atlanta, GA (RSG-02-151-01-CCE to C.E.) and the National Cancer Institute, Bethesda, MD (1P01CA124570-01A1 to C.E.). M.G.P. is a predoctoral fellow in the Cleveland Clinic Lerner Research Institute and is also a graduate student of the Integrated Biomedical Sciences Graduate Program of The Ohio State University. C.E. is a recipient of the Doris Duke Distinguished Clinical Scientist Award and is the Sondra J. and Stephen R. Hardis Endowed Chair of Cancer Genomic Medicine at the Cleveland Clinic.
References
- 1.Zbuk K.M., Eng C. Cancer phenomics: RET and PTEN as illustrative models. Nat. Rev. Cancer. 2007;7:35–45. doi: 10.1038/nrc2037. [DOI] [PubMed] [Google Scholar]
- 2.Zhou X.P., Waite K.A., Pilarski R., Hampel H., Fernandez M.J., Bos C., Dasouki M., Feldman G.L., Greenberg L.A., Ivanovich J. Germline PTEN promoter mutations and deletions in Cowden/Bannayan-Riley-Ruvalcaba syndrome result in aberrant PTEN protein and dysregulation of the phosphoinositol-3-kinase/Akt pathway. Am. J. Hum. Genet. 2003;73:404–411. doi: 10.1086/377109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eng C. PTEN: one gene, many syndromes. Hum. Mutat. 2003;22:183–198. doi: 10.1002/humu.10257. [DOI] [PubMed] [Google Scholar]
- 4.Maehama T., Dixon J.E. PTEN: a tumour suppressor that functions as a phospholipid phosphatase. Trends Cell Biol. 1999;9:125–128. doi: 10.1016/s0962-8924(99)01519-6. [DOI] [PubMed] [Google Scholar]
- 5.Myers M.P., Pass I., Batty I.H., Van der Kaay J., Stolarov J.P., Hemmings B.A., Wigler M.H., Downes C.P., Tonks N.K. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc. Natl. Acad. Sci. USA. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stambolic V., Suzuki A., de la Pompa J.L., Brothers G.M., Mirtsos C., Sasaki T., Ruland J., Penninger J.M., Siderovski D.P., Mak T.W. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 7.Waite K.A., Eng C. BMP2 exposure results in decreased PTEN protein degradation and increased PTEN levels. Hum. Mol. Genet. 2003;12:679–684. [PubMed] [Google Scholar]
- 8.Weng L.P., Smith W.M., Dahia P.L., Ziebold U., Gil E., Lees J.A., Eng C. PTEN suppresses breast cancer cell growth by phosphatase activity-dependent G1 arrest followed by cell death. Cancer Res. 1999;59:5808–5814. [PubMed] [Google Scholar]
- 9.Marsh D.J., Kum J.B., Lunetta K.L., Bennett M.J., Gorlin R.J., Ahmed S.F., Bodurtha J., Crowe C., Curtis M.A., Dasouki M. PTEN mutation spectrum and genotype-phenotype correlations in Bannayan-Riley-Ruvalcaba syndrome suggest a single entity with Cowden syndrome. Hum. Mol. Genet. 1999;8:1461–1472. doi: 10.1093/hmg/8.8.1461. [DOI] [PubMed] [Google Scholar]
- 10.Calin G.A., Dumitru C.D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan J.A., Krichevsky A.M., Kosik K.S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 12.Iorio M.V., Ferracin M., Liu C.G., Veronese A., Spizzo R., Sabbioni S., Magri E., Pedriali M., Fabbri M., Campiglio M. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 13.Metzler M., Wilda M., Busch K., Viehmann S., Borkhardt A. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer. 2004;39:167–169. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- 14.Michael M.Z., van Holst S.M.O.C., Pellekaan N.G., Young G.P., James R.J. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 15.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 16.Meng F., Henson R., Lang M., Wehbe H., Maheshwari S., Mendell J.T., Jiang J., Schmittgen T.D., Patel T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 17.Calin G.A., Liu C.G., Sevignani C., Ferracin M., Felli N., Dumitru C.D., Shimizu M., Cimmino A., Zupo S., Dono M. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl. Acad. Sci. USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S.T., Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C[T]) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Marsh D.J., Coulon V., Lunetta K.L., Rocca-Serra P., Dahia P.L., Zheng Z., Liaw D., Caron S., Duboue B., Lin A.Y. Mutation spectrum and genotype-phenotype analyses in Cowden disease and Bannayan-Zonana syndrome, two hamartoma syndromes with germline PTEN mutation. Hum. Mol. Genet. 1998;7:507–515. doi: 10.1093/hmg/7.3.507. [DOI] [PubMed] [Google Scholar]
- 21.Minaguchi T., Waite K.A., Eng C. Nuclear localization of PTEN is regulated by Ca(2+) through a tyrosil phosphorylation-independent conformational modification in major vault protein. Cancer Res. 2006;66:11677–11682. doi: 10.1158/0008-5472.CAN-06-2240. [DOI] [PubMed] [Google Scholar]
- 22.Pezzolesi M.G., Zbuk K.M., Waite K.A., Eng C. Comparative genomic and functional analyses reveal a novel cis-acting PTEN regulatory element as a highly conserved functional E-box motif deleted in Cowden syndrome. Hum. Mol. Genet. 2007;16:1058–1071. doi: 10.1093/hmg/ddm053. [DOI] [PubMed] [Google Scholar]
- 23.Sarquis M.S., Agrawal S., Shen L., Pilarski R., Zhou X.P., Eng C. Distinct expression profiles for PTEN transcript and its splice variants in Cowden syndrome and Bannayan-Riley-Ruvalcaba syndrome. Am. J. Hum. Genet. 2006;79:23–30. doi: 10.1086/504392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teresi R.E., Zbuk K.M., Pezzolesi M.G., Waite K.A., Eng C. Cowden syndrome-affected patients with PTEN promoter mutations demonstrate abnormal protein translation. Am. J. Hum. Genet. 2007;81:756–767. doi: 10.1086/521051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waite K.A., Eng C. Protean PTEN: form and function. Am. J. Hum. Genet. 2002;70:829–844. doi: 10.1086/340026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freeman D., Lesche R., Kertesz N., Wang S., Li G., Gao J., Groszer M., Martinez-Diaz H., Rozengurt N., Thomas G. Genetic background controls tumor development in PTEN-deficient mice. Cancer Res. 2006;66:6492–6496. doi: 10.1158/0008-5472.CAN-05-4143. [DOI] [PubMed] [Google Scholar]
- 27.Nelen M.R., Padberg G.W., Peeters E.A., Lin A.Y., van den Helm B., Frants R.R., Coulon V., Goldstein A.M., van Reen M.M., Easton D.F. Localization of the gene for Cowden disease to chromosome 10q22–23. Nat. Genet. 1996;13:114–116. doi: 10.1038/ng0596-114. [DOI] [PubMed] [Google Scholar]
- 28.Okahara F., Ikawa H., Kanaho Y., Maehama T. Regulation of PTEN phosphorylation and stability by a tumor suppressor candidate protein. J. Biol. Chem. 2004;279:45300–45303. doi: 10.1074/jbc.C400377200. [DOI] [PubMed] [Google Scholar]
- 29.Marsh D.J., Dahia P.L., Zheng Z., Liaw D., Parsons R., Gorlin R.J., Eng C. Germline mutations in PTEN are present in Bannayan-Zonana syndrome. Nat. Genet. 1997;16:333–334. doi: 10.1038/ng0897-333. [DOI] [PubMed] [Google Scholar]
- 30.Eccles D.M., Russell S.E., Haites N.E., Atkinson R., Bell D.W., Gruber L., Hickey I., Kelly K., Kitchener H., Leonard R. Early loss of heterozygosity on 17q in ovarian cancer. The Abe Ovarian Cancer Genetics Group. Oncogene. 1992;7:2069–2072. [PubMed] [Google Scholar]
- 31.Lin Y.W., Sheu J.C., Liu L.Y., Chen C.H., Lee H.S., Huang G.T., Wang J.T., Lee P.H., Lu F.J. Loss of heterozygosity at chromosome 13q in hepatocellular carcinoma: identification of three independent regions. Eur. J. Cancer. 1999;35:1730–1734. doi: 10.1016/s0959-8049(99)00205-1. [DOI] [PubMed] [Google Scholar]
- 32.Wang G., van der Walt J.M., Mayhew G., Li Y.J., Zuchner S., Scott W.K., Martin E.R., Vance J.M. Variation in the miRNA-433 binding site of FGF20 confers risk of Parkinson disease by overexpression of alpha-synuclein. Am. J. Hum. Genet. 2008;82:283–289. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


