Abstract
Human apolipoprotein A-IV (apoA-IV) is a 46-kDa exchangeable plasma protein with many proposed functions. It is involved in chylomicron assembly and secretion, protection from atherosclerosis through a variety of mechanisms, and inhibition of food intake. There is little structural basis for these proposed functions due to the lack of a solved three-dimensional structure of the protein by x-ray crystallography or NMR. Based on previous studies, we hypothesized that lipid-free apoA-IV exists in a helical bundle, like other apolipoprotein family members and that regions near the N and C termini may interact. Utilizing a homobifunctional lysine cross-linking agent, we identified 21 intramolecular cross-links by mass spectrometry. These cross-links were used to constrain the building of a sequence threaded homology model using the I-TASSER server. Our results indicate that lipid-free apoA-IV does indeed exist as a complex helical bundle with the N and C termini in close proximity. This first structural model of lipid-free apoA-IV should prove useful for designing studies aimed at understanding how apoA-IV interacts with lipids and possibly with unknown protein partners.
Apolipoprotein A-IV (apoA-IV)2 is a 46-kDa plasma protein with myriad proposed functions. In humans, it is primarily produced in the small intestine in response to lipid-rich meals and seems to be involved in the assembly and secretion of chylomicrons (1, 2). Although the apoA-IV knock-out mouse displayed relatively normal lipid absorption on a normal chow diet (3), the importance of apoA-IV during periods of high lipid intake is beginning to become evident. For example, apoA-IV has been shown to facilitate the assembly of very large chylomicrons during high lipid stress in a cell culture model of lipoprotein secretion (4), possibly by regulating the transit rate of the nascent particles through the endoplasmic reticulum (5). The protein has also been postulated to play many roles in the reverse cholesterol transport system (6), a process important for protection from cardiovascular disease whereby excess cholesterol is removed from the vessel wall. ApoA-IV can accept cholesterol similarly to apoA-I via ABCA1 (ATP-binding cassette transporter A1) (7). It can activate lecithin:cholesterol acyltransferase and cholesterol ester transfer protein (8, 9) and bind to hepatocellular membranes (10, 11) possibly to deliver high density lipoprotein cholesterol to the liver. ApoA-IV has also been shown to possess anti-inflammatory and antioxidative properties (12–15) and may even help modulate food intake (16).
Like many of the exchangeable apolipoproteins, apoA-IV can exist in at least two states in plasma, lipid-bound and lipid-free. It is associated with newly secreted chylomicrons in the intestinal lymph, but it rapidly disassociates from these upon the initiation of lipolysis in the plasma compartment (17). Thus, the majority of human plasma apoA-IV is found distributed between high density lipoproteins and a lipid-free (or lipid-poor) fraction. The actual distribution between these in plasma is controversial, with different groups obtaining widely differing estimates, probably due to the separation technique used (18). The 376-amino acid sequence is dominated by 22-mer amino acid repeats predicted to form the amphipathic α-helices that are a hallmark of the apolipoprotein family (19). Being the longest exchangeable apolipoprotein, apoA-IV has the most repeats at 13 (20). The first repeat is predicted to be a G* helix with characteristics similar to those found in globular proteins. The remaining helices are predicted to be either A- or Y-type amphipathic α-helices (differing in the distribution of charged residues in the polar face (21)) and are characteristic of lipid-interacting proteins. Despite the prevalence of these repeats in apoA-IV, it exhibits a more labile lipid binding affinity than other apolipoproteins. This was originally thought to be due to an overall lower hydrophobic moment of the apoA-IV repeats versus those in apoA-I, for example (1). However, our recent work has demonstrated that a single amino acid mutation in a region outside of these repeats can dramatically transform apoA-IV into a more effective lipid binder than apoA-I. Using a series of biophysical and functional experiments, we provided evidence that a region near position 333 in the C terminus of apoA-IV may participate in an intramolecular interaction that masks a lipid-binding site located in the N-terminal portion of the protein (22). We have hypothesized that this interaction may toggle the protein between the lipid-free and lipid-bound states, potentially to modulate its function. Indeed, after its roles in chylomicron metabolism are apparently accomplished, lipid-free plasma apoA-IV does not seem to be immediately catabolized in circulation.
To derive a more complete structural context for how this interaction may modulate apoA-IV lipid affinity, we have set out to derive a structural model for lipid-free apoA-IV. Since an x-ray crystal or NMR structure is not yet available, we have employed cross-linking chemistry, mass spectrometry, and homology modeling techniques to derive the first model of its structure. The resulting low resolution model supports our proposed intramolecular interaction and provides a basis for hypothesizing how apoA-IV modulates its contact with lipid.
MATERIALS AND METHODS
ApoA-IV Cross-linking and Trypsin Digestion—Recombinant human apoA-IV was expressed as described previously in BL21 Escherichia coli (23). Protein was purified using the pET30 vector's polyhistidine tag and nickel affinity chromatography. His tag was cleaved with IgA protease (Mobitec), and the protein was separated from tag by nickel chromatography. The recombinant apoA-IV was identical in primary sequence to human apoA-IV Type I (Gln at position 360) with the addition of the dipeptide, Thr-Pro, at the N terminus after cleavage of the tag. The addition of these two residues results in a slight residue numbering change versus the native protein. For example, the cross-link Lys75 × Lys79 actually corresponds to residues Lys73 and Lys77 being linked in wild-type apoA-IV. In this paper, every mention of amino acid numbers refers to native apoA-IV. Recombinant human apoA-IV has previously been compared with plasma-purified apoA-IV and found to be identical in terms of secondary structure and lipid binding (24). Proteins were dialyzed into phosphate-buffered saline (PBS), pH 7.8, and concentration was determined by the Markwell-modified Lowry method (25). Proteins for mass spectrometry were prepared similarly as previously described for similar studies on apoA-I (26). Protein was cross-linked at >1 mg/ml (23–92 μm) in PBS at 4 °C. The cross-linker used in these experiments was bis(sulfosuccinimidyl) suberate (BS3) (Pierce). Preparation and addition of the BS3 stock solution (6.5 mg/ml in PBS) to the protein solution was done within 1 min in order to minimize the competing hydrolysis reaction. Proteins were cross-linked at a molar ratio of 10:1 (BS3/apoA-IV) or 50:1 for the initial cross-linking protein concentration experiment (Fig. 1A). The reaction was carried out for periods up to 16 h at 4 °C with brief vortexing every 15 min for the first 1 h. The reaction was quenched by the addition of Tris-HCl to a final concentration of 100 mm. To ensure that the 16-h reaction time did not result in cross-links from low abundance conformations of apoA-IV, we performed a time course cross-linking experiment and found 20 of 21 cross-link masses at 30 min, 19 of 21 at 2 h, and 20 of 21 at 4 h (data not shown). The cross-linked proteins were dialyzed against 10 mm ammonium bicarbonate buffer (pH 8.1) to remove any unbound cross-linker. After lyophilization, the proteins were resuspended in 200 μl of standard Tris salt buffer and 3 m guanidine hydrochloride. Monomeric and dimeric cross-linked apoA-IV were separated in the same buffer by gel filtration chromatography using a tandem gel filtration column set-up (Superose 6-Superdex 200; GE Healthcare) controlled by a fast protein liquid chromatography system with a flow rate of 0.6 ml/min. Fractions corresponding to monomer and dimer as determined by Coomassie-stained SDS-PAGE analysis were pooled separately and dialyzed against ammonium bicarbonate. The samples were concentrated by ultrafiltration (membrane molecular weight cut-off 30,000; Millipore Corp.) to 1 mg/ml and digested with sequencing grade trypsin (catalog number V5111; Promega) at 2.5% (w/w) enzyme/apoA-IV at 37 °C overnight. The next morning, 2.5% more trypsin was added, and the protein was digested for an additional 2 h. 50–100-μg aliquots of the digested protein samples were lyophilized and stored at -20 °C until used for mass spectrometric analysis. Three separate preparations of cross-linked lipid-free apoA-IV were made on three separate days and used for collecting the five mass spectra described in this paper.
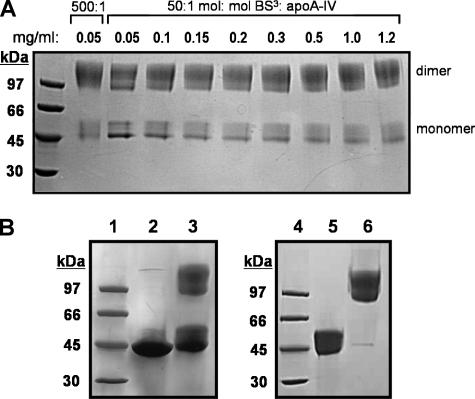
FIGURE 1.
Cross-linking lipid-free apoA-IV. A, the effect of concentration on the extent of apoA-IV dimerization. Lipid-free apoA-IV was diluted to concentrations from 0.05 to 1.2 mg/ml and cross-linked with 50:1 molar ratio BS3/apoA-IV and 500:1 molar ratio BS3/apoA-IV for the lowest concentration only. Samples were incubated for 16 h at 4 °C. The reaction was quenched, and the entire sample was analyzed by SDS-PAGE. B, cross-linking and separation of monomeric and dimeric apoA-IV for mass spectrometry analysis. Lipid-free apoA-IV was cross-linked in PBS solution using a 10:1 molar ratio of BS3 cross-linker at a concentration of 1 mg/ml in PBS for 16 h at 4 °C. After quenching, 10 μg of each sample was analyzed by SDS-PAGE. Lane 1, low molecular weight standards; lane 2, apoA-IV without BS3; lane 3, apoA-IV plus BS3. Cross-linked lipid-free apoA-IV was then purified by fast protein liquid chromatography gel filtration. Fractions corresponding to monomeric and dimeric apoA-IV were pooled separately, and 10 μg of each was run on an SDS-polyacrylamide gel. Lane 4, low molecular weight standards; lane 5, cross-linked monomeric apoA-IV; lane 6, cross-linked dimeric apoA-IV. All gels were visualized with Coomassie Blue stain.
CD Spectroscopy—Human recombinant apoA-IV was prepared as described above for cross-linking. An additional sample was treated alongside without the cross-linking agent as a control. After quenching the cross-linking reaction, the proteins were dialyzed against 20 mm phosphate buffer (pH 7.8). Protein concentration was determined using the modified Markwell-Lowry method, and sample solutions were diluted to 100 μg/ml with 20 mm phosphate buffer. Concentrations were verified by A280 measurements after sample dilution. Spectra were collected on a Jasco J-715 spectropolarimeter in a 1-mm path cell as an average of three scans. The scans were collected at a speed of 50 nm/min with a 0.5-nm step size and 0.5-s response from 260 to 190 nm. Bandwidth was set to 1 mm, and slit width was 500 μm. Mean residual ellipticity was calculated as described by Woody (27) using 115.3 as the mean residual weight for apoA-IV. Fractional helical content was calculated using the formula of Chen et al. (28) and the mean residual ellipticity at 222 nm.
Fluorescence Spectroscopy—The same or similar samples used for the CD experiments above were used to monitor Trp fluorescence. Fluorescence emission spectra of unmodified apoA-IV and BS3-cross-linked apoA-IV were recorded at room temperature on a Photon Technology International Quanta-master spectrometer. The emission spectra of unmodified and cross-linked apoA-IV were collected from 305 to 380 nm using the Trp excitation wavelength of 295 nm to minimize Tyr fluorescence. The background due to buffer was subtracted from each sample spectrum. Significance was determined using a Student's t test (p < 0.05 was considered significant).
Mass Spectrometry—Mass spectrometric measurements were performed on a Sciex/Applied Biosystems QSTAR XL mass spectrometer equipped with an electrospray ionizer and a quadropole time-of-flight dual analyzer equipped with an on-line capillary HPLC (Agilent 1100). Tryptic peptides derived from the cross-linked monomeric and dimeric samples described above were resuspended in 0.1% trifluoroacetic acid in water (2 μg/μl). 30 pmol were injected into the HPLC and separated on a C18 capillary reversed phase column (500 μm × 15 cm; Vydac). The tryptic peptide elution was carried out by application of an acetonitrile gradient of 0–40% in 60 min at a flow rate of 6.0 μl/min, which was optimized for the separation of tryptic peptides from human apoA-I. The eluting peaks were subjected to subsequent mass spectrometric detection in the range 300–1800 m/z. Automated tandem mass spectrometry (MS/MS) sequencing was carried out between 100 and 2000 m/z in Q2 pulsing mode. The instrument was externally calibrated using a CsI and [Glu1]Fibrinopeptide B (Sigma) just prior to each set of runs.
Data Analysis—For each mass spectrum collected, a mass list was generated using the AnalystQS software (Applied Biosystems, Foster City, CA). The completeness of the computer-generated mass list was verified by manual peak selection across the entire mass spectrum for the first experiment. In order to identify unmodified peptides, peptides containing a hydrolyzed cross-linker, and intrapeptide cross-links, the mass list was analyzed using GPMAW (available on the World Wide Web). Potential cross-linked peptide pairs (interpeptide cross-links) were identified using a manually constructed spreadsheet containing the masses of each possible peptide pair plus the mass of the remaining spacer arm of the cross-linker. For some analyses, a program developed in our laboratory, CrossID, was used to identify all four peptide sets, unmodified, hydrolyzed, intrapeptide cross-links, and interpeptide cross-links. Once a given mass was identified as a putative cross-linked peptide pair, the identity was confirmed by manual evaluation of the MS/MS sequence evidence. The inclusion criteria for interpeptide cross-links were as follows: 1) the mass must have appeared in at least 2 replicate experiments, and 2) the corresponding MS/MS spectrum must exhibit at least three predicted fragments ions for each of the peptides present in the putative cross-linked pair. This did not include the last y-series ion of each peptide, which has a 50% chance of being present in any case because trypsin cleaves only after Arg and Lys. In most cases, we identified over 50% of all possible fragment ions for a given peptide pair and found convincing MS/MS evidence from more than one mass spectrum for a given peptide. For intrapeptide cross-links, MS/MS evidence was typically limited due to the proximity of the cross-linker to both ends of the peptide. Therefore, intrapeptide cross-links were included if they were identified by mass within 15 ppm of the theoretical mass in at least three of five mass spectra and the actual computer peak selections were accurate.
Homology Modeling—The structure of apoA-I (Protein Data Bank code 2A01) was used to model the first 241 residues of apoA-IV (identities = 24%, positives = 49%). The modeling was performed using the Molecular Operating Environment (MOE 2006.08; The Chemical Computing Group Inc.). Ten intermediate models were created; each was subjected to an energy minimization procedure to reduce poor van der Waals contacts. The most energy-favored intermediate was retained for further consideration. Six distance restraints within the region, determined by cross-linking and mass spectrometry experiments, were then applied to bring the amino groups in the side chains of two participating lysine residues within 12 Å. This distance restraint was chosen despite being slightly longer than the spacer arm length of BS3 (11.4 Å), because the lysine side chain is flexible. An initial structural model of the C-terminal residues (residues 242–378) was generated using the I-TASSER server (29). I-TASSER was ranked as the number 1 server in the recent Critical Assessment of Techniques for Protein Structure Prediction (CASP7) competition for homology modeling and threading. The I-TASSER combines the methods of threading, ab initio modeling, and structural refinement to build reliable models. Protein structures 1av1_A (human apoA-I), 1ya9_A (mouse apoE), 1u4q_A (chicken brain α spectrin), 2fji_1 (exocyst subunit Sec6p), and 1ee4_A (yeast karyopherin (importin) α) were chosen by I-TASSER as the templates in the modeling. We found that the raw structure model from I-TASSER did not meet the experimental distance constraints. We then broke the model into several major secondary structure elements (i.e. β strands and α helices) and merged them onto the model of the N terminus (residues 1–241) guided by relevant cross-linking distance restraints. The composite model was subjected to a refining procedure. First, all backbone atoms belonging to helices were fixed, and the model was subjected to an energy minimization in MOE using AMBER-99 force field. The minimization was terminated when the root mean square gradient fell below 0.01. A second minimization was performed after distance restraints obtained from cross-link experiments were applied to the model with the distance restraint weight set to 100. A final minimization was done in MOE after all atoms were unfixed and restraints were removed. This final step, along with the lysine side chain allowance, permitted some cross-links having distances greater than 11.4 Å.
Template Homology—ApoA-IV and apoA-I share 24% identities and 49% positives without any gap in the aligned sequences. This level of identity falls within the so-called “twi-light zone.” In other words, a true homology is likely but not guaranteed. We are confident that these two proteins are truly homologous, because both proteins have an apolipoprotein AI/A4/E domain and are functionally related. In addition, their sequences contain several 22-residue repeats that form a set of α helices. Moreover, our modeling was guided by the distance constraints determined by cross-links and mass spectrometry experiments. It is worth pointing out that there is no clear line of the minimal degree of homology that can result in the same protein fold and function. For example, Vitreoscilla stercoraria hemoglobin and Perkinsus marinus hemoglobin share only 8% sequence identity, but they share identical overall fold and function. However, it is widely believed that functionally related proteins with a certain degree of sequence identity most likely share same protein fold.
Disulfide Formation—As described previously (22), a double cysteine mutant was designed with introduced cysteines at positions 16 and 336 (both normally serine). The protein was expressed in bacterial cells, purified, reduced with dithiothreitol, and denatured with 3 m guanidine. The protein was allowed to refold, under reducing conditions, by dialysis against standard Tris salt buffer plus 5 mm dithiothreitol. After the guanidine was removed, the dithiothreitol was removed by further dialysis against standard Tris salt buffer alone. The refolded protein was allowed to oxidize for several days, and then protein was analyzed by reducing and nonreducing SDS-PAGE and visualized using Coomassie Blue stain.
Limited Proteolysis—Chymotrypsin was added to lipid-free apoA-IV in PBS at 275:1 apoA-IV/chymotrypsin (w/w). At various times, the cleavage was quenched with 1.4 mm phenylmethylsulfonyl fluoride (final) and boiled for 3 min. 10 μg of each sample was analyzed by SDS-PAGE and visualized with Coomassie Blue stain. A small amount (3 pmol in 0.75 μl) was analyzed by electrospray ionization-liquid chromatography/mass spectrometry. Large peptide products and intact apoA-IV were identified using the Bayesian protein reconstruct function of AnalystQS and comparing the resultant experimental mass with a theoretical chymotryptic peptide list generated by GPMAW. The identity of one particular protein band, along with the intact apoA-IV band, was further confirmed by MALDI-TOF analysis of the trypsin-digested gel band.
RESULTS
Overall Strategy—In the current study, we set out to determine a molecular model of lipid-free apoA-IV in solution. To accomplish this, we combined the techniques of chemical cross-linking, mass spectrometry, and homology modeling. The cross-linker used, BS3, primarily reacts with the free amines of proximal lysine residues but can also react with the free N-terminal amine of the protein. After cross-linking, the protein is then exhaustively trypsinized, and the resulting linked peptides are identified by mass spectrometry. BS3 has a spacer arm length of 11.4 Å, so the underlying assumption is that the side chain ε-amino groups of the two residues involved lie within that distance in the native lipid-free structure. We then modeled the protein by comparing the sequence to homologous proteins of known structure and constrained the model using the cross-linking data obtained.
Conditions for Cross-linking—Our initial intention was to model the simplest form of apoA-IV, the lipid-free monomer. However, apoA-IV is known to exist as a mixture of monomers and dimers in solution (30). Self-association is a common feature of the exchangeable apolipoproteins, with the extent of oligomerization typically increasing with concentration (i.e. apoA-I). Therefore, we began by cross-linking apoA-IV across a range of concentrations from 50 μg/ml to 1.2 mg/ml in an effort to find conditions that will favor the monomeric form. Fig. 1A shows a Coomassie-stained SDS-polyacrylamide gel showing apoA-IV at each concentration after cross-linking with BS3 at a ratio of 50:1. ApoA-IV indeed existed as ∼60% dimer (∼97 kDa) and 40% monomer (∼45 kDa). Although there appeared to be a slight increase in the ratio of monomer to dimer as apoA-IV concentration decreased, substantial dimeric apoA-IV remained even at very low concentrations. This is in contrast to apoA-I, which primarily exists as a monomer below 0.1 mg/ml. Since it was not possible to study the monomer in relative isolation, we elected to cross-link apoA-IV at concentrations above 1 mg/ml. This was chosen in order to limit hydrolysis of the cross-linker in excess aqueous volume. For all subsequent experiments, apoA-IV was cross-linked using a lower 10:1 BS3/apoA-IV ratio. We then analyzed both the monomer and dimer after separation by gel filtration chromatography post-cross-linking (26).
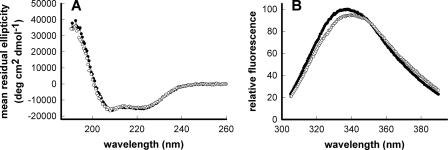
To make certain that the cross-linker did not alter the apoA-IV structure, we performed far UV-CD and Trp fluorescence measurements to determine any changes in helical content and/or conformation of apoA-IV. Fig. 2A shows the CD spectra of apoA-IV pre- and post-cross-linking. Both samples exhibited the classical spectral shape of a highly α-helical protein with two minima at 208 and 222 nm. The shape and amplitude of the curves were highly similar. Calculations described under “Materials and Methods” showed that the α-helical content of apoA-IV was 43.3% under native conditions and 41.1% after BS3 cross-linking, a difference that is well within the expected error for this technique. Fig. 2B shows the emission spectra of the single Trp residue at position 12 in apoA-IV. Maximum emission of apoA-IV with and without cross-linker was 340.8 ± 0.9 and 336.8 ± 1.3 nm, respectively (p < 0.05). Although these were statistically different, this difference is minor compared with Trp residues in highly exposed environments (about 352 nm). Taken together, these results indicate that the structure of apoA-IV was not significantly disturbed by the addition of BS3. Thus, we were confident that the distance constraints identified by cross-linking faithfully represent the native three-dimensional structure of lipid-free apoA-IV.
FIGURE 2.
Structural characterization of lipid-free apoA-IV with and without BS3 cross-linker. A, far UV-circular dichroism. Lipid-free apoA-IV with and without BS3 cross-linker was diluted to 100 μg/ml in phosphate buffer and analyzed by circular dichroism from 190 to 260 nm. Curves show data from three accumulations in a single experiment. B, tryptophan fluorescence. Samples similar to those used in A were excited at 295 nm, and Trp emission was monitored. Closed circles, unmodified apoA-IV; open circles, apoA-IV cross-linked with BS3. Curves shown are the average of three replicates from one of two similar experiments.
Fig. 1B (left) shows a Coomassie-stained SDS-polyacrylamide gel of lipid-free apoA-IV with and without cross-linker under the final conditions we chose, 10:1 BS3/apoA-IV. First, in lane 2, it is clear that the recombinant human apoA-IV we used was highly pure (about 95%). Second, in lane 3, cross-linking of apoA-IV showed that, in solution, apoA-IV exists as monomers and dimers, consistent with Fig. 1A. The protein bands in the cross-linked sample were more diffuse than might be expected for the unmodified protein, due to various locked conformations and random addition of the cross-linking agent. We then separated the cross-linked monomer from the dimer by gel filtration chromatography. Fig. 1B (right) shows purified cross-linked monomeric apoA-IV (lane 5) and dimeric apoA-IV (lane 6). These samples were independently digested with trypsin for mass spectrometry.
Mass Spectrometry—The mass spectrometric analysis of the monomeric sample generated the following general types of identified peptides: 1) unmodified tryptic peptides that did not contain an available Lys side chain for cross-linking, 2) intrapeptide cross-linked peptides containing an internal cross-link between two Lys residues on the same tryptic peptide, and 3) interpeptide linkages connecting Lys residues on two peptides separated by at least one tryptic cleavage site. In the case of the dimeric apoA-IV sample, we anticipated finding interpeptide linkages connecting Lys residues on peptides originating from two separate apoA-IV molecules. Theoretically, these should be present in the dimeric but not in the monomeric sample.
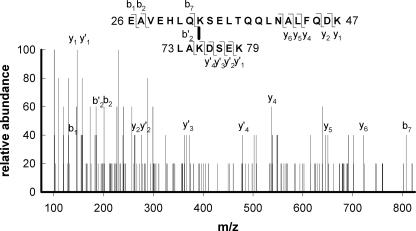
To verify the identity of putative cross-linked peptides, we employed MS/MS to collect peptide sequence data for each peptide pair. On average, for the interpeptide cross-links we screened, we identified ∼60% of all possible MS/MS b- and y-series fragment ions for the peptide pair. Fig. 3 shows an example of MS/MS data obtained for one such interpeptide cross-link. The availability of the MS/MS data virtually eliminated the possibility of misidentifying a particular peptide pair.
FIGURE 3.
An example of MS/MS characterization of the interpeptide cross-link Lys33–Lys75. The sequence of the cross-linked peptides 26–47 and 73–79 of the putatively assigned mass are shown in the inset above using their single letter amino acid abbreviations. The cross-link is shown as a vertical black bar between the two lysines involved. The b- and y-series fragment ions resulting from the +5-charged parent ion are designated.
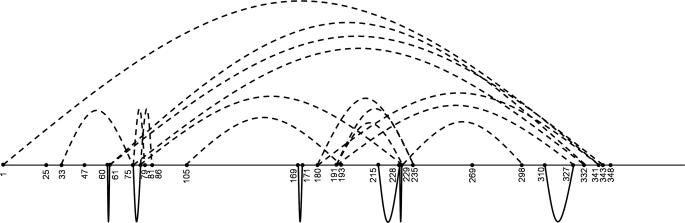
We performed three independent cross-linking experiments on similarly prepared protein samples and assembled a list of identified cross-links (see “Materials and Methods”) within monomeric lipid-free apoA-IV. Table 1 lists the intrapeptide cross-links, and Table 2 lists the interpeptide cross-links. Unfortunately, we were unable to detect interpeptide (i.e. intermolecular) cross-links in the dimeric apoA-IV sample that were unique from those found in the monomeric sample (see “Discussion”). In all, we confidently identified six intrapeptide and 15 interpeptide cross-links. A schematic representation illustrating the locations of the cross-links in Tables 1 and 2 is shown in Fig. 4. Several cross-links, particularly the intrapeptide ones, joined Lys residues in close proximity with respect to the linear sequence of the protein. However, most of the interpeptide cross-links joined sites that were far apart in the sequence with many reaching from near the N terminus all the way to the C-terminal regions of apoA-IV.
TABLE 1.
Intrapeptide peptide cross-links
| Cross-link | Peptide | Theoretical mass | Experimental mass | ppm | Distancea |
|---|---|---|---|---|---|
| Da | Da | Å | |||
| 75 × 79 | 73–81 | 1168.6701 | 1168.6855 | –13 | 14.10 |
| 228 × 229 | 216–235 | 2508.3046 | 2508.3373 | –13 | 3.93 |
| 310 × 327 | 309–330 | 2583.3444 | 2583.3838 | –15 | 20.74 |
| 60 × 61 | 48–72 | 2965.5548 | 2965.5696 | –5 | 3.87 |
| 215 × 228 | 204–229 | 3155.5961 | 3155.5965 | –0.1 | 19.30 |
| 169 × 171 | 152–180 | 3398.7315 | 3398.7774 | –14 | 6.32 |
Distance measurement calculated between the two cross-linked amino acid backbones using PyMOL
TABLE 2.
Interpeptide Cross-links
| Cross-link | Peptide 1 | Peptide 2 | Theoretical mass | Experimental mass | ppm | Distancea |
|---|---|---|---|---|---|---|
| Da | Da | Å | ||||
| 75 × 81 | 73–79 | 80–86 | 1742.9678 | 1742.9850 | –10 | 19.00 |
| 79 × 86 | 76–81 | 82–92 | 2200.1478 | 2200.1463 | 0.7 | 5.67 |
| 75 × 332 | 73–79 | 331–341 | 2246.1478 | 2246.1825 | –15 | 21.42 |
| 191 × 229 | 183–193 | 229–235 | 2278.2078 | 2278.2462 | –17 | 15.45 |
| 180 × 235 | 172–182 | 230–237 | 2367.2378 | 2367.2837 | –19 | 16.66 |
| 191 × 235 | 183–193 | 230–237 | 2377.2478 | 2377.2971 | –21 | 17.74 |
| 79 × 229 | 76–86 | 229–235 | 2399.2661 | 2399.3094 | –18 | 22.31 |
| 61 × 343 | 61–72 | 342–348 | 2439.2578 | 2439.2969 | –16 | 11.51 |
| 61 × 332 | 61–72 | 331–341 | 2895.5178 | 2895.5380 | –7 | 21.56 |
| 180 × 341 | 172–182 | 331–343 | 3169.6161 | 3169.6417 | –8 | 21.57 |
| 229 × 298 | 229–235 | 289–308 | 3360.6978 | 3360.7613 | –19 | 9.29 |
| 105 × 193 | 95–111 | 192–202 | 3369.8278 | 3369.8606 | –10 | 20.40 |
| 33 × 75 | 26–47 | 73–79 | 3495.8078 | 3495.8805 | –21 | 21.99 |
| 191 × 327 | 183–193 | 311–330 | 3636.8778 | 3636.9412 | –17 | 23.16 |
| 1 × 341 | 1–25 | 333–343 | 4271.0378 | 4271.1164 | –18 | 11.82 |
Distance measurement calculated between the two cross-linked amino acid backbones using PyMOL
FIGURE 4.
Schematic representation of interpeptide and intrapeptide cross-links identified within cross-linked monomeric apoA-IV. The horizontal line represents linear amino acid sequence with all lysines and the N terminus numbered. Interpeptide cross-links are shown as curved dotted lines above the line, and intrapeptide cross-links are shown as curved solid lines below the line.
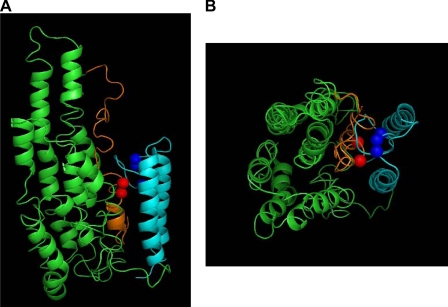
Homology Modeling—The cross-linking information described above was used to constrain the building of an all-atom, three-dimensional model of the lipid-free apoA-IV monomer. We utilized sequence-threading techniques using homologous proteins of known structure as described under “Materials and Methods.” The template proteins used for the assembly of the final model are listed under “Materials and Methods.” The template proteins were selected based on homology and by their ability to satisfy the distance constraints provided by the cross-links (26). The resulting model is shown in Fig. 5. The model indicates that lipid-free apoA-IV in solution adopts a helical bundle consisting of a total of 13 stretches of α-helix varying in length from 4 to 26 amino acids. These helices are punctuated with varying lengths of random coil. The bundle consists of nine major turns, resulting in 10 basically parallel segments composed of all 13 helices. The residues that are predicted to be helical in each of these segments are listed in Table 3. Overall, the helical bundle measures ∼37 Å across the shorter horizontal axis, 49 Å across the longer horizontal axis, and 77 Å tall. The C-terminal two helices, which form a smaller distinct set of helices, are about 40 Å tall and shown in blue. The extreme N terminus, shown in orange, is buried underneath the C-terminal helices.
FIGURE 5.
Homology model of lipid-free apoA-IV. ApoA-IV was sequence-threaded with proteins of known structure that share homology with regions of apoA-IV. Residues 1–241 were modeled using the full-length apoA-I crystal structure (2A01) using the Molecular Operating Environment. The remaining sequence was modeled using fragments of various other protein structures using the I-TASSER server, which uses threading, ab initio modeling, and structural refinement. The resulting model was broken into major secondary structure components and reassembled using the distance constraints obtained by cross-linking and mass spectrometry. The model is shown in a side view (A) and top-down view (B). The ribbon diagram of the model is shown with the majority of the protein colored green. The N-terminal 39 amino acids, encoded by a separate exon, are colored orange with amino acids Trp12 and Phe15 shown as red spheres. The C-terminal 66 amino acids are colored light blue, with residues Phe334, Phe335, and Phe338 shown as blue spheres.
TABLE 3.
Homology model-predicted helices
| Segment | Residues | Helices | Helical residues | Percentage of helix |
|---|---|---|---|---|
| % | ||||
| 1 | 1–41 | 1 | 7–12 | 12 |
| 2 | 42–89 | 2 | 53–69 | 34 |
| 3 | 90–138 | 3, 4 | 95–101 and 107–132 | 65 |
| 4 | 139–186 | 5, 6, 7 | 142–152, 163–170, and 174–179 | 47 |
| 5 | 187–213 | 8 | 208–211 | 12 |
| 6 | 214–240 | 9 | 219–240 | 81 |
| 7 | 241–273 | 10 | 261–271 | 31 |
| 8 | 274–307 | 11 | 279–286 | 21 |
| 9 | 308–335 | 12 | 313–334 | 78 |
| 10 | 351–378 | 13 | 354–372 | 67 |
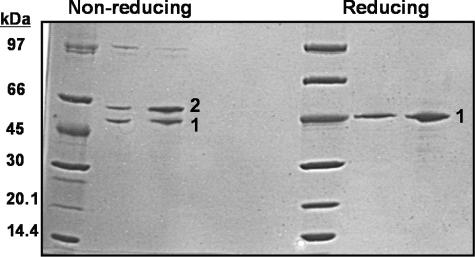
Disulfide Formation Experiment—In order to test the hypothesis that the N and C termini of apoA-IV (specifically amino acids 12, 15, and 334) reside near each other in the native lipid-free conformation, a double cysteine mutant of apoA-IV with cysteines at positions 16 and 336 was produced and refolded under reducing conditions. Reducing and nonreducing lanes of protein standards and mutant apoA-IV were run on an SDS-polyacrylamide gel (Fig. 6). Under reducing conditions (right), apoA-IV migrated as a single band at the 45-kDa protein standard. Under nonreducing conditions (left), some of the monomeric apoA-IV migrated slower (indicated by the number 2). This gel migration shift did not occur with WT protein. This indicates that the Cys residues were close together in the native structure of apoA-IV and could form a disulfide bond. This connection resulted in a change in SDS-PAGE migration of apoA-IV. In previous work, the intramolecular disulfide bond between these two residues was confirmed by mass spectrometry (22).
FIGURE 6.
Disulfide formation in double cysteine apoA-IV mutant. A double cysteine mutant of apoA-IV was produced with cysteines replacing amino acids Ser16 and Ser336. The left half of the SDS-polyacrylamide gel was run with nonreducing loading buffer, and the right half was run with reducing loading buffer. The number 1 on both sides indicates the migration of monomeric apoA-IV with no disulfide (either because the sample was reduced or because one had not formed). The number 2 in the nonreducing section indicates a slower migrating monomeric apoA-IV due to an internal disulfide between amino acids 16 and 336.
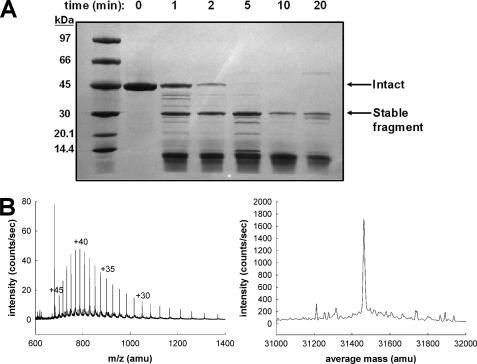
Limited Proteolysis—As an independent test of the homology model, we used limited proteolysis to determine regions of proteolytic sensitivity. Fig. 7A shows a Coomassie-stained SDS-polyacrylamide gel of the proteolysis of apoA-IV over a time range from 1 to 20 min. As intact apoA-IV was proteolyzed, one band around 31 kDa persisted as a stable fragment out to 20 min. This band was determined by electrospray ionization-liquid chromatography/mass spectrometry (Fig. 7B) to be 31463.3 Da, which matched very well with amino acids 64–335 (Mr 31461.4 Da). The identity of this fragment was confirmed by peptide mapping using MALDI-TOF mass spectrometry (83% sequence coverage) of the ∼31 kDa band cut and digested directly from the gel. Importantly, no peptides from the regions of 1–63 and 336–376 were seen in the sample.
FIGURE 7.
Limited proteolysis of lipid-free apoA-IV. Recombinant apoA-IV in solution was digested for indicated time periods with chymotrypsin. After quenching with phenylmethylsulfonyl fluoride, samples were analyzed by mass spectrometry. A, Coomassie-stained SDS-polyacrylamide gel of 10 μg of apoA-IV after various periods of proteolysis. B, mass spectra of the apoA-IV stable fragment identified to be amino acids 64–335. Left, m/z spectrum of elution time 64–65.3 min showing multiple charge states of the mass 31,463.3 Da. Several charge states are noted, but intervening ones are evident in the spectrum. Right, deconvolved spectrum of the above charge states showing the molecular weight evidence for this mass.
DISCUSSION
This study resulted in the first molecular model of full-length human apolipoprotein A-IV (apoA-IV). Structural models of several other members of the exchangeable apolipoprotein family have been reported, including full-length lipophorin III (by NMR (31)), apoA-I (by x-ray crystallography (32) and homology modeling (26)), and a fragment of apolipoprotein E (by x-ray crystallography (33)). A unifying theme among these models is the presence of a four-helix bundle of amphipathic helices, aligned with the hydrophobic faces interacting inwardly, constituting the majority of the protein structure. Surprisingly, our model predicts that apoA-IV forms a more complex helical bundle composed of nine or 10 helical segments as opposed to four. At 376 amino acids, apoA-IV is the longest of the exchangeable apolipoproteins. If it were to adopt a four-helical bundle organization, the helical segments would be substantially longer than those found in the structure of apoA-I. However, the cross-linking pattern unequivocally rules this out. It appears that shorter helical runs averaging about 40 Å in length are maintained in apoA-IV, apparently requiring a cluster of more than four helices to properly sequester hydrophobic residues into the core of the bundle.
One interesting feature of the homology model is the relative proximity of the N- and C-terminal regions of apoA-IV. We previously showed evidence for an intramolecular interaction between the regions around residues 12 and 15 in the N terminus and residue 334, which lies some 42 residues in from the C terminus (22). ApoA-IV lipid binding can be manipulated by point mutations in the N and C termini of the protein (23, 34). Fluorescence resonance energy transfer occurs between the N-terminal tryptophan (Trp12) and an acceptor probe at amino acid 336. Also, a disulfide bond forms between introduced cysteines at positions 16 and 336 without disrupting the secondary structure compared with WT (Fig. 6). We have suggested that this direct interaction could represent a conformational switch mechanism that converts the protein from a relatively poor lipid binder to a highly avid one. The homology model independently places these three residues within a radius of about 9 Å. Interestingly, Weinberg (35) predicted that human apoA-IV may contain two short regions of β sheet between residues 6–15 and 331–336, exactly corresponding with our proposed intramolecular interaction (22, 34). Since the quality and resolution of our homology model is heavily dependent on a limited number of known structural templates, our model lacks the resolution to confirm the presence or absence of these short β-sheets. However, these observations lead to the intriguing hypothesis that these two regions may form a “β-clasp” that holds these two regions together in order to modulate apoA-IV lipid affinity. A similar idea has previously been proposed for apoA-I (36).
The experimentally determined α-helical content of lipid-free apoA-IV varies considerably in the literature: 35% (37), 40% (23), 43% (38), 54% (39), 60% (22), and 68% (40). The reason for this range is not clear, but it may relate to the known sensitivity of apoA-IV conformation to even slight changes in buffer pH (39). Our final homology model puts the α-helical content of apoA-IV at 45%. This matches well with our experimentally derived value of 43% based on CD measurements (Fig. 2A) performed under similar conditions and on the same protein preparations as the cross-linking reactions used to derive the model. This value also fits relatively well with a consensus average from the literature of about 50% helicity.
In their classic studies, Segrest et al. (20) predicted regions of α-helical content in apoA-IV using a variety of algorithms. In Fig. 8, we compared the theoretical helical residues from this analysis with those predicted by our sequence threaded model. It is clear that significant differences exist between the two schemes. First, the algorithm approach predicted apoA-IV to be 85% helical, significantly higher than experimental estimates, although this may be more realistic in lipid-bound forms of apoA-IV. Another clear difference relates to the location of proline residues with respect to the helical segments. Due to its cyclic nature, proline has long been thought to delineate the beginnings and ends of α-helices. However, our model predicts that three of the 12 prolines in apoA-IV reside within a helix and that only two mark the beginning of an α-helix. This is consistent with the lack of observed helical punctuation by proline found in the crystal structure of apoA-I, in which six of 10 proline residues were within a helical segment (32).
FIGURE 8.
Comparison of helical regions predicted by the homology model with previously published secondary structure predictions for apoA-IV. The sequence of apoA-IV is aligned in rows according to the helices predicted by Segrest et al. (20). Helical regions in the current homology model are in red text. Lysines are underlined.
Our model is in agreement with several important pieces of experimental structural information generated over the years by Weinberg and colleagues (39, 41). For example, the lone Trp residue is known to be in a relatively sequestered, hydrophobic environment (39) (Fig. 2B). In the homology model, Trp12 is buried under the C-terminal domain composed of the two helices that span residues 313–372. Additionally, it was noted that the majority of Tyr residues in apoA-IV were buried, but two were relatively exposed (41). Our model supports these data in that four Tyr residues are buried and three are exposed, one of which is Tyr140 that was not addressed in Weinberg's original study (41). He also showed that nonvicinal Tyr residues can participate in energy transfer with the single Trp. Again, our model supports this assertion in that three of the seven Tyr residues are within 22 Å of Trp12 with two more being within 28 Å. Furthermore, limited proteolysis of lipid-free apoA-IV (Fig. 7) revealed a large fragment corresponding to amino acids 64–335 that was relatively resistant to proteolytic cleavage. This fragment corresponds to the last C-terminal helix being cleaved from the apoA-IV molecule at the predicted turn between helices 12 and 13 of our model. The exposed N-terminal 63 amino acids might be cleaved subsequently, consistent with the sequestered N terminus in our model and the literature.
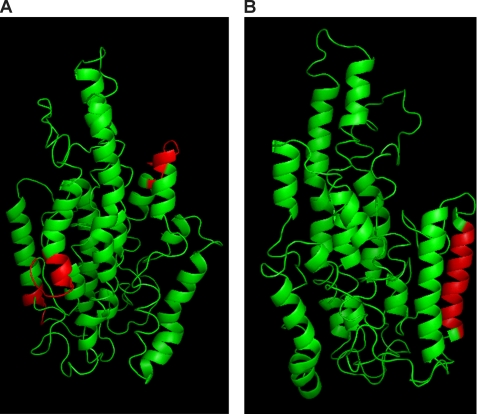
An issue that we had hoped to clarify in this study was the site of interaction mediating apoA-IV dimerization. Weinberg (35) has theorized that the dimerization occurs via hydrophobic forces and, since only monomer and dimer exist, this interaction probably occurs between two identical sites on each participating molecule. Based on sequence comparisons with rat apoA-IV, which does not dimerize, candidate interacting sites were predicted, which include the regions from 267–277, 367–372, and 62–72. Fig. 9A shows that each of these sites is predicted to be on the surface of the molecule, exposed to the aqueous environment. Residues 62–72 and 367–372 are particularly intriguing, because they are directly adjacent to each other in the structure. Unfortunately, we were unable to define the site of interaction in this study, because we could not identify intermolecular cross-links (i.e. those that appeared in the dimer sample that did not appear in the monomer). This could have resulted for two reasons. First, it may be that we were simply unable to detect the relevant cross-linked peptides, possibly because their characteristics may not lend themselves to optimal ionization in the mass spectrometer. A second explanation, which we favor, is that cross-links that form intramolecularly in the monomer are identical to those cross-links that occur intermolecularly in the dimer. For example, consider a cross-link that occurs intramolecularly between Lys residues A1 and B1 in the monomer. If, upon dimerization, Lys A1 on one molecule now can cross-link with the corresponding Lys on the second molecule of apoA-IV, Lys B2, then the cross-link pattern detected by mass spectrometry would be identical between the monomeric and dimeric samples, as we observed. This scenario may be particularly plausible, since two identical interaction sites are probably at work. It may be that the dimerization does not result in a significant conformational change in either participating monomer.
FIGURE 9.
Correlation of experimental data from the literature with surface exposed regions in the apoA-IV homology model. A, putative self-association sites predicted by Weinberg (35); 62–72, 267–277, and 367–372 are colored red (see “Discussion”). B, unique EQQQ repeat domain located in the C terminus of apoA-IV; residues 354–369 are colored red.
Using apoA-IV sequence homology among species, Weinberg found that residues 1–50, including the N-terminal G* helix, 314–338 including a class Y helix, and 354–370 including a unique EQQQ repeat domain are highly conserved (42) among mammals. Much speculation has surrounded the role of this unique EQQQ repeat. In baboons, it is extended by five repeats, and a polymorphism exists in chimpanzees in which one repeat is deleted (43). Finally, the most common human polymorphism, Q360H, which exhibits various structural and functional differences from wild-type apoA-IV, is in this region (44). Our model shows this 16-amino acid stretch as part of the last helix in apoA-IV residing on the exterior of the molecule perfectly exposed for interaction, possibly with a receptor or an unknown protein partner, as we have proposed previously. Lu et al. (4) found that deletion of this region drastically increases the size of chylomicrons secreted from cells in culture, suggesting an inhibitory role for this region. Fig. 9B highlights this stretch of amino acids in red.
Although our model matches well with the preponderance of experimental evidence, we caution that it is limited in resolution to the general organization of secondary structural elements. Obviously, information on specific side chain interactions and fine details of secondary structure must await successful studies with higher resolution techniques, such as NMR or x-ray crystallography. However, we believe that this model provides new insight and a structural framework for generating hypotheses and experiments designed to understand the structure and dynamics of this important protein. Looking forward, it will be important to understand the structural adaptations as the protein binds lipids both in high density lipoprotein-sized particles and much larger chylomicrons. These studies are currently under way in our laboratory.
Acknowledgments
We thank Dr. Apryll Stalcup for use of the spectropolarimeter for CD measurements, Kelly Rask and Dr. Ken Greis (University of Cincinnati GRI Proteomics Facility) for MALDI-TOF gel band identification, and Loren Faulkner for help with manuscript revision.
This work was supported, in whole or in part, by National Institutes of Health Grants HL67093 and HL82734 (to W. S. D.). This work was also supported by a predoctoral fellowship from the Great Rivers Affiliate of the American Heart Association (to M. R. T.) and a University of Cincinnati graduate research fellowship (to M. R. T.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: apo, apolipoprotein; PBS, phosphate-buffered saline; BS3, (bis)sulfosuccinimidyl suberate; HPLC, high performance liquid chromatograph; MS, mass spectrometry; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight.
References
- 1.Green, P. H., Glickman, R. M., Riley, J. W., and Quinet, E. (1980) J. Clin. Invest. 65 911-919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalogeris, T. J., Rodriguez, M. D., and Tso, P. (1997) J. Nutr. 127 537S-543S [DOI] [PubMed] [Google Scholar]
- 3.Weinstock, P. H., Bisgaier, C. L., Hayek, T., Aalto-Setala, K., Sehayek, E., Wu, L., Sheiffele, P., Merkel, M., Essenburg, A. D., and Breslow, J. L. (1997) J. Lipid Res. 38 1782-1794 [PubMed] [Google Scholar]
- 4.Lu, S., Yao, Y., Cheng, X., Mitchell, S., Leng, S., Meng, S., Gallagher, J. W., Shelness, G. S., Morris, G. S., Mahan, J., Frase, S., Mansbach, C. M., Weinberg, R. B., and Black, D. D. (2006) J. Biol. Chem. 281 3473-3483 [DOI] [PubMed] [Google Scholar]
- 5.Gallagher, J. W., Weinberg, R. B., and Shelness, G. S. (2004) J. Lipid Res. 45 1826-1834 [DOI] [PubMed] [Google Scholar]
- 6.Stein, O., Stein, Y., Lefevre, M., and Roheim, P. S. (1986) Biochim. Biophys. Acta 878 7-13 [DOI] [PubMed] [Google Scholar]
- 7.Remaley, A. T., Stonik, J. A., Demosky, S. J., Neufeld, E. B., Bocharov, A. V., Vishnyakova, T. G., Eggerman, T. L., Patterson, A. P., Duverger, N. J., Santamarina-Fojo, S., and Brewer, H. B., Jr. (2001) Biochem. Biophys. Res. Commun. 280 818-823 [DOI] [PubMed] [Google Scholar]
- 8.Steinmetz, A., and Utermann, G. (1985) J. Biol. Chem. 260 2258-2264 [PubMed] [Google Scholar]
- 9.Main, L. A., Ohnishi, T., and Yokoyama, S. (1996) Biochim. Biophys. Acta 1300 17-24 [DOI] [PubMed] [Google Scholar]
- 10.Ghiselli, G., Crump, W. L., III, and Gotto, A. M., Jr. (1986) Biochem. Biophys. Res. Commun. 139 122-128 [DOI] [PubMed] [Google Scholar]
- 11.Weinberg, R. B., and Patton, C. S. (1990) Biochim. Biophys. Acta 1044 255-261 [DOI] [PubMed] [Google Scholar]
- 12.Qin, X., Swertfeger, D. K., Zheng, S., Hui, D. Y., and Tso, P. (1998) Am. J. Physiol. 274 H1836-H1840 [DOI] [PubMed] [Google Scholar]
- 13.Ostos, M. A., Conconi, M., Vergnes, L., Baroukh, N., Ribalta, J., Girona, J., Caillaud, J. M., Ochoa, A., and Zakin, M. M. (2001) Arterioscler. Thromb. Vasc. Biol. 21 1023-1028 [DOI] [PubMed] [Google Scholar]
- 14.Recalde, D., Ostos, M. A., Badell, E., Garcia-Otin, A. L., Pidoux, J., Castro, G., Zakin, M. M., and Scott-Algara, D. (2004) Arterioscler. Thromb. Vasc. Biol. 24 756-761 [DOI] [PubMed] [Google Scholar]
- 15.Vowinkel, T., Mori, M., Krieglstein, C. F., Russell, J., Saijo, F., Bharwani, S., Turnage, R. H., Davidson, W. S., Tso, P., Granger, D. N., and Kalogeris, T. J. (2004) J. Clin. Invest. 114 260-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujimoto, K., Cardelli, J. A., and Tso, P. (1992) Am. J. Physiol. 262 G1002-G1006 [DOI] [PubMed] [Google Scholar]
- 17.Weinberg, R. B., and Spector, M. S. (1985) J. Lipid Res. 26 26-37 [PubMed] [Google Scholar]
- 18.Ezeh, B., Haiman, M., Alber, H. F., Kunz, B., Paulweber, B., Lingenhel, A., Kraft, H. G., Weidinger, F., Pachinger, O., Dieplinger, H., and Kronenberg, F. (2003) J. Lipid Res. 44 1523-1529 [DOI] [PubMed] [Google Scholar]
- 19.Elshourbagy, N. A., Walker, D. W., Boguski, M. S., Gordon, J. I., and Taylor, J. M. (1986) J. Biol. Chem. 261 1998-2002 [PubMed] [Google Scholar]
- 20.Segrest, J. P., Jones, M. K., De Loof, H., Brouillette, C. G., Venkatachalapathi, Y. V., and Anantharamaiah, G. M. (1992) J. Lipid Res. 33 141-166 [PubMed] [Google Scholar]
- 21.Segrest, J. P., De Loof, H., Dohlman, J. G., Brouillette, C. G., and Anantharamaiah, G. M. (1990) Proteins 8 103-117 [DOI] [PubMed] [Google Scholar]
- 22.Tubb, M. R., Silva, R. A. G. D., Pearson, K. J., Tso, P., Liu, M., and Davidson, W. S. (2007) J. Biol. Chem. 282 28385-28394 [DOI] [PubMed] [Google Scholar]
- 23.Pearson, K., Saito, H., Woods, S. C., Lund-Katz, S., Tso, P., Phillips, M. C., and Davidson, W. S. (2004) Biochemistry 43 10719-10729 [DOI] [PubMed] [Google Scholar]
- 24.Duverger, N., Murry-Brelier, A., Latta, M., Reboul, S., Castro, G., Mayaux, J. F., Fruchart, J. C., Taylor, J. M., Steinmetz, A., and Denefle, P. (1991) Eur. J. Biochem. 201 373-383 [DOI] [PubMed] [Google Scholar]
- 25.Markwell, M. A., Haas, S. M., Bieber, L. L., and Tolbert, N. E. (1978) Anal. Biochem. 87 206-210 [DOI] [PubMed] [Google Scholar]
- 26.Silva, R. A., Hilliard, G. M., Fang, J., Macha, S., and Davidson, W. S. (2005) Biochemistry 44 2759-2769 [DOI] [PubMed] [Google Scholar]
- 27.Woody, R. W. (1995) Methods Enzymol. 246 34-71 [DOI] [PubMed] [Google Scholar]
- 28.Chen, Y. H., Yang, J. T., and Martinez, H. M. (1972) Biochemistry 11 4120-4131 [DOI] [PubMed] [Google Scholar]
- 29.Zhang, Y. (2007) Proteins Struct. Funct. Bioinformatics 69 108-117 [Google Scholar]
- 30.Weinberg, R. B., and Spector, M. S. (1985) J. Biol. Chem. 260 14279-14286 [PubMed] [Google Scholar]
- 31.Fan, D., Zheng, Y., Yang, D., and Wang, J. (2003) J. Biol. Chem. 278 21212-21220 [DOI] [PubMed] [Google Scholar]
- 32.Ajees, A. A., Anantharamaiah, G. M., Mishra, V. K., Hussain, M. M., and Murthy, H. M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 2126-2131 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Wilson, C., Wardell, M. R., Weisgraber, K. H., Mahley, R. W., and Agard, D. A. (1991) Science 252 1817-1822 [DOI] [PubMed] [Google Scholar]
- 34.Pearson, K., Tubb, M. R., Tanaka, M., Zhang, X. Q., Tso, P., Weinberg, R. B., and Davidson, W. S. (2005) J. Biol. Chem. [DOI] [PubMed]
- 35.Weinberg, R. B. (1987) Biochim. Biophys. Acta 918 299-303 [DOI] [PubMed] [Google Scholar]
- 36.Lagerstedt, J. O., Budamagunta, M. S., Oda, M. N., and Voss, J. C. (2007) J. Biol. Chem. 282 9143-9149 [DOI] [PubMed] [Google Scholar]
- 37.Dvorin, E., Mantulin, W. W., Rohde, M. F., Gotto, A. M., Jr., Pownall, H. J., and Sherrill, B. C. (1985) J. Lipid Res. 26 38-46 [PubMed] [Google Scholar]
- 38.Weinberg, R. B., and Jordan, M. K. (1990) J. Biol. Chem. 265 8081-8086 [PubMed] [Google Scholar]
- 39.Weinberg, R. B., and Spector, M. S. (1985) J. Biol. Chem. 260 4914-4921 [PubMed] [Google Scholar]
- 40.Jonas, A., Steinmetz, A., and Churgay, L. (1993) J. Biol. Chem. 268 1596-1602 [PubMed] [Google Scholar]
- 41.Weinberg, R. B. (1988) Biochemistry 27 1515-1521 [DOI] [PubMed] [Google Scholar]
- 42.Weinberg, R. B. (1994) J. Lipid Res. 35 2212-2222 [PubMed] [Google Scholar]
- 43.Kamboh, M. I., Kelly, L. J., Ahn, Y. I., and Ferrell, R. E. (1994) Human Biol. 66 625-638 [PubMed] [Google Scholar]
- 44.Lohse, P., Kindt, M. R., Rader, D. J., and Brewer, H. B., Jr. (1990) J. Biol. Chem. 265 10061-10064 [PubMed] [Google Scholar]