Abstract
Lumbar-disc degeneration (LDD) is a polygenic disease. Susceptibility genes reported so far are mainly extracellular matrix proteins. D14 allele of asporin (ASPN) is associated with osteoarthritis (OA). Candidate-gene association studies showed that the D14 allele is also significantly associated with LDD in Chinese and Japanese individuals. Meta-analysis showed that individuals harboring a D14 allele had higher risk with a summary odds ratio of 1.70 (p = 0.000013). ASPN expression in vertebral discs increased with age and degeneration. Our results indicate ASPN is a LDD gene in Asians, and common risk factors may be considered for OA and LDD.
Main Text
Lumbar-disc degeneration (LDD) is a major cause of low back pain. Approximately 80% of the global population experience low back pain in their life time, presenting a huge medical and economical burden to society.1 There is a significant genetic contribution in LDD. Twin studies demonstrated 74% heritability on the basis of magnetic resonance imaging (MRI) of the spine.2–4 Also, genetic association studies have identified a number of risk factors (see recent review).5 The majority of these risk factors for LDD result from changes in sequences of genes encoding extracellular matrix (ECM) proteins expressed in the nucleus pulposus (inner structure) and annulus fibrosus (outer layer) of the disc, such as type IX collagen (COL9A2 and COL9A3),6–8 aggrecan (AGC1),9 and cartilage intermediate layer protein (CILP).10 These findings lead us to the hypothesis that LDD is caused by changes in the structural integrity of the intervertebral disc.
Recently, an extracellular matrix protein, asporin (ASPN), was shown to be associated with osteoarthritis (OA) of the knee.11–13 ASPN belongs to the small leucine-rich proteoglycan (SLRP) family. It contains a unique aspartate (D) residues repeat in the N terminus, which is a polymorphic region in the gene with alleles that contains D repeats ranging from 9–20 residues.14 The D14 polymorphism with 14 D residues is the risk allele identified for OA.11 Functional studies demonstrated that ASPN inhibited in vitro chondrogenesis and expression of Col2a1 and Agc1 through inhibition of TGF-β signaling, with a stronger inhibitory effect for asporin D14 over others.11 Given that OA and LDD are both degenerative diseases of skeletal joint regions, and that many of the genes expressed in cartilage are also expressed in the intervertebral disc, we hypothesize that ASPN is an excellent candidate as a susceptibility gene to LDD.
We tested for association of ASPN with LDD in two large Asian cohorts of Chinese and Japanese ethnicities. The Chinese cohort consisted of 1055 individuals recruited from the general population. LDD status was assessed by MRI, with severity of LDD graded with the Schneiderman's classification.15 Because age is a factor in LDD, we used a sliding-window method for an adjustment.16 In brief, the method normalizes the LDD score by (1) logarithmic transformation of the quantitative variable to reduce skewness, (2) definition of an age band for each individual as the age of the individual plus or minus 5 years, and (3) standardization of each individual's quantitative variable value by first subtracting the mean trait value and then dividing by the standard deviation of the trait, in the individual's age band. This normalization of LDD scores allows us to divide the cohort into two groups; individuals with normalized scores above and below the median were classified as cases (527) and controls (528), respectively. Differences in allele or genotype frequencies between groups were assessed by odds ratios and chi-square tests. The Japanese cohort is an extension of one previously published for an association with CILP.10 The cohort consisted of 608 controls and 745 cases. All LDD cases had a history of sciatica for longer than 3 months, and LDD confirmed with MRI graded with the Schneiderman's classification.15 Disc herniation was present in all cases.
We analyzed the ASPN D-repeat polymorphisms. Similar to previous findings for the association study in OA,11 the most common allele is D13, and there were no significant differences between the case (LDD) and control groups in both cohorts (Table 1). However, the D14 allele was overrepresented in the case groups (Table 1). The D14 allele is a risk allele with an odds ratio of 1.49 and 1.69 for the Chinese and Japanese cohorts, respectively (Table 1). With the D14 genotype in a dominant model, individuals carrying at least one D14 allele are at risk with an odds ratio of 1.66 and 1.79 for the Chinese and Japanese cohorts, respectively (Table 2).
Table 1.
Distribution of the D Repeat Alleles of ASPN in Chinese and Japanese Cohorts
| Cohort | Allele |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D9 | D10 | D11 | D12 | D13 | D14 | D15 | D16 | D17 | D18 | D19 | D20 | Total | |||
| Chinese | |||||||||||||||
| Case | Count | 1 | 0 | 4 | 213 | 652 | 98 | 36 | 38 | 9 | 3 | 0 | 0 | 1054 | |
| Case | % | 0.1 | 0 | 0.4 | 20.2 | 61.9 | 9.3 | 3.4 | 3.6 | 0.9 | 0.3 | 0 | 0 | 100 | |
| Control | Count | 0 | 1 | 4 | 236 | 666 | 68 | 30 | 37 | 10 | 3 | 0 | 1 | 1056 | |
| Control | % | 0 | 0.1 | 0.4 | 22.3 | 63.1 | 6.4 | 2.8 | 3.5 | 0.9 | 0.3 | 0 | 0.1 | 100 | |
| Japanese | |||||||||||||||
| Case | Count | 0 | 0 | 1 | 230 | 888 | 116 | 50 | 137 | 64 | 2 | 2 | 0 | 1490 | |
| Case | % | 0 | 0 | 0.1 | 15.4 | 59.6 | 7.8 | 3.4 | 9.2 | 4.3 | 0.1 | 0.1 | 0 | 100 | |
| Control | Count | 0 | 0 | 1 | 167 | 793 | 58 | 56 | 88 | 51 | 1 | 1 | 0 | 1216 | |
| Control | % | 0 | 0 | 0.1 | 13.7 | 65.2 | 4.8 | 4.6 | 7.2 | 4.2 | 0.1 | 0.1 | 0 | 100 | |
Statistical analyses for the D14 allele: Chinese cohort (OR = 1.49, 95%CI = 1.08-2.06, p = 0.015), Japanese cohort (OR = 1.69, 95% CI = 1.22-2.33, p = 0.0015), and meta-analysis (OR = 1.58, 95% CI = 1.26-1.99, p = 0.000081).
Table 2.
Association between the Presence of the ASPN D14 Allele and LDD
| Cohort | Case D14+/D14− (%D14+) | Control D14+/D14− (%D14+) | p Value | OR (95% CI) |
|---|---|---|---|---|
| Chinese | 94/433 (17.8) | 61/467 (11.6) | 0.0039 | 1.66 (1.17-2.35) |
| Japanese | 112/633 (15.0) | 55/553 (9.0) | 0.00087 | 1.78 (1.26-2.51) |
| Meta-Analysis | 0.000013 | 1.70 (1.35-2.20) |
“D14+” refers to subjects harboring one or two D14 alleles. “D14−” refers to subjects harboring no D14 alleles.
To integrate the results of the two populations, we performed a meta-analysis (random-effect model) by using the software “MIX.”17 This program uses Excel as a calculation and programming platform and has been validated against two major software packages, STATA and Comprehensive Meta-Analysis Version 2. This meta-analysis showed a summary odds ratio of 1.58 (95% CI = 1.26–1.99, p = 0.00081) (Table 1) for the allele frequency, and an overall odds ratio of 1.70 (95% CI = 1.35–2.20, p = 0.000013) (Table 2) for the D14 genotype in a dominant model, strongly supporting the D14 allele of ASPN as a risk factor for LDD. In the Chinese cohort, we also found a correlation of the D14 allele with the number of degenerated discs in an individual (Table 3).
Table 3.
Correlation of the Number of Degenerative Lumbar Discs and ASPN D14 Allele in the Chinese Cohort
| Individuals (%) |
||
|---|---|---|
| Number of Degenerated Lumbar Disc | D14+ | D14− |
| 0 | 21 (14) | 205 (23) |
| 1 | 45 (29) | 278 (31) |
| 2 | 43 (28) | 230 (26) |
| 3 | 26 (17) | 113 (13) |
| 4 | 11 (7) | 54 (6) |
| 5 | 9 (6) | 20 (2) |
“D14+” refers to subjects harboring one or two D14 alleles. “D14−” refers to subjects harboring no D14 alleles; p = 0.0010.
To assess ASPN in the pathogenesis of LDD, we investigated its expression in the intervertebral discs by using microarray analysis of genes expressed in cells isolated from the nucleus pulposus and annulus fibrosus, from “normal” individuals of various ages with no evidence of disc degeneration. The expression of ASPN, normalized to the average expression in 2- and 22-year-old samples, was similar and slightly above average. However, it was upregulated 3- to 5-fold in 46-year-old nucleus pulposus and annulus fibrosus cells, with a corresponding downregulation of COL2A1 and AGC1 expression with age (Figure 1A). Real-time RT-PCR also showed an upregulated ASPN expression in degenerative discs (Figure 1B). Although the sample size is small to make definitive conclusions, there is a trend for a reduced expression of COL2A1 and AGC1 in degenerate samples, with decreased COL2A1 expression in annulus fibrosus and decreased AGC1 expression in nucleus pulposus and end-plate tissues (Figure 1B).
Figure 1.
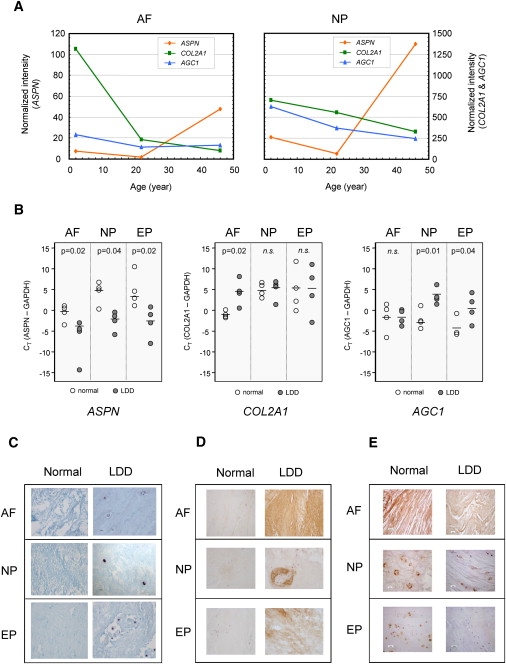
Expression of ASPN, COL2A1, AGC1, and TGF-β1 in Normal and LDD Discs
(A) Relative expression levels of ASPN, COL2A1, and AGC1 in disc cells from microarray analysis with the CODELINK plateform.
(B) Dot plots of real-time RT-PCR analysis of ASPN, COL2A1, and AGC1 expression in normal (n = 4, age 16–39) and LDD tissues (n = 4, age 21–49). The bar represents the average value in each group. A p value is given for a statistical difference between normal and LDD groups, and n.s. denotes nonsignificance.
(C) ASPN in situ hybridization.
(D and E) Immunostaining of (D) ASPN and (E) TGF-β1, in “normal” nondegenerative (35-year-old) and LDD (40-year-old) disc tissues. The following abbreviations are used: AF, annulus fibrosus; NP, nucleus pulposus; and EP, end plate.
ASPN expression was further evaluated by in situ hybridization, comparing “normal” nondegenerative (16–35 years) and degenerative (20–49 years) discs obtained from surgery. ASPN mRNA was clearly detectable in cells from degenerative disc samples, but only traces appeared in normal samples (Figure 1C). Immunostaining with an antibody specific for ASPN also showed a stronger staining for all degenerative discs analyzed compared to “normal” nondegenerative tissues (Figure 1D). In “normal” nondegenerative intervertebral discs, TGF-β1 can be detected in the cartilage end plate, nucleus pulposus, and annulus fibrosus (Figure 1E), in regions where ASPN is also localized. Interestingly, there is a trend for a lower staining intensity in LDD disc samples when compared with “normal” nondegenerative samples (Figure 1E).
Given that previous in vitro studies have demonstrated ASPN can exert a negative effect on TGF-β signaling through direct interaction,11,18 and that we have shown ASPN is upregulated with age and degeneration, it is reasonable to suggest that an inhibitory effect on cell function would occur if ASPN is overexpressed in intervertebral disc tissues, and individuals with the D14 allele could be compromised further and would become more susceptible to LDD. The significance of ASPN in the regulation of TGF-β signaling for intervertebral disc function is not clear, but our data together with previous finding for an association of CILP10 to LDD suggest that factors that alter TGF-β signaling may contribute to the pathogenesis of LDD.
Of particular relevance is the phenotype definition for LDD in the two cohorts, in which the definition in the Chinese cohort is independent of symptoms relating to pain, whereas the Japanese cohort is dependent on painful disc herniations. This suggests that the ASPN association is relevant across a broader spectrum of the disease. In the Chinese cohort, there is a trend for an association between ASPN and severity of LDD. A smaller p value for an association of the D14 allele in the Japanese cohort, in which the phenotype definition correlated with severe LDD, supported this observation. The significance of ASPN in LDD in non-Asian populations awaits replication studies of additional large cohorts.
The finding that the same alleles predispose to cartilage degeneration in different cartilage types is of particular interest, suggesting the presence of common pathomechanisms for degenerative “cartilage diseases.” Similar scenario may be applicable to other susceptibility genes of OA and LDD. Thus, systematic comparative association analyses of susceptibility genes for OA, LDD, and other degenerative cartilage diseases will be informative, with the possibility of developing common therapeutic and preventative treatments.
Acknowledgments
We thank Ms. Pei Yu for coordinating blood collection and assistance in MRI and questionnaire acquisition. This work was supported by grants from the University Grants Committee of Hong Kong (AoE/M-04/04) and grant-in-aid from Ministry of Education, Culture, Sports and Science of Japan (Contract grant number: 19209049).
References
- 1.Andersson G.B. Epidemiological features of chronic low-back pain. Lancet. 1999;354:581–585. doi: 10.1016/S0140-6736(99)01312-4. [DOI] [PubMed] [Google Scholar]
- 2.Battie M.C., Videman T., Gibbons L.E., Fisher L.D., Manninen H., Gill K. 1995 Volvo Award in clinical sciences. Determinants of lumbar disc degeneration. A study relating lifetime exposures and magnetic resonance imaging findings in identical twins. Spine. 1995;20:2601–2612. [PubMed] [Google Scholar]
- 3.Matsui H., Kanamori M., Ishihara H., Yudoh K., Naruse Y., Tsuji H. Familial predisposition for lumbar degenerative disc disease. A case-control study. Spine. 1998;23:1029–1034. doi: 10.1097/00007632-199805010-00013. [DOI] [PubMed] [Google Scholar]
- 4.Sambrook P.N., MacGregor A.J., Spector T.D. Genetic influences on cervical and lumbar disc degeneration: a magnetic resonance imaging study in twins. Arthritis Rheum. 1999;42:366–372. doi: 10.1002/1529-0131(199902)42:2<366::AID-ANR20>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 5.Chan D., Song Y., Sham P., Cheung K.M. Genetics of disc degeneration. Eur. Spine J. 2006;15(Suppl 1):317–325. doi: 10.1007/s00586-006-0171-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Annunen S., Paassilta P., Lohiniva J., Perala M., Pihlajamaa T., Karppinen J., Tervonen O., Kroger H., Lahde S., Vanharanta H. An allele of COL9A2 associated with intervertebral disc disease. Science. 1999;285:409–412. doi: 10.1126/science.285.5426.409. [DOI] [PubMed] [Google Scholar]
- 7.Jim J.J., Noponen-Hietala N., Cheung K.M., Ott J., Karppinen J., Sahraravand A., Luk K.D., Yip S.P., Sham P.C., Song Y.Q. The TRP2 allele of COL9A2 is an age-dependent risk factor for the development and severity of intervertebral disc degeneration. Spine. 2005;30:2735–2742. doi: 10.1097/01.brs.0000190828.85331.ef. [DOI] [PubMed] [Google Scholar]
- 8.Paassilta P., Lohiniva J., Goring H.H., Perala M., Raina S.S., Karppinen J., Hakala M., Palm T., Kroger H., Kaitila I. Identification of a novel common genetic risk factor for lumbar disk disease. JAMA. 2001;285:1843–1849. doi: 10.1001/jama.285.14.1843. [DOI] [PubMed] [Google Scholar]
- 9.Kawaguchi Y., Osada R., Kanamori M., Ishihara H., Ohmori K., Matsui H., Kimura T. Association between an aggrecan gene polymorphism and lumbar disc degeneration. Spine. 1999;24:2456–2460. doi: 10.1097/00007632-199912010-00006. [DOI] [PubMed] [Google Scholar]
- 10.Seki S., Kawaguchi Y., Chiba K., Mikami Y., Kizawa H., Oya T., Mio F., Mori M., Miyamoto Y., Masuda I. A functional SNP in CILP, encoding cartilage intermediate layer protein, is associated with susceptibility to lumbar disc disease. Nat. Genet. 2005;37:607–612. doi: 10.1038/ng1557. [DOI] [PubMed] [Google Scholar]
- 11.Kizawa H., Kou I., Iida A., Sudo A., Miyamoto Y., Fukuda A., Mabuchi A., Kotani A., Kawakami A., Yamamoto S. An aspartic acid repeat polymorphism in asporin inhibits chondrogenesis and increases susceptibility to osteoarthritis. Nat. Genet. 2005;37:138–144. doi: 10.1038/ng1496. [DOI] [PubMed] [Google Scholar]
- 12.Valdes A.M., Loughlin J., Oene M.V., Chapman K., Surdulescu G.L., Doherty M., Spector T.D. Sex and ethnic differences in the association of ASPN, CALM1, COL2A1, COMP, and FRZB with genetic susceptibility to osteoarthritis of the knee. Arthritis Rheum. 2007;56:137–146. doi: 10.1002/art.22301. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura T., Shi D., Tzetis M., Rodriguez-Lopez J., Miyamoto Y., Tsezou A., Gonzalez A., Jiang Q., Kamatani N., Loughlin J. Meta-analysis of association between the ASPN D-repeat and osteoarthritis. Hum. Mol. Genet. 2007;16:1676–1681. doi: 10.1093/hmg/ddm115. [DOI] [PubMed] [Google Scholar]
- 14.Lorenzo P., Aspberg A., Onnerfjord P., Bayliss M.T., Neame P.J., Heinegard D. Identification and characterization of asporin. a novel member of the leucine-rich repeat protein family closely related to decorin and biglycan. J. Biol. Chem. 2001;276:12201–12211. doi: 10.1074/jbc.M010932200. [DOI] [PubMed] [Google Scholar]
- 15.Schneiderman G., Flannigan B., Kingston S., Thomas J., Dillin W.H., Watkins R.G. Magnetic resonance imaging in the diagnosis of disc degeneration: Correlation with discography. Spine. 1987;12:276–281. doi: 10.1097/00007632-198704000-00016. [DOI] [PubMed] [Google Scholar]
- 16.Virtanen I.M., Song Y.Q., Cheung K.M.C., Ala-Kokko L., Karppinen J., Ho D.W.H., Luk K.D.K., Yip S.P., Leong J.C.Y., Cheah K.S.E. Phenotypic and population differences in the association between CILP and lumbar disc disease. J. Med. Genet. 2007;44:285–288. doi: 10.1136/jmg.2006.047076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bax L., Yu L.M., Ikeda N., Tsuruta H., Moons K.G. Development and validation of MIX: Comprehensive free software for meta-analysis of causal research data. BMC Med. Res. Methodol. 2006;6:50. doi: 10.1186/1471-2288-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakajima M., Kizawa H., Saitoh M., Kou I., Miyazono K., Ikegawa S. Mechanisms for asporin function and regulation in articular cartilage. J. Biol. Chem. 2007;282:32185–32192. doi: 10.1074/jbc.M700522200. [DOI] [PubMed] [Google Scholar]


