Abstract
Similar to many flavivirus types including Dengue and yellow fever viruses, the nonstructural NS3 multifunctional protein of West Nile virus (WNV) with an N-terminal serine proteinase domain and an RNA triphosphatase, an NTPase domain, and an RNA helicase in the C-terminal domain is implicated in both polyprotein processing and RNA replication and is therefore a promising drug target. To exhibit its proteolytic activity, NS3 proteinase requires the presence of the cofactor encoded by the upstream NS2B sequence. During our detailed investigation of the biology of the WNV helicase, we characterized the ATPase and RNA/DNA unwinding activities of the full-length NS2B-NS3 proteinase-helicase protein as well as the individual NS3 helicase domain lacking both the NS2B cofactor and the NS3 proteinase sequence and the individual NS3 proteinase-helicase lacking only the NS2B cofactor. We determined that both the NS3 helicase and NS3 proteinase-helicase constructs are capable of unwinding both the DNA and the RNA templates. In contrast, the full-length NS2B-NS3 proteinase-helicase unwinds only the RNA templates, whereas its DNA unwinding activity is severely repressed. Our data suggest that the productive, catalytically competent fold of the NS2B-NS3 proteinase moiety represents an essential component of the RNA-DNA substrate selectivity mechanism in WNV and, possibly, in other flaviviruses. Based on our data, we hypothesize that the mechanism we have identified plays a role yet to be determined in WNV replication occurring both within the virus-induced membrane-bound replication complexes in the host cytoplasm and in the nuclei of infected cells.
The genus Flavivirus viruses are responsible for significant human disease and mortality. The World Health Organization estimates annual human cases of more than 50 million, 200,000, and 50,000, for Dengue virus types 1–4 (DV1–4),2 yellow fever virus, and Japanese encephalitis virus, respectively. There is no specific therapy available for any flavivirus infection. The discovery of potent therapeutic compounds against the medically important flaviviruses is urgently needed.
WNV, an emerging disease in the United States, is a mosquito-borne pathogen that causes central nervous system damage in humans, including encephalitis and acute paralysis in severe cases (1–3). WNV is a prototype member of the Flaviviruses that currently comprise over 70 viruses including Kunjin virus (an Australian variant of WNV), yellow fever virus, Japanese encephalitis virus, and DV. Following infection of the host, the flavivirus positive-strand 11-kb RNA genome is transcribed into a negative-strand RNA. The daughter genomic RNA is then synthesized using a negative RNA strand template. The genomic WNV RNA encodes a polyprotein precursor, which consists of three structural proteins (C, capsid; prM, membrane, and E, envelope) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) arranged in the order C-prM-E-NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5. The precursor is inserted into the endoplasmic reticulum membrane and processed by the host cell and viral proteinases to transform the precursor into individual, functionally potent proteins.
Similar to many flavivirus types including Kunjin, DV, yellow fever virus, and Japanese encephalitis virus, the full-length NS3 peptide sequence of WNV represents a multifunctional protein in which the N-terminal 184 residues encode serine proteinase (NS3pro) and the C-terminal 440 residues code for an RNA triphosphatase, an NTPase, and an RNA helicase (NS3hel) (4). Because of its enzymatic activities, NS3 is implicated in both the polyprotein processing and RNA replication (5, 6). The RNA triphosphatase activity likely contributes to RNA capping (7, 8). The NTP-helicase activity unwinds the viral RNA 3′ region secondary structure, separates nascent RNA strands from the template (9–11), and facilitates the initiation of the viral replication (12).
Because the activity of NS3pro is required for several internal cleavages of the viral polyprotein precursor, NS3pro is essential for the viral replication (13). NS3pro alone, however, is inert, and it has an aberrant fold and structure (14–16). The presence of the upstream virus-encoded NS2B cofactor is required for NS3pro to exhibit its functional activity (17, 18). The folding and the spatial structure of the NS3 proteinase domain alone are significantly different from those of the two-component NS2B-NS3 proteinase complex (14, 15, 17, 18).
Because both the plus and minus strands of template RNA are highly structured, the viral replicating machinery requires the unwinding of the RNA secondary structure in the template RNAs. This important function is performed by NS3hel. NS3hel is a member of the DEAH/D-box family within the helicase superfamily 2 (5, 6, 19). NS3hel has seven sequence motifs that are conserved in the helicase superfamily 2 and that are involved in nucleic acid binding and ATP hydrolysis (20). Motifs I and II, known also as Walker A and Walker B, directly bind the ATP substrate and divalent cations and are conserved in all helicase types. Flaviviruses with impaired NS3pro or NS3hel or both are unable to replicate, making the NS3pro-hel biological system a promising drug target for antiviral therapy (21–27).
In the process of replication, the NS3pro-hel interacts with the RNA-dependent RNA polymerase, the product of the viral NS5 gene (28). The NS5 RNA polymerase is responsible for replication of the viral genome within putative complexes comprising both viral and as-yet-unidentified host proteins (28–30). Until recently, flaviviral replication was believed to occur within virus-induced membrane-bound replication complexes in the host cytoplasm. In cells infected with WNV, Japanese encephalitis virus, and DV, a significant proportion (20%) of the total NS5 RNA polymerase activity, however, is resident, jointly with NS3, within the nucleus (31, 32).
Little, however, is known about the unwinding mechanism of flavivirus helicases, including the specificity for DNA and RNA duplex and the requirement for a 3′ or 5′ overhang. In addition, previous biochemical studies of flavivirus helicases were of truncations that included only the helicase domain of NS3 but not the proteinase region of NS3 and the NS2B cofactor. This insufficient knowledge makes the discovery of novel, NS3-targeting anti-virals exceedingly difficult.
We hypothesize that the performance of the WNV NS3 helicase domain alone is distinct from that of the full-length NS2B-NS3 proteinase-helicase protein that unwinds the RNA substrates and processes the viral polyprotein precursor in the infected host cell. Here, we demonstrate that the individual NS3hel domain is capable of unwinding both the DNA and the RNA templates. In contrast, the activity of the full-length NS2B-NS3pro-hel protein is restricted to unwinding only the RNA templates. These findings suggest the existence of a regulatory switch mechanism in WNV, and possibly in other flaviviruses, that allows the virus to specifically focus on the unwinding of the viral RNA templates and the follow-on transcription.
MATERIALS AND METHODS
Protein Expression and Purification—Cloning of the DNA sequence encoding the wild-type two-component NS2B-NS3pro from WNV strain NY99 has been described (33–35). The 48-amino acid residue central portion of NS2B (residues 1423–1470 of the WNV polyprotein precursor) and the NS3pro (residues 1505–1689 of the WNV polyprotein precursor) sequences are linked by a flexible GGGGSGGGG linker. The WNV autocatalytic site-deficient NS2B-NS3pro K48A and the proteolytically inert NS2B-NS3pro H51A constructs were also described earlier (34, 36). The DNA sequence of the individual NS3hel domain was amplified by PCR using the WNV cDNA template, and the 5′-GCCGGATTCGAACCTGAGATGCTG-3′ direct and the 5′-TCAATGGTGATGGTGATGATGTCCCGAGGCGAAGTCCTTGAACGCC-3′ reverse primers (the inserted C-terminal His6 tag is underlined). The NS2B-NS3pro-hel constructs were prepared by linking the NS2B-NS3pro K48A, NS2B-NS3pro H51A, and NS3pro H51A with the NS3hel construct to generate the full-length NS2B-NS3pro-hel and NS3pro-hel sequences, respectively (34). The constructs were then cloned in the pET101/D-TOPO expression vector (Invitrogen). The authenticity of the constructs was verified by DNA sequencing.
Competent Escherichia coli BL21 CodonPlus (DE3)-RIL cells (Stratagene) were transformed with the individual recombinant pET101/D-TOPO vectors. Transformed cells were grown in 2 liters of LB broth at 37 °C to reach A600 = 0.8. The protein expression was then induced at 18 °C using 0.8 mm isopropyl β-d-thiogalactoside for an additional 16 h. The cells were collected by centrifugation, resuspended in 40 ml of Buffer A (50 mm Tris-HCl, pH 7.8, 200 mm NaCl) supplemented with the Complete Proteinase Inhibitors mixture (Roche Applied Sciences) and 1 mg/ml lysozyme (Sigma) and snap frozen at -80 °C. After thawing, the cells were disrupted by sonication (6 × 30 s with a 60 s interval). The cell debris was removed by centrifugation (19,000 × g; 30 min). The collected supernatant fraction was filtered using a 0.45-μm filter (Whatman). The NS2B-NS3pro-hel, NS3pro-hel, and NS3hel constructs were each purified from the supernatant fraction using HiTrap Co2+-chelating chromatography (GE Healthcare). The His6-tagged constructs were eluted with a 15–500 mm gradient of imidazole concentrations. When indicated, the isolated samples were additionally purified by FPLC on a Mono Q 5/50 column (GE Healthcare) equilibrated in 20 mm Tris-HCl buffer, pH 7.8, containing 100 mm NaCl and 1 mm dithiothreitol. The constructs were eluted using a linear gradient of 0.1–1 m NaCl. The fractions were analyzed using SDS-PAGE followed by Coomassie staining and also by Western blotting with a His6 antibody (Clontech) and an AbD05320 monoclonal antibody. The AbD05320 antibody was raised against the NS3pro domain through the collaboration of our laboratory with MorphoSys (Martinsried/Planegg, Germany).
ATPase Activity Assay—The ATPase activity of the constructs was measured at 37 °C for 5–60 min in wells of a 96-well plate using the NTPase assay colorimetric system (Innova Biosciences). In addition to the NTPase assay components, the reactions (50 μl each) contained Buffer A and the purified protein samples (0.04 nm). The resulting fluorescence was measured at A620 on a SpectraFluor fluorescence reader (Tecan). The rate of ATP hydrolysis was quantified using a standard curve and analyzed using Prism 5 software (GraphPad). Each sample was measured in triplicate.
DNA and RNA Substrates—The high performance liquid chromatography-purified DNA oligonucleotides D1 (5′-GCCTCGCTGCCGTCGCCA-3′), D2 (5′-TGGCGACGGCAGCGAGGCTTTTTTTTTTTTTTTTTTTT-3′), and D3 (5′-TTTTTTTTTTTTTTTTTTTTTGGCGACGGCAGCGAGGC-3′) and ribooligonucleotides R1 (5′-GCCUCGCUGCCGUCGCCA-3′) and R2 (5′-UGGCGACGGCAGCGAGGCUUUUUUUUUUUUUUUUUUUU-3′) (Eurogentec) were used. The D1 and the R1 (50 pmol each) were 5′-labeled using T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (PerkinElmer Life Sciences). The labeled products were separated from the free label using a Micro Bio-Spin 6 column (Bio-Rad). To generate duplex DNA, the labeled D1 (50 pmol) was mixed with either D2 or D3 (100 pmol) in 20 mm Tris-HCl buffer, pH 7.8, containing 150 mm NaCl and 0.1 mm EDTA. To generate the RNA duplex, the labeled R1 (50 pmol) was mixed with R2 (100 pmol) in 10 mm HEPES buffer, pH 7.8, containing 150 mm NaCl and 0.1 mm EDTA. The samples were boiled for 1 min and then slowly cooled to 20 °C. The duplex formation was confirmed by analyzing the samples in a 10% polyacrylamide gel followed by radioautography.
Helicase Activity Assay—The assays were performed at 37 °C in the reactions (200 μl) containing 20 mm Tris-HCl buffer, pH 7.8, supplemented with 25 mm NaCl, 3 mm MgCl2, 2 mm dithiothreitol, 20 μg/ml bovine serum albumin, 5 mm ATP, 5 nm DNA or RNA oligonucleotide preformed duplex and a purified protein sample (500 nm). One unit/μl RNasin (Promega) was additionally included in the RNA reactions, and only RNase-free reagents including water were used. In 5–60 min, the 20-μl aliquots were withdrawn from the reactions. The reactions were stopped by adding 6 μl of 50 mm EDTA in 50 mm Tris-HCl buffer, pH 7.8, containing 50% glycerol, and 0.01 mg/ml bromphenol blue to the withdrawn aliquots. Single-stranded oligonucleotides were separated from the duplex substrate using nondenaturing 10% polyacrylamide gel and the TBE buffer system (0.1 m Tris, 0. 1 m boric acid, 2 mm EDTA, pH 8.3). The gels were autoradiographed using an FLA-5100 imaging system (Fujifilm). The images were analyzed, and the signal intensity was digitized using Multi Gauge software (Fujifilm).
Protein N-terminal Microsequencing—The purified protein samples (4 μg each) were separated using a 4–20% polyacrylamide gradient gel, followed by the transfer of the protein bands onto a polyvinylidene difluoride membrane and N-terminal microsequencing of the resulting bands. Microsequencing was performed at ProSeq (Boxford, MA).
Proteinase Activity Assay—The proteinase assay was performed in triplicate in wells of a 96-well plate using 10 mm Tris-HCl buffer, pH 8.0, containing 20% glycerol and 0.005% Brij 35. Fluorogenic proteinase peptide substrate pyroglutamyl-arginyl-threonyl-lysyl-arginine 4-methyl-coumaryl-7-amide (Pyr-RTKR-AMC) was purchased from the Peptide Institute (Osaka, Japan). The final concentrations of the substrate and the enzyme used in the 0.1-ml reactions were 24 μm and 50 nm, respectively. Initial reaction velocity was monitored continuously at λex 360 nm and λem 460 nm on a Spectramax Gemini EM fluorescence spectrophotometer (Molecular Devices). The values of Km and kcat were derived from a double reciprocal plot of 1/V0 versus 1/[S], using the Lineweaver-Burk transformation: 1/V0 = Km/Vmax × 1/[S] + 1/Vmax, where V0 is the initial velocity of the substrate hydrolysis, [S] is the substrate concentration, Vmax is the maximum rate of hydrolysis, and Km is the Michaelis-Menten constant. The concentration of active proteinase was measured, using a fluorescence assay, by titration against a standard aprotinin solution of a known concentration (35).
RESULTS
Recombinant Constructs—To determine a potential role of the NS2B-NS3pro sequence regions in the regulation of the ATPase and unwinding activities of the NS3hel enzyme, we constructed the full-length soluble NS2B-NS3pro-hel protein. In this construct, the hydrophilic 48-amino acid central part of the natural membrane-tethered NS2B cofactor was linked via a flexible GGGGSGGGG linker to the 1505–2123 sequence of the WNV polyprotein precursor, which encodes NS3pro-hel. The N-terminal and the C-terminal sequences of the NS2B cofactor represent transmembrane segments, and therefore, they were not included in our constructs.
To facilitate the analysis of the full-length construct and to prevent its self-cleavage at the 48KG ↓ GGGSGGGG site (the linker sequence is underlined, and the cleavage site is shown by an arrow), we mutated Lys48 into Ala and generated the autolytic site-deficient NS2B-NS3pro-hel K48A mutant. To inactivate the proteolytic activity of the NS3pro domain and the self-cleavage of the NS2B-NS3pro-hel construct, we also mutated the catalytically essential active site His51 into Ala and, as a result, obtained the catalytically inert NS2B-NS3pro-hel H51A mutant.
To determine in detail the potential effect of the NS2B cofactor and the NS3pro domain on the functionality of NS3hel, we also generated the truncated NS3pro-hel H51A construct lacking the NS2B cofactor and the individual NS3hel domain construct (residues 1676–2123 of the WNV polyprotein precursor) that was lacking the NS2B-NS3pro sequence. To facilitate the isolation of the recombinant proteins from E. coli cells, the constructs were C-terminally tagged with a His6 tag. The sequence and the structure of the WNV constructs with the relative positions of the mutations are shown in Fig. 1.
FIGURE 1.
Constructs of the NS2-NS3 proteinase-helicase of WNV. A, the structure of the WNV polyprotein precursor (GenBank™ entry YP_001527877). The numbering starts from the N-terminal methionine. The 48-residue central portion of NS2B (residues 49–96 of the NS2B sequence; see supplemental Fig. S1) was linked with the 184-residue NS3pro sequence (residues 1505–1689 of the polyprotein precursor sequence) via a GGGGSGGGG linker. The Ala residue substituted for the essential His51 in the H51A inert mutant. The Ala substituted for Lys48 in the autolytic site-deficient K48A mutant. The K48A and H51A NS2B-NS3pro constructs were linked to the NS3hel sequence (residues 171–625 of the NS3 sequence) to restore the natural sequence of NS3 and to obtain the full-length NS2B-NS3pro-hel K48A (autolytic site-deficient) and NS2B-NS3pro-hel H51A (proteolytically inert) mutant constructs. To construct NS3pro-hel, the NS2B portion was truncated in the wild-type NS2B-NS3pro-hel construct. The N-terminal Met171 was used for the translation initiation. The Leu85 NS3pro-hel species we identified represents the proteolytic fragment of the NS3pro-hel construct because of the aberrant proteolysis of the latter by E. coli proteinases. The constructs were C-terminally tagged with a His6 tag. B, the N-terminal amino acid sequence of NS2B-NS3pro-hel. The positions of the K48A and H51A mutants are indicated by the residue number and the vertical arrow. The N-terminal Leu85 of the 60-kDa proteolytic fragments of the NS2B-NS3pro-hel that we identified is numbered and underlined. The linker sequence is italicized. The initiation methionine (not shown) was added to the N terminus of the NS3pro-hel construct. C, model of membrane-bound WNV NS2B-NS3pro-hel. The N- and C-terminal lobes of NS3pro are shown in green and cyan, and the NS2B cofactor is in magenta. The NS3pro Asp-His-Ser catalytic triad is shown as black dots. The relative positions of the N and C termini of the NS2B cofactor are arbitrary but consistent with the lengths of the linker regions (shown as dotted lines). Because the junction region between NS2B and NS3pro can be auto-proteolyzed, the C-terminal region of NS2B is placed into the NS3pro active site. Ser49 and Lys96 are the N- and C-terminal residues, respectively, of the NS2B cofactor used in our constructs.
Isolation of the Recombinant Constructs—After induction of the expression of the recombinant constructs in E. coli using isopropyl β-d-thiogalactoside, the majority of the produced protein appeared in a soluble form. Chromatography on a Co2+-chelating column yielded a high purity individual 51-kDa NS3hel construct (Fig. 2A). In contrast, both the NS2B-NS3pro-hel H51A and the NS3pro-hel H51A samples appeared to be partially degraded (Fig. 2A). The NS2B-NS3pro-hel H51A construct was represented by a major 75-kDa protein and two minor 60- and 51-kDa forms. Similar degraded species were observed in NS2B-NS3pro-hel K48A that we also purified.
FIGURE 2.
Purified recombinant constructs. A, NS2B-NS3pro-hel H51A (H51A), NS3pro-hel H51A (NS3pro-hel), and NS3hel were purified using a Co2+-chelating column. To separate the low molecular mass proteolytic fragments, the original H51A and NS2B-NS3pro-hel K48A (K48A) samples were additionally fractionated using a Mono Q FPLC column resulting in the purified H51A and K48A. The samples were separated by gel electrophoresis and stained with Coomassie Blue. B, Western blotting of the purified samples. Analysis of the NS2B-NS3pro-hel H51A (H51A) construct purified on a Co2+-chelating column, the full-length intact H51A (purified H51) and the Leu85 NS3pro-hel proteolytic fragment (both purified by FPLC), and the NS2B-NS3pro K48A construct (control) by Western blotting with the AbD05230 antibody (raised against the NS3pro). C, the relative position of the 1 and 2 cleavage sites and the N-terminal sequence of the proteolytic fragments of NS2B-NS3pro-hel H51A. The cleavage sites are indicated by the arrows. The determined N-terminal sequence of the Leu85 and Met171 fragments is shown. Because of the cleavage at site 2, the Met171 fragment represents the individual NS3hel domain. D, the relative position of the cleavage sites in the structural model of NS2B-NS3pro-hel. The structure of NS2B-NS3pro was determined earlier (Protein Data Bank accession number 2IJO) (17). The arrows indicate the cleavage sites (shown also in yellow).
Because of the extensive proteolysis of the aberrantly folded NS3pro domain of the 66-kDa NS3pro-hel H51A construct that lacked the NS2B cofactor, the intact NS3pro-hel H51A was not available, and only its 60- and 51-kDa degradation forms commencing from Leu85 and Met171 (see below) were purified (Fig. 2A).
Western blotting with the antibody against the His6 tag demonstrated the presence of the C-terminal tag in the 75-, 60-, and 51-kDa species, thus suggesting that proteolysis within the N-terminal region led to the generation of the 60- and 51-kDa proteins.
To separate the full-length intact proteins from their proteolytic fragments, we chromatographed the NS2B-NS3pro-hel H51A and NS2B-NS3pro-hel K48A samples using FPLC on a Mono Q column. As a result, we purified the corresponding full-length constructs (Fig. 2A). The original and the FPLC-purified samples were analyzed by Western blotting with the monoclonal antibody AbD05320 raised against the NS3pro domain (Fig. 2B). As expected, the AbD05320 antibody readily interacted with the NS2B-NS3pro control and also with the full-length 75-kDa NS2B-NS3pro-hel H51A construct and its 60-kDa degradation product. The antibody, however, did not bind the 51-kDa protein, thus confirming the absence of the NS3pro domain (Fig. 2B).
We also determined the N-terminal sequence of the 60- and 51-kDa proteins by N-terminal microsequencing. This sequence study demonstrated that because of the cleavage of the 75-kDa construct at the PWK ↓ LQH87 site (cleavage site 1), the 60-kDa protein commenced from Leu85 and consisted of the C-terminal portion of the NS3pro domain sequence and the intact NS3hel domain (Fig. 2C). Because of the cleavage at GER ↓ MDE173 (cleavage site 2), the 51-kDa species commenced from Met171 and represented the individual NS3hel domain. Because the atomic resolution structure of NS2B-NS3pro (Protein Data Bank accession number 2IJO) is available, we identified the cleavage site positions in the NS2B-NS3pro structure. Both cleavage sites are exposed to the molecule surface, suggesting that they can be readily accessed by the E. coli proteinases. The PWK ↓ LQH site is located in the 80–95 loop that links the N-terminal and the C-terminal lobes of the NS3pro domain, and GER ↓ MDE is positioned in the extended C-end terminal portion of the NS3pro sequence (Fig. 2D). The H51A mutation fully inactivated the proteolytic activity and the ability of self-cleavage of NS2B-NS3pro-hel. Because of this, we are confident that the recombinant protein was cleaved by the E. coli proteinases.
ATPase Activity of the NS3hel and NS2B-NS3pro-hel—Both RNA and DNA helicases require the hydrolysis of ATP to accomplish their unwinding activity (6). In agreement, the unwinding activity of both NS2B-NS3pro-hel H51A and NS3hel was observed only in the presence of ATP (5 mm) in the reactions, whereas a nonhydrolyzable ATP analog (AMP-PNP), CTP, GTP, and UTP were inert (Fig. 3A). Concentrations of ATP below 5 mm were unable to support efficiently the unwinding activity of both the NS2B-NS3pro-hel H51A and the NS3hel constructs (Fig. 3B). As a result, a 5 mm concentration of ATP was used in the unwinding reactions we analyzed further. The ATPase activity of both NS2B-NS3pro-hel H51A and NS3-hel did not require the binding of the enzyme to its nucleotide substrate. Therefore, the ATPase activity was observed in the reactions that did not include the duplex nucleotide substrates. Both constructs efficiently hydrolyzed ATP. The specific ATPase activity of NS2B-NS3pro-hel H51A was higher when compared with that of NS3hel (Fig. 3C).
FIGURE 3.
Unwinding and ATPase activity of NS2B-NS3pro-hel and NS3hel. A, ATP is required for the unwinding activity of NS3hel. The dsDNA unwinding activity was measured in the presence of CTP, APT, GTP, UTP, and AMP-PNP (5 mm each). The reaction time was 10 min. B, high concentrations of ATP are required for the unwinding activity. The indicated concentrations of ATP were added to the reactions, and the dsRNA and dsDNA unwinding activities of NS2B-NS3pro-hel H51A and NS3hel, respectively, were determined. The reaction time was 30 min. C, ATPase activity of the constructs. NS2B-NS3pro-hel H51A, NS3hel, and NS2B-NS3pro K48A (control) were incubated for the indicated time with ATP (5 mm). D, a high enzyme-substrate ratio is required for the unwinding activity. The unwinding activity was measured at the indicated NS3hel-dsDNA enzyme-substrate ratio. The reaction time was 10 min. E, NS3hel exhibits 3′ to 5′ unwinding activity. The DNA duplexes with either 3′-protruding or 5′-protruding strands were incubated with NS3hel for the indicated time. The asterisk indicates a [32P]-labeled oligonucleotide. The experiments shown in C and E were repeated four-five times. The means of triplicate values from representative experiments are shown.
Both NS3hel and NS2B-NS3pro-hel H51A Exhibit 3′ to 5′ Unwinding Activity—The helicase activity assay we used utilized double-stranded DNA substrates with a 3′-single-stranded terminus, formed by annealing a 38-mer DNA to a 32P-labeled 18-mer oligonucleotide. In a similar fashion, the RNA duplexes were prepared and then used to measure the RNA unwinding activity of the WNV helicase samples. In agreement with earlier studies of the flaviviral helicases (29, 37), the purified samples of the full-length WNV NS2B-NS3pro-hel H51A and the individual WNV NS3hel domain demonstrated high unwinding activity. The efficient unwinding of the duplexes required the presence in the reactions of a high, millimolar range concentration of ATP and a significant, 100-fold molar excess of the helicase constructs relative to the nucleotide duplex substrate (Fig. 3D). Similar to the DV helicase (37), the unwinding activity of WNV NS2B-NS3pro-hel H51A and NS3hel requires the presence of a 3′-overhang oligonucleotide duplex. In contrast, both WNV constructs were inert against the 5′-overhang duplexes (Fig. 3E) and the blunt end duplexes (data not shown). We conclude that WNV NS3 is an ATPase with 3′ to 5′ helicase activity.
Both NS3hel and NS2B-NS3pro-hel Efficiently Unwind dsRNA—The unwinding activity of the WNV constructs was stimulated by adding the unlabeled R1 oligonucleotide to the reactions (Fig. 4A). In the absence of the unlabeled R1 oligonucleotide excess, the constructs did not demonstrate RNA unwinding. According to our tests, a 50 nm final concentration of the unlabeled R1 oligonucleotide (a 10-fold molar excess relative to the labeled duplex) was optimal for the unwinding activity. In contrast, DNA oligonucleotides did not demonstrate this stimulatory action. An additional increase in the unlabeled R1 oligonucleotide concentrations did not significantly increase the unwinding activity. In our unwinding reactions, this 50 nm level of ssRNA was roughly equimolar to that of the correctly folded helicase protein, thus suggesting that at least one molecule to ssRNA is required to optimally stimulate the unwinding activity of helicase against dsRNA. Therefore, a 50 nm final concentration of the unlabeled R1 oligonucleotide was used in our follow-on studies. Both NS2B-NS3pro-hel H51A and NS3hel demonstrated similarly high RNA unwinding activity under these experimental conditions. In our control reactions, in which the helicase constructs were not added to the samples, we did not observe any significant nonenzymatic unwinding leading to the dissociation of the RNA and DNA duplexes in the presence of 50 nm R1 oligonucleotide. Accordingly, we conclude that NS2B-NS3pro-hel and NS3hel exhibit a similar RNA unwinding activity and that this activity is enhanced in the presence of the ssRNA oligonucleotide in the reactions.
FIGURE 4.
Unwinding activity of NS2B-NS3pro-hel H51A and NS3hel. A, the NS2B-NS3pro-hel H51A and NS3hel constructs (left and right panels, respectively) were coincubated for the indicated time with the dsRNA substrate and 5 mm ATP and with and without the single-stranded oligonucleotide R1 (50 nm; a 10-fold molar excess over the dsRNA substrate). B, the NS2B-NS3pro-hel H51A and NS3hel constructs (left and right panels, respectively) were coincubated for the indicated time with the dsDNA substrate and 5 mm ATP and with and without the single-stranded oligonucleotide R1 (50 nm; a 10-fold molar excess over the dsDNA substrate). No protein, a control reaction lacking NS2B-NS3pro-hel H51A to demonstrate the insignificant accumulation of the ssDNA because of the presence of the single-stranded oligonucleotide R1 (50 nm) in the sample. C, the unwinding efficiency of dsRNA and dsDNA by NS2B-NS3pro-hel H51A and NS3hel (left and right panels, respectively). NS2B-NS3pro-hel H51A and NS3hel were coincubated for the indicated time with dsDNA and dsRNA and with and without the single-stranded oligonucleotide R1 (50 nm). The experiments were repeated a minimum of five times using individual batches of the purified enzymes. The means of triplicate values from a representative experiment are shown. D, dsRNA remains intact in the presence of NS2B-NS3pro-hel H51A and NS3hel. The NS2B-NS3pro-hel H51A and NS3hel constructs were coincubated for the indicated time with the dsRNA substrate. ATP was not included in the reactions to preclude unwinding.
NS3hel, but Not NS2B-NS3pro-hel, Unwinds dsDNA—In the absence of the unlabeled ssRNA, we did not observe any unwinding of the DNA duplex by the intact 75-kDa full-length NS2B-NS3pro-hel H51A (Fig. 4B). The insignificant level of DNA unwinding by NS2B-NS3pro-hel H51A we observed in the presence of the R1 oligonucleotide (50 mm) is the result of a nonenzymatic strand exchange because a similar level of unwinding was detected in the absence of the enzyme in the reactions. In contrast, NS3hel was highly active in the DNA unwinding reactions both in the presence and the absence of the unlabeled R1 oligonucleotide excess. The addition of the unlabeled R1 oligonucleotide to the reactions did not affect the unwinding rate of the DNA duplex by NS3hel. The data suggest that NS3hel exhibits both the DNA and the RNA unwinding activity whereas the unwinding capacity of NS2B-NS3pro-hel is limited to RNA (Fig. 4C).
RNase activity, if present in our samples, may degrade dsRNA and significantly affect our RNA unwinding results. To confirm the absence of any significant RNase activity in our samples, we coincubated the purified NS2B-NS3pro-hel H51A and NS3hel constructs with dsRNA. To preclude unwinding, ATP was not included in the reactions. Fig. 4D clearly shows that dsRNA remained predominantly intact in the reactions. Thus, we concluded that RNase contamination was minimal in the purified protein samples we used in our study.
To determine whether the excess of the single-stranded R1 (50 nm) affects reannealing, we coincubated the individual single-stranded labeled R1 and unlabeled R2 with and without the unlabeled R1 excess. The helicase samples were not included in the reactions (supplemental Fig. S2). There was an insignificant, ∼3%, reannealing of ssRNA that resulted in the formation of dsRNA. In the presence of the unlabeled R1 excess, this reannealing was completely blocked. Accordingly, we concluded that reannealing, especially in the presence of the R1 excess, did not affect the unwinding reactions.
NS3pro-hel Exhibits the DNA Unwinding Activity—Our results imply that the presence of a properly folded and functional NS2B-NS3pro domain is required for the selective unwinding activity of tNS2B-NS3pro-hel. This suggestion agrees with the earlier data that the presence of the NS2B cofactor is essential for the proteolytic activity and the productive folding of the flaviviral NS2B-NS3pro constructs (17, 18). To test whether our suggestion is correct, we characterized the 60-kDa Leu85 NS3pro-hel construct that lacks the NS2B cofactor and represents the C-terminal portion of the NS3pro domain sequence and the intact NS3hel domain (Figs. 1A and 2). Consistent with our belief, the Leu85 NS3pro-hel construct acquired the ability to unwind the DNA duplex as efficiently as NS3hel (Fig. 5A).
FIGURE 5.
The DNA unwinding activity emerges in the proteolytic fragments of NS2B-NS3pro-hel and NS3pro-hel. A, the DNA unwinding activity of the Leu85 proteolytic fragment of NS3pro-hel H51A. The purified Leu85 fragment and NS3hel were each coincubated for the indicated time with the dsDNA substrate and 5 mm ATP. B, the proteolytic fragment of NS2B-NS3pro-hel H51A has the DNA unwinding activity. Top panel, Western blot analysis with the His6 antibody of the intact purified NS2B-NS3pro-hel H51A sample aliquot and the same 10-μg sample aliquot after a 48-h incubation at 37 °C with and without NS2B-NS3pro K48A (1 μg). Bottom panel, both the intact purified NS2B-NS3pro-hel H51A sample and the same sample after a 48-h incubation at 37 °C were coincubated for the indicated time with the dsDNA substrate and 5 mm ATP.
To further corroborate our conclusions, we specifically induced the proteolysis of the 75-kDa NS2B-NS3pro-hel by the residual E. coli proteinases by incubating the purified sample for 48 h at 37 °C. As a result, we generated low amounts of the proteolyzed form in the NS2B-NS3pro-hel sample. As demonstrated by Western blotting, the C-terminal His6 tag was present in this proteolytic fragment, suggesting that the fragment resulted because of the proteolysis of the N-terminal region of NS2B-NS3pro-hel. The presence of this proteolytic fragment of NS2B-NS3pro-hel correlated with the ability of the sample to unwind dsDNA in a manner similar to NS3hel (Fig. 5B).
In sum, we concluded that the correct, productive fold of the N-terminal NS2B-NS3pro portion was essential for the RNA selectivity of the full-length NS2B-NS3pro-hel construct. It is well established now that the NS2B cofactor is required for the proteolytic activity of NS3pro to emerge and that NS3pro alone is catalytically inactive. Based on these earlier data and our current results, we hypothesize that the NS3pro-hel constructs lacking the NS2B cofactor will acquire the ability to unwind dsDNA and that the performance of these constructs will be similar to that of the individual NS3hel domain.
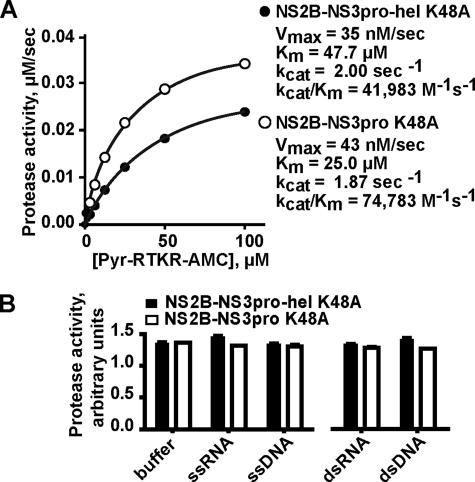
NS3hel Domain Does Not Significantly Affect the Proteolytic Activity of NS2B-NS3pro—Because we determined that the NS2B-NS3pro domain affects the functionality of the NS3hel domain, we next asked whether this effect is reciprocal and whether the NS3hel domain affects the functionality of NS2B-NS3pro. For this purpose, we determined the cleavage kinetics of the Pyr-RTKR-AMC fluorescence peptide substrate by the autolytic site-deficient, proteolytically potent NS2B-NS3pro-hel K48A and NS2B-NS3pro K48A constructs. Both constructs efficiently cleaved the Pyr-RTKR-AMC fluorescence peptide substrate. There was no significant difference in the kinetics parameters including Vmax, kcat, and kcat/Km of the cleavage reactions that employed the NS2B-NS3pro-hel K48A and NS2B-NS3pro K48A constructs (Fig. 6A). In agreement, the individual proteinase domain (NS2B-NS3pro K48A) and the full-length intact NS3 construct (NS2B-NS3pro-hel K48A) demonstrated a similar cleavage efficiency of protein substrates including myelin basic protein (data not shown). To support these data, we determined whether dsRNA, dsRNA, ssDNA, and ssRNA affect the proteolytic activity of NS2B-NS3pro-hel K48A and NS2B-NS3pro K48A. Our measurements showed, however, that there was no effect of oligonucleotides on the proteolytic activity of the constructs (Fig. 6B).
FIGURE 6.
The NS3hel part does not significantly affect the functionality of the NS2B-NS3pro part. A, kinetics of the Pyr-RTKR-AMC cleavage by NS2B-NS3pro-hel K48A and NS2B-NS3pro K48A. The samples were coincubated with increasing concentrations of the substrate and the Vmax, Km, kcat, and kcat/Km parameters of the cleavage reactions were calculated. B, nucleotides do not affect the proteolytic activity of NS2B-NS3pro-hel K48A and NS2B-NS3pro K48A. The proteolytic activity of the samples (50 nm each) was determined against Pyr-RTKR-AMC in the presence of ssRNA, ssDNA, dsRNA, and DNA dsDNA (200 nm each). The reactions also contained 10 mm ATP. The experiments shown in A and B were repeated three times. The means of triplicate values from a representative experiment are shown.
DISCUSSION
In our effort to investigate the biology of the WNV helicase in more detail, we cloned cDNA encoding the full-length NS2B-NS3 proteinase-helicase complex (NS2B-NS3pro-hel) as well as the individual NS3 helicase domain (NS3hel) lacking the NS2B cofactor and the NS3 proteinase sequence, the individual NS3 proteinase-helicase (NS3pro-hel) without the NS2B cofactor and the two-component NS2B-NS3 proteinase (NS2B-NS3pro) lacking the helicase sequence. To prevent the autocatalytic processing of the constructs, the essential active site His51 residue of NS3pro was mutated into Ala (H51A), which inactivated the proteolytic activity of the proteinase domain. An additional mutation we used in our studies was K48A in the junction region between the NS2B cofactor and the NS3pro sequence. This mutation resulted in the autolytic site-deficient, proteolytically potent NS2B-NS3pro-hel constructs.
We determined that following the expression in E. coli cells the recombinant NS2B-NS3pro-hel constructs were proteolytically degraded generating, in addition to the intact full-length 75-kDa protein, the 60- and 51-kDa C-terminal species. Because of the cleavage at GER ↓ MDE173 in the extended C-terminal portion of the NS3pro sequence, the 51-kDa species commenced from Met171 and represented the individual NS3hel domain. Because of the cleavage of the NS2B-NS3pro-hel construct at the PWK ↓ LQH87 site within the 80–95 loop that links the N-terminal and the C-terminal lobes of the NS3pro domain, the 60-kDa fragment commenced at Leu85 and consisted of the C-terminal portion of the NS3pro domain sequence and the intact NS3hel domain.
Our biochemical studies identified that the full-length construct was active only against the RNA duplex, whereas the individual NS3hel domain construct was capable of unwinding both the RNA and DNA duplexes. The RNA unwinding activity of the constructs was optimally stimulated in the presence of a 10-fold ssRNA excess relative to the level of the dsRNA substrate in the reactions.
These results suggest that the presence of the functional NS2B-NS3pro moiety significantly affects the functionality of the helicase in the full-length NS2B-NS3pro-hel construct, thus making its functionality clearly distinct from that of the individual NS3hel sequence and of the NS3pro-hel sequence that lacks only the NS2B cofactor. Because of the autolytic cleavage of the NS2B-NS3 junction region, the cofactor-less NS3pro-hel species are frequently observed in virus-infected mammalian cells (31, 38–42).
In contrast, the proteolytic capacity of NS2B-NS3pro was similar to that of the full-length NS2B-NS3pro-hel, suggesting that the NS3hel domain had only a limited effect on the proteolytic activity of NS2B-NS3pro.
To support these novel data, we evaluated the substrate specificity of the NS3pro-hel lacking the NS2B cofactor sequence. The folding of the proteolytically inert NS3pro alone is clearly distinct from the productive folding of the two-component NS2B-NS3pro protein. Based on these data, we suspected that the helicase activity of NS3pro-hel would be similar to that of the NS3hel alone. Our experimental results proved that our suspicion was correct because the several different length NS3pro-hel species we purified and studied demonstrated the high unwinding activity of the DNA duplex. We conclude that if the NS3pro part is folded incorrectly, the NS3hel domain has a broad substrate specificity. The correct folding and the activity of NS3pro require the presence of the NS2B cofactor. The correctly folded NS2B-NS3pro domain restricts the substrate specificity of the NS3hel, and these restrictions result in the switch to RNA.
The similar studies of the analogous NS3 and NS4A proteins from HCV also suggest that there is a significant interference of the cofactor-proteinase moiety with the NS3hel functionality and that this interference also affects the unwinding activity of the helicase against the DNA and RNA substrates (43–45). In HCV, the protease domain is also required for the efficient RNA unwinding by NS3hel (46–48).
It is tempting to hypothesize that in the course of WNV replication within virus-induced, membrane-bound replication complexes in the host cytoplasm, the strict RNA unwinding activity of WNV NS3 is required for coordinating the helicase activity with the activity of the NS5 RNA-dependent RNA polymerase and avoiding interference by the host DNA. In turn, when a significant fraction of NS3 that is no longer associated with the membrane NS2B cofactor relocalizes jointly with NS5 to the nucleus in infected cells (31, 32), the capacity of this viral helicase to unwind DNA opens a wide spectrum of possibilities beyond its role in viral RNA replication. Further studies are warranted to elucidate the precise mechanisms and the functional role of the NS3 protein in viral replication in vivo.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants CA83017 and CA77470 (to A. Y. S.), RR020843 (to J. W. S. and A. Y. S.), and AI055789 (to R. C. L. and A. Y. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: DV, Dengue fever virus; ds, double-stranded; ss, single-stranded; NS3pro, NS3 proteinase; NS3hel, NS3 helicase; Pyr-RTKR-AMC, pyroglutamyl-arginyl-threonyl-lysyl-arginine 4-methyl-coumaryl-7-amide; WNV, West Nile virus; FPLC, fast protein liquid chromatography; AMP-PNP, β,γ-iminoadenosine 5′-triphosphate.
References
- 1.Hayes, E. B., Komar, N., Nasci, R. S., Montgomery, S. P., O'Leary, D. R., and Campbell, G. L. (2005) Emerg. Infect. Dis. 11 1167-1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sejvar, J. J. (2004) Neurology 63 206-207 [DOI] [PubMed] [Google Scholar]
- 3.Solomon, T., and Ravi, V. (2003) Lancet Infect. Dis. 3 189-190 [DOI] [PubMed] [Google Scholar]
- 4.Padmanabhan, R., Mueller, N., Reichert, E., Yon, C., Teramoto, T., Kono, Y., Takhampunya, R., Ubol, S., Pattabiraman, N., Falgout, B., Ganesh, V. K., and Murthy, K. (2006) Novartis Found. Symp. 277 74-86 [DOI] [PubMed] [Google Scholar]
- 5.Li, H., Clum, S., You, S., Ebner, K. E., and Padmanabhan, R. (1999) J. Virol. 73 3108-3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gorbalenya, A. E., Koonin, E. V., Donchenko, A. P., and Blinov, V. M. (1989) Nucleic Acids Res. 17 4713-4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wengler, G., and Wengler, G. (1993) Virology 197 265-273 [DOI] [PubMed] [Google Scholar]
- 8.Mastrangelo, E., Milani, M., Bollati, M., Selisko, B., Peyrane, F., Pandini, V., Sorrentino, G., Canard, B., Konarev, P. V., Svergun, D. I., de Lamballerie, X., Coutard, B., Khromykh, A. A., and Bolognesi, M. (2007) J. Mol. Biol. 372 444-455 [DOI] [PubMed] [Google Scholar]
- 9.Cui, T., Sugrue, R. J., Xu, Q., Lee, A. K., Chan, Y. C., and Fu, J. (1998) Virology 246 409-417 [DOI] [PubMed] [Google Scholar]
- 10.Borowski, P., Niebuhr, A., Mueller, O., Bretner, M., Felczak, K., Kulikowski, T., and Schmitz, H. (2001) J. Virol. 75 3220-3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo, D., Xu, T., Hunke, C., Gruber, G., Vasudevan, S. G., and Lescar, J. (2008) J. Virol. 82 173-183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frick, D. N. (2003) Drug News Perspect. 16 355-362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahl, G., Sandstrom, A., Akerblom, E., and Danielson, U. H. (2007) FEBS J. 274 5979-5986 [DOI] [PubMed] [Google Scholar]
- 14.Murthy, H. M., Clum, S., and Padmanabhan, R. (1999) J. Biol. Chem. 274 5573-5580 [DOI] [PubMed] [Google Scholar]
- 15.Murthy, H. M., Judge, K., DeLucas, L., and Padmanabhan, R. (2000) J. Mol. Biol. 301 759-767 [DOI] [PubMed] [Google Scholar]
- 16.Yusof, R., Clum, S., Wetzel, M., Murthy, H. M., and Padmanabhan, R. (2000) J. Biol. Chem. 275 9963-9969 [DOI] [PubMed] [Google Scholar]
- 17.Aleshin, A. E., Shiryaev, S. A., Strongin, A. Y., and Liddington, R. C. (2007) Protein Sci. 16 795-806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erbel, P., Schiering, N., D'Arcy, A., Renatus, M., Kroemer, M., Lim, S. P., Yin, Z., Keller, T. H., Vasudevan, S. G., and Hommel, U. (2006) Nat. Struct. Mol. Biol. 13 372-373 [DOI] [PubMed] [Google Scholar]
- 19.Luking, A., Stahl, U., and Schmidt, U. (1998) Crit. Rev. Biochem. Mol. Biol. 33 259-296 [DOI] [PubMed] [Google Scholar]
- 20.Benarroch, D., Selisko, B., Locatelli, G. A., Maga, G., Romette, J. L., and Canard, B. (2004) Virology 328 208-218 [DOI] [PubMed] [Google Scholar]
- 21.Maga, G., Gemma, S., Fattorusso, C., Locatelli, G. A., Butini, S., Persico, M., Kukreja, G., Romano, M. P., Chiasserini, L., Savini, L., Novellino, E., Nacci, V., Spadari, S., and Campiani, G. (2005) Biochemistry 44 9637-9644 [DOI] [PubMed] [Google Scholar]
- 22.Nall, T. A., Chappell, K. J., Stoermer, M. J., Fang, N. X., Tyndall, J. D., Young, P. R., and Fairlie, D. P. (2004) J. Biol. Chem. 279 48535-48542 [DOI] [PubMed] [Google Scholar]
- 23.Johnston, P. A., Phillips, J., Shun, T. Y., Shinde, S., Lazo, J. S., Huryn, D. M., Myers, M. C., Ratnikov, B. I., Smith, J. W., Su, Y., Dahl, R., Cosford, N. D., Shiryaev, S. A., and Strongin, A. Y. (2007) Assay Drug Dev. Technol. 5 737-750 [DOI] [PubMed] [Google Scholar]
- 24.Ray, D., and Shi, P. Y. (2006) Recent Patents Anti-Infect Drug Disc. 1 45-55 [DOI] [PubMed] [Google Scholar]
- 25.Keller, T. H., Chen, Y. L., Knox, J. E., Lim, S. P., Ma, N. L., Patel, S. J., Sampath, A., Wang, Q. Y., Yin, Z., and Vasudevan, S. G. (2006) Novartis Found. Symp. 277 102-119 [PubMed] [Google Scholar]
- 26.Sampath, A., Xu, T., Chao, A., Luo, D., Lescar, J., and Vasudevan, S. G. (2006) J. Virol. 80 6686-6690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu, T., Sampath, A., Chao, A., Wen, D., Nanao, M., Luo, D., Chene, P., Vasudevan, S. G., and Lescar, J. (2006) Novartis Found. Symp. 277 87-101 [DOI] [PubMed] [Google Scholar]
- 28.Kapoor, M., Zhang, L., Ramachandra, M., Kusukawa, J., Ebner, K. E., and Padmanabhan, R. (1995) J. Biol. Chem. 270 19100-19106 [DOI] [PubMed] [Google Scholar]
- 29.Lindenbach, B., Thiel, H. J., and Rice, C. M. (2007) in Fields Virology, (Knipe, D. M., and Howley, P. M., eds) 5th Edition, pp. 1101-1152, Lippincott-Raven Publishers, Philadelphia, PA
- 30.Liu, W. J., Sedlak, P. L., Kondratieva, N., and Khromykh, A. A. (2002) J. Virol. 76 10766-10775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uchil, P. D., Kumar, A. V., and Satchidanandam, V. (2006) J. Virol. 80 5451-5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brooks, A. J., Johansson, M., John, A. V., Xu, Y., Jans, D. A., and Vasudevan, S. G. (2002) J. Biol. Chem. 277 36399-36407 [DOI] [PubMed] [Google Scholar]
- 33.Shiryaev, S. A., Aleshin, A. E., Ratnikov, B. I., Smith, J. W., Liddington, R. C., and Strongin, A. Y. (2007) Protein Expr. Purif. 52 334-339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiryaev, S. A., Kozlov, I. A., Ratnikov, B. I., Smith, J. W., Lebl, M., and Strongin, A. Y. (2007) Biochem. J. 401 743-752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiryaev, S. A., Ratnikov, B. I., Chekanov, A. V., Sikora, S., Rozanov, D. V., Godzik, A., Wang, J., Smith, J. W., Huang, Z., Lindberg, I., Samuel, M. A., Diamond, M. S., and Strongin, A. Y. (2006) Biochem. J. 393 503-511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shiryaev, S. A., Ratnikov, B. I., Aleshin, A. E., Kozlov, I. A., Nelson, N. A., Lebl, M., Smith, J. W., Liddington, R. C., and Strongin, A. Y. (2007) J. Virol. 81 4501-4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yon, C., Teramoto, T., Mueller, N., Phelan, J., Ganesh, V. K., Murthy, K. H., and Padmanabhan, R. (2005) J. Biol. Chem. 280 27412-27419 [DOI] [PubMed] [Google Scholar]
- 38.Arias, C. F., Preugschat, F., and Strauss, J. H. (1993) Virology 193 888-899 [DOI] [PubMed] [Google Scholar]
- 39.Bera, A. K., Kuhn, R. J., and Smith, J. L. (2007) J. Biol. Chem. 282 12883-12892 [DOI] [PubMed] [Google Scholar]
- 40.Cahour, A., Falgout, B., and Lai, C. J. (1992) J. Virol. 66 1535-1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chambers, T. J., Grakoui, A., and Rice, C. M. (1991) J. Virol. 65 6042-6050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kummerer, B. M., and Rice, C. M. (2002) J. Virol. 76 4773-4784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frick, D. N., Rypma, R. S., Lam, A. M., and Gu, B. (2004) J. Biol. Chem. 279 1269-1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuang, W. F., Lin, Y. C., Jean, F., Huang, Y. W., Tai, C. L., Chen, D. S., Chen, P. J., and Hwang, L. H. (2004) Biochem. Biophys. Res. Commun. 317 211-217 [DOI] [PubMed] [Google Scholar]
- 45.Zhang, C., Cai, Z., Kim, Y. C., Kumar, R., Yuan, F., Shi, P. Y., Kao, C., and Luo, G. (2005) J. Virol. 79 8687-8697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beran, R. K., Serebrov, V., and Pyle, A. M. (2007) J. Biol. Chem. 282 34913-34920 [DOI] [PubMed] [Google Scholar]
- 47.Gu, B., Pruss, C. M., Gates, A. T., and Khandekar, S. S. (2005) Protein Pept. Lett. 12 315-321 [DOI] [PubMed] [Google Scholar]
- 48.Pang, P. S., Jankowsky, E., Planet, P. J., and Pyle, A. M. (2002) EMBO J. 21 1168-1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.