Abstract
The sexual cycle of Plasmodium is required for transmission of malaria from mosquitoes to mammals, but how parasites induce the expression of genes required for the sexual stages is not known. We disrupted the Plasmodium yoelii gene encoding high mobility group nuclear factor hmgb2, which encodes a DNA-binding protein potentially implicated in transcriptional regulation of malaria gene expression. We investigated its function in vivo in the vertebrate and invertebrate hosts. Δpyhmgb2 parasites develop into gametocytes but have drastic impairment of oocyst formation. A global transcriptome analysis of the Δpyhmgb2 parasites identified ∼30 genes whose expression is down-regulated in the Δpyhmgb2 parasites. These genes are conserved in all malaria species, and more than 90% of these genes show a peak of mRNA expression at the gametocyte stage. Surprisingly, the transcripts coding for the Plasmodium berghei orthologues of those genes are stored and translated in the ookinete stage. Therefore, sexual stage protein expression appears to be both transcriptionally and translationally regulated with Plasmodium HMGB2 acting as an important regulator of malaria sexual stage gene expression.
Plasmodium species, the causative agents of malaria, are the most deadly members of the apicomplexan phylum. As with other members of the Apicomplexa, Plasmodium has a multifaceted life cycle encompassing an asexual phase within the intermediate host and a sexual cycle in the mosquito. Gametocytogenesis and gametogenesis consist of an ensemble of processes leading to the differentiation of erythrocytic forms of the parasite into male or female gametocytes. These complex and coordinated steps ensure the transmission of the parasite to the invertebrate host and are required for transmission of malaria to mammalian hosts. Gametocytes form in mammalian host, are taken up by the mosquito during a blood meal, transform into gametes in the mosquito midgut, and then fuse to form a zygote. Zygotes differentiate into ookinetes that penetrate the midgut wall and differentiate into oocysts, in which sporozoites will develop.
Starting from the well controlled vertebrate milieu, the parasite must adapt to the mosquito environment where temperature fluctuates and nutrients are limited. Gametocytogenesis requires growth arrest and a differentiation step that will prepare the parasite for these changes. Sexual differentiation is accompanied by a tightly controlled gene expression program during which ∼200–300 mRNAs are specifically expressed or predominantly expressed in sexual forms of the parasite (1–4). Transcriptional control of gene expression is implicated as a crucial step during this process with sex-specific transcript expression in gametocytes controlled by the sequences in the 5′-flanking regions (5).
For some sexual stage genes post-translational processes mediate gene expression. The genes coding for P25 and P28 ookinete proteins (PY00522 and PY00523, respectively) are transcribed at the gametocyte stage, but their translation is delayed until the gamete and zygote stages (6). A group of mRNAs with a common 3′-untranslated region motif are specifically transcribed at the gametocyte stage, but their translation is repressed until the parasite requires the presence of the proteins they encode (4). Recently, an RNA helicase called DOZI (7) was identified in Plasmodium berghei that participates in the storage and translation repression of a large number of transcripts. Therefore, sexual differentiation in Plasmodium requires fine tuning of both transcriptional and post-transcriptional processes to regulate gene expression.
The mechanisms controlling the transition from the asexual erythrocytic stage into a gametocyte-specific gene expression program are unknown. Initial analysis of the genomes of Plasmodium species revealed few canonical transcriptional regulators (8). On the other hand, the RNA polymerase II machinery (9) is conserved within Apicomplexa, and chromatin modification plays an important role in gene regulation (10). The finding that certain DNA motifs are recurrent in the promoters of Apicomplexa and bind to nuclear factors (11–13) suggests that unrecognized transcription factors exist. Recently, a novel family of AP2 domain plant-like transcription factors was identified in the genomes of the Apicomplexa (14). In plants these transcription factors bind to specific DNA motifs and regulate a variety of biological responses (15), but the function of apicomplexan AP2 proteins has not been experimentally validated.
Two proteins containing a high mobility group box (HMGB)3 are highly conserved among Plasmodium species (16). HMGB box proteins are small transcriptional regulators that bind DNA in a nonsequence-specific fashion and bend it, leading to the recruitment of RNA polymerase II and the induction of transcription (17). HMGB proteins also actively participate in chromatin remodeling by increasing nucleosome sliding and accessibility of the chromatin (17). The two Plasmodium falciparum HMGB proteins, HMGB1 (PFL0145c) and HMGB2 (MAL8P1.72), have the biochemical characteristics of the HMGB box family of transcriptional regulators (16). Interestingly, these HMGB are differentially expressed. HMGB1 is expressed during the erythrocytic asexual cycle, whereas HMGB2 is slightly expressed at the schizont stage and highly expressed at the gametocyte stage (2, 16).
In this study, we evaluated the role of the HMGB2 protein in regulation of sexual stage gene expression. We disrupted hmgb2 in the rodent malaria model Plasmodium yoelii and investigated its function in vivo in its two hosts, the mouse and the mosquito Anopheles stephensi. Our results indicate that PyHMGB2 plays a central and conserved role in regulating gene expression during the sexual cycle of Plasmodium.
EXPERIMENTAL PROCEDURES
P. yoelii Life Cycle—P. yoelii YM lethal strain was maintained in naive BALB/c or Swiss Webster mice. For mosquito infection experiments, 4–5-day-old A. stephensi were fed for 15 min on anesthetized Swiss Webster mice infected with either wild-type (WT) or Δpyhmgb2 (KO) P. yoelii parasites. Prior to feeding, the abundance and the sex ratio of the gametocyte-stage parasites were checked on Giemsa-stained blood smears. At day 7 post-infection, midguts were monitored for infection, and oocysts were counted by either counting fluorescent GFP-positive KO parasite oocysts (see below) or after mercurochrome staining (WT and KO parasite lines). Salivary glands from mosquitoes on day 14 or 15 post-infective blood meal were dissected to recover sporozoites.
Exflagellation experiments were performed as described in Ref. 18, and exflagellation centers were counted at 9 min post-induction. Male and female gametocytes were identified by morphology on Giemsa smears. In vitro culture of ookinetes was performed as described in Ref. 19. Ookinete numbers were counted with a hemocytometer under a phase-contrast microscope and normalized to the original gametocyte content of the culture. Statistical analysis was performed using an unpaired t test available using the GraphPad Prism software package (GraphPad software, San Diego).
P. yoelii Transfection, Plasmid Construction, and Genotyping—A fragment of the pyhmgb2 gene (PY07077) amplified from YM strain genomic DNA using primers 5′-ggatccATTACATGTTATGATCTTCTAC-3′ and 5′-gcggccgcCAATGCTCTCTTTGGAGCTAATG-3′ was inserted into the pMD205-GFP vector in the BamHI and NotI restriction sites. An SpeI restriction site was created by site-directed mutagenesis (Stratagene, La Jolla, CA) at nucleotide position 258 of the cloned fragment (position 3152 of the MALPY02526 contig sequence) using the following primer, 5′-TACAACTTTAGAACAAGACTAGTACTGTTTTGAAACAACTCA-3′. Prior to transfection, the targeting plasmid was linearized using an SpeI site created in the pyhmgb2 open reading frame. Transfections were performed as described previously (20, 21). Cloning was performed after limiting dilution and infection of 20 BALB/c mice. A WT nonrecombinant clone was passaged in parallel with KO clones so that mouse and mosquito passage numbers were comparable for all experiments. Diagnostic PCR was performed using the primers T7 (5′-TAATACGACTCACTATAGGG-3′) and A (5′-ATCGATTAGTAGAATATGTAATATA-3′) as indicated in Fig. 1.
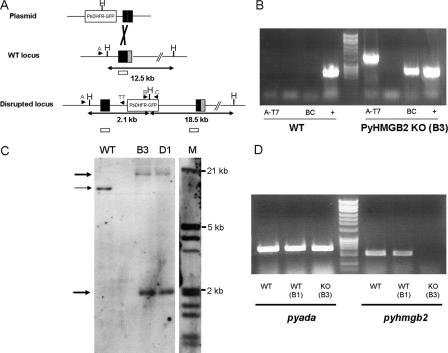
FIGURE 1.
A, schematic diagram of pyhmgb2 gene disruption strategies. Sites of recombination are indicated in black. The HindIII restriction sites utilized in the Southern blot are indicated by the capital letter H. The position of the Southern probe is represented by an empty bar. Predicted size of the fragments is indicated below each locus. Arrows show the position of primers used in the diagnostic PCR. B, diagnostic PCR on genomic DNA extracted from Δpyhmgb2 clone B3 and WT control parasite. Primer set A-T7 yielded a fragment of 1.2-kb size. Primer B-C amplified a 0.7-kb fragment of the GFP gene. Amplification of the pyada gene (0.6 kb) is used as a PCR-positive control (+). The data are representative of the two clones B3 and D1. C, Southern blot analysis of the HindIII-digested gDNA for the WT, B3, and D1 clones. The sizes of the marker (M) are indicated on the right side of the figure. The thin arrow indicates the band expected for the WT locus. The bold arrows indicate the two bands expected for a disrupted locus. D, RT-PCR analysis of the pyada and pyhmgb2 transcript expression for a WT control (WT), a WT clone (B1) cloned at the same time as the KO clone (B3). Amplification of the pyada gene is used as a positive control (left panel). The RT-PCR shows that no expression of pyhmgb2 (right panel) could be detected in the KO clone (B3). Data are representative of the two clones B3 and D1.
A probe containing a portion of the pyhmgb2 gene was amplified by PCR and digoxigenin (DIG)-labeled using the DIG random prime labeling kit (Roche Applied Science). Southern blot was performed after running 1 μg of HindIII-digested genomic DNA on a 0.8% agarose gel and passive transfer on a nylon membrane and revealed using an anti-DIG antibody coupled to peroxidase (Roche Applied Science).
Infectivity and Survival Assays—Five BALB/c or Swiss Webster mice per group were infected intravenously using 5 × 105 and 1 × 106 parasites, respectively. Beginning 24 h after injection, a thin smear of blood was prepared daily and stained with Giemsa reagent. For each sample, at least 1000 red blood cells were examined by microscopy, and the percentage of parasitized erythrocytes was calculated. Mouse viability was monitored every 12 h. Statistical analysis was performed using the GraphPad Prism software (GraphPad software, San Diego).
Microarray Design and Experiments—Two mice per group were infected with 1 million purified schizonts from the WT and two clones (B3 and D1) of the Δpyhmgb2 lines. When parasitemia reached 10–20% and equivalent gametocytemia, total RNA was extracted from the WT and the B3 and D1 clones of the KO parasite lines using the TRIzol method on saponinlysed parasites. For each group, RNA originating from the two mice was pooled. Total RNA integrity was checked using the Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA) at the Albert Einstein College of Medicine shared sequencing facility.
P. yoelii microarrays were produced in the Molecular Genomics Core Facility, Drexel University College of Medicine, under the direction of L. Bergman. The spotted microarray contained 7,080 65-mer oligonucleotides (probes). Each probe was spotted twice on the array. The array has been developed and previously evaluated with hybridizations of total RNA extracted from blood stages. Probe sequences were blasted against annotated P. yoelii genes downloaded from PlasmoDB (version 5.2). In total, 6,451 probes were specifically mapped to 6,428 annotated P. yoelii genes with a perfect match (22).
To prepare each probe, 10 μg of P. yoelii RNA was reverse-transcribed into cDNA in the presence of aminoallyl-dUTP (FairPlay microarray labeling kit; Stratagene, La Jolla, CA). The cDNA was then fluorescently labeled by reaction with monofunctional, normal N-hydroxysuccinimide-activated Cy3 or Cy5 dye (Amersham Biosciences). The Cy dye-labeled cDNA was purified, and the yield and specific activity of each probe were determined by absorption spectroscopy. Pairs of Cy3- and Cy5-labeled probes were pooled and hybridized to the P. yoelii microarrays for 14–16 h at 60 °C in a 60-μl mixture. Following hybridization and washing, slides were scanned using a GenePix 4000A microarray laser scanner (Axon Instruments Inc., Union City, CA), and the fluorescence intensity of each DNA feature was determined at 532 nm (Cy3) and at 635 nm (Cy5). Data for each gene were obtained from replicate features (n = 2). On a second set of arrays, the assignments of Cy3 and Cy5 for labeling of the paired cDNA probes were reversed (standard dye flip). Gene expression data were acquired and analyzed using the GenePixPro 5.0 and Acuity 3.0 Microarray Informatics software (Axon Instruments). Two biological replicates with the proper dye swap controls were performed resulting in a total of eight hybridization signals from each parasite line for each gene represented on the microarray. Data have been deposited to GEO data base (GSE9952) together with the microarray platform (GPL5381).
Microarray Normalization and Statistical Analysis—The .gpr files were loaded using the Bioconductor limma package. Quality assessment of the microarrays was performed using the package arrayQuality. The data were then normalized to remove systematic technical variation. Two-channel normalization was performed using the function maNorm in the package marray. Three types of normalization were performed as follows: (i) using the median enabling global median location normalization; (ii) using a global normalization using the scatter-plot smoother loess; (iii) a two-dimension spatial normalization using the loess function. Selection of differentially expressed genes was performed using the package limma that is based on the fitting of the expression data for each gene to a linear model. Empirical Bayes were then used to borrow information across genes making the analyses stable even for experiments with a small number of arrays (23). In this method, the basic statistic used for significance analysis is the moderated t statistic. Moderated t statistic leads to p values with increased degrees of freedom reflecting the greater reliability associated with smoothed standards errors (24). A p value cutoff to 0.05 was set leading to a false discovery rate of 5%.
RT-PCR and Real Time Quantitative RT-PCR—One microgram of DNase-treated total RNA was utilized to produce cDNA following the instructions of the first strand cDNA kit (Invitrogen) with oligo(dT) as a primer. PCR was performed during 30 cycles using the following primers: pyada forward (5′-ATGATGGAAATTCCAACTGAAGA-3′) and pyada reverse (5′-TTAAAAGTACAACGCTTTAAGCTCG-3′); pyhmgb2 forward (5′-CCTATCAAGTGTGTTGTATATGTCTTC-3′) and pyhmgb2 reverse (5′-TCTTCTATTTCTTTGGAATAACGAAC-3′). Real time quantitative PCR was performed on the 7300 ABI apparatus using the Power Sybr (Applied Biosystems, Foster City, CA) mastermix in a 20-μl volume according to the manufacturer's instructions. PCR primers were designed using the Primerexpress software (Applied Biosystems, Foster City, CA) to amplify regions of 100–150 nucleotides. Each experiment was performed at least two times in duplicate. A list of other primers used for verification of microarray results by real time quantitative RT-PCR is available in the supplemental material.
Western Blotting—Ookinete cultures of WT and KO parasites containing a similar number of ookinetes (∼1 × 106) were harvested by centrifugation and extracted with SDS-PAGE loading buffer. The extracts were size-fractionated on SDS-12% polyacrylamide gels, and proteins were transferred to polyvinylidene difluoride membrane. The membrane was incubated overnight at 4 °C with mouse monoclonal anti-Pys21 (Pys21 is also known as Pys28 and is a member of the P28 family that includes Pbs21, Pfs28, and Pvs28) and anti-Pys25 antibodies (gift from Dr. T. Tsuboi, Ehime University, Japan (25)), diluted 1/500 and an anti-PyADA antibody diluted 1/1000 in a phosphate-buffered saline, 5% milk solution. The membrane was then incubated 1 h at room temperature with a 1/10,000 dilution of peroxidase-conjugated anti-mouse antibody. After extensive washing, signal was revealed using a chemiluminescent substrate, according to the manufacturer's instructions (ECL, Amersham Biosciences). Relative amounts of proteins were determined by densitometry using the Quantiscan software (Densylab, Microvision Instruments, Evry, France). Relative expression of Pys25 and Pys21 (or Pys28) in wild-type and KO was normalized by comparing the level of expression of the PyADA protein in each sample.
RESULTS
Disruption of the Pyhmgb2 Locus—Disruption of the pyhmgb2 locus was performed after integration of a plasmid containing a segment of the pyhmgb2 gene and the selectable marker cassette PbDHFR-TS fused to the GFP gene. The integration of this plasmid at the pyhmgb2 locus created two copies of the truncated gene after a single crossover event (Fig. 1A). Two independent pyhmgb2 knock-out clones (Δpyhmgb2; clones B3 and D1) were verified by PCR of genomic DNA (Fig. 1B and data not shown) and Southern blot using a pyhmgb2 gene probe (Fig. 1C). Disruption of the pyhmgb2 locus resulted in a total loss of detectable pyhmgb2 transcript expression by RT-PCR for the two clones (Fig. 1D and data not shown), whereas the transcript of an unrelated gene (pyada (PY020760) Fig. 1D, right panel) was not affected. Real time quantitative RT-PCR also confirmed that no residual expression of pyhmgb2 gene could be detected.
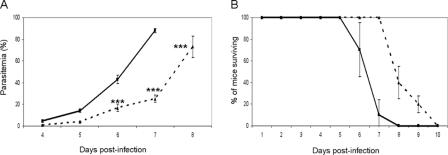
The PyHMGB2 KO Parasites Are Slightly Attenuated during the Asexual Erythrocytic Cycle—We studied the effect of the disruption of the pyhmgb2 locus on the asexual erythrocytic stage by examining growth of the parasite (Fig. 2A) as well as survival curves of mice infected by the WT or the Δpyhmgb2 parasite strains (Fig. 2B). Δpyhmgb2 parasite strains were able to proliferate normally, and parasitemia was readably detectable after day 4. However, there was a delay in the onset of parasitemia in the Δpyhmgb2-infected mice when compared with the WT parasite strain (Fig. 2A), and the mice infected by the Δpyhmgb2 parasite strains survived longer, dying at days 8–9 post-infection, whereas mice infected with the WT parasite strain died 6–8 days after infection (Fig. 2B). The delay in the onset of parasitemia and the change in the kinetics of death of the mice were reproducible in experiments with both BALB/c and Swiss Webster mice (data not shown).
FIGURE 2.
A, parasitemia (percentage of infected erythrocytes ± S.D.) was monitored on Giemsa smears for the WT (solid line) and the Δpyhmgb2 (dashed line) parasite lines during days 4–8 of the infection. ***, statistically significant in a two-way analysis of variance test (p < 0.001). B, survival curves for mice infected by WT (straight line) and the Δpyhmgb2 (dashed line) parasite lines. The average of three independent experiments (five mice per group) with standard deviation is reported. Mice were monitored twice a day until all died. The difference between the survival curve is statistically significant as considered by the log-rank (Mantel-Cox) test (p value = 0.0023) or Gehan-Breslow-Wilcoxon test (p value = 0.0039).
The Δpyhmgb2 Parasites Form Gametocytes and Ookinetes—We examined whether the Δpyhmgb2 parasites could complete the sexual cycle. The number of gametocytes on Giemsa-stained blood smears was similar in mice infected with Δpyhmgb2 parasite strains compared with the WT parasite after 2 days of infection (Table 1). There were no significant differences in the level of mature gametocytes between the Δpyhmgb2 parasite strains and the WT. The parasites were able to undergo gametocyte exflagellation (Table 1), but Δpyhmgb2 strains produced a decreased number of ookinetes when compared with the WT (Table 1).
TABLE 1.
Δpyhmgb2 parasites form in vitro ookinetes less efficiently
| Gametocytemiaa | Exflagellationb | Ookinete formation (in vitro)c | |
|---|---|---|---|
| % | % | ||
| Experiment 1 | |||
| WT | 0.36 | 57 ± 12 | 100 |
| KO | 0.25 | 63 ± 14 | 51.6d |
| Experiment 2 | |||
| WT | 0.56 | 43 ± 16 | 100 |
| KO | 0.49 | 39 ± 12 | 63.2d |
Percentage of gametocytes per 100 red blood cells is shown. Two mice were infected with each strain.
Exflagellation is the number of exflagellation centers per 10,000 red blood cells divided by the number of male gametocytes per 10,000 red blood cells multiplied by 100.
Ookinetes were produced by in vitro culture as described under “Experimental Procedures.” Efficiency of ookinete formation is expressed as the percentage of the WT control.
Data are statistically significant in a non-paired t test (p value <0.05).
The Δpyhmgb2 Parasites Have Dramatically Impaired Oocyst Production—We fed mosquitoes on mice infected with Δpyhmgb2 and WT parasite strains bearing similar numbers of gametocytes and asexual stages. Eight days after feeding, mosquito midguts were dissected, and the number of oocysts was scored using light microscopy. The number of infected mosquitoes for each strain was similar in each experiment (Table 2). In three independent experiments, the mean number of oocysts per Δpyhmgb2-infected mosquito was ∼10% of the mean oocyst number seen in WT parasite-infected mosquitoes (Table 2). The oocysts produced by Δpyhmgb2-infected mosquitoes were able to complete the life cycle and produce viable sporozoites that were infectious to mice. The blood of the mice infected by those sporozoites was collected 1 week after infection. Using real time quantitative PCR, we showed that these parasites do not express the pyhmgb2 transcript and have the same genotype as the Δpyhmgb2 parent (data not shown), indicating that no reversion of the disrupted allele had occurred.
TABLE 2.
Δpyhmg2 in vivo oocyst production is impaired
| Mean number of oocystsa | Oocyst prevalenceb | |
|---|---|---|
| Experiment 1 | ||
| WT | 65.5 ± 24 | 94.1 |
| KO | 7.8 ± 4.9c | 81.6 |
| Experiment 2 | ||
| WT | 92.1 ± 38 | 100.0 |
| KO | 15.7 ± 7.8c | 82.0 |
| Experiment 3 | ||
| WT | 64 ± 19 | 48.0 |
| KO | 4.3 ± 3.2c | 48.0 |
The mean number of oocysts per infected mosquito ± S.D. is shown.
Percentage of mosquito midguts containing oocysts is shown. 50 midguts were dissected for each experiment.
Data are statistically significant in a non-paired t test (p value <0.0001).
Microarray Comparison of Δpyhmgb2 and Wild-type Gene Expression—To examine the global expression profile of Δpyhmgb2 parasites, we compared the transcriptome of Δpyhmgb2 to the WT strain using a long oligonucleotide microarray designed to represent all the annotated genes of P. yoelii. These arrays have been previously validated to accurately reflect blood stage and liver stage P. yoelii gene expression (22, 26). We harvested total RNA from a mixture of synchronized late erythrocytic stage parasites with similar numbers of gametocytes. Two independent clones of the Δpyhmgb2 parasites were compared with the WT strain. Of the 6,428 annotated P. yoelii genes spotted on the array, only 32 genes showed differential expression in the KO (Table 3). It is interesting to note that we found only down-regulated genes in the Δpyhmgb2 parasites. There was no detectable difference in expression of the transcript for Pyhmgb1 (PY05184).
TABLE 3.
Microarray analysis of ΔPyhmgb2 clones reveals down-regulation of conserved sexual stage genes
| Annotation | FCa | Transcriptomics of P. falciparumb | Transcriptomics of P. bergheic | Proteomics of P. bergheid | Orthologue of P. falciparume | |
|---|---|---|---|---|---|---|
| PY07077 | High mobility group protein 2 | 6.16 | Gametocyte and schizonts | Gametocyte | MAL8P1.72 | |
| PY03130 | STOP repeat protein | 4.22f | Gametocyte | Gametocyte | Ookinete | PFI0460w |
| PY00522 | Ookinete surface antigen-like protein Pys25 | 4.16f | Gametocyte | Gametocyte | Ookinete and oocysts | PF10_0303 |
| PY00523 | Ookinete surface antigen-like protein Pys21 (Pys28) | 4.13f | Gametocyte | Ookinete and oocysts | PF10_0302 | |
| PY00860 | Hypothetical protein | 3.04 | Gametocyte and schizonts | Gametocyte | Ookinete | PF14_0522 |
| PY04557 | Hypothetical protein | 2.61 | Gametocyte | Gametocyte | Ookinete | PF13_0220 |
| PY05340 | Hypothetical protein | 2.39 | Gametocyte and schizonts | Ookinete | PF08_0073 | |
| PY00380 | Hypothetical protein | 2.38 | Gametocyte | Ookinete | PFL2320w | |
| PY01580 | LCCL domain-containing protein CCp2 | 2.33 | Gametocyte and schizonts | Gametocyte | Gametocyte and ookinete | PF14_0532 |
| PY00220 | SAC3/GANP/Nin1/mts3/eIF-3 p25 domain protein | 2.07 | PFF0125c | |||
| PY01624 | PfFNPA, a protein in the gametocyte-specific PfCCp family | 2.06 | Gametocyte | Gametocyte | Gametocyte and ookinete | PF14_0491 |
| PY03088 | Pfs77 protein-related; articulin | 1.97f | Gametocyte | Gametocyte | Ookinete | PF13_0226 |
| PY06201 | Acid protease | 1.91 | Merozoite and sporozoite | Gametocyte | PFL1660c | |
| PY02871 | Hypothetical protein | 1.87 | Gametocyte and schizonts | Gametocyte | Ookinete | PF11_0422 |
| PY01183 | Zinc finger domain protein | 1.86 | Gametocyte | Gametocyte | PF13_0314 | |
| PY07861 | Hypothetical protein | 1.85 | ||||
| PY04502 | Articulin | 1.84 | Gametocyte and schizonts | Gametocyte | Gametocyte and ookinete | PF08_0033 |
| PY02014 | Hypothetical protein | 1.78 | ||||
| PY01757 | Four tandem kelch domains followed by a BTB/POZ domain | 1.77 | Gametocyte | |||
| PY02893 | Adenosylhomocysteinase | 1.73f | Schizonts | Asexual and ookinete | PFE1050w | |
| PY01887 | Hypothetical protein | 1.71 | Gametocyte | Gametocyte | PF08_0024 | |
| PY02308 | Hypothetical protein | 1.68 | Gametocyte and schizonts | Gametocyte | Ookinete | PFD0520c |
| PY01032 | Hypothetical protein | 1.66 | Gametocyte | Gametocyte | PF08_0023 | |
| PY06780 | Hypothetical protein | 1.65 | Gametocyte | PFB0177c | ||
| PY00469 | Eukaryotic aspartyl protease Plasmepsin | 1.64 | Gametocyte | PF10_0329 | ||
| PY01137 | Articulin | 1.63 | Gametocyte | Gametocyte | Ookinete | PFL1030w |
| PY07430 | Calcium/calmodulin-stimulated cyclic nucleotide phosphodiesterase, putative | 1.63 | Gametocyte | PF14_0672 | ||
| PY04297 | P. falciparum CPW-WPC domain | 1.62f | Gametocyte | PF13_0168 | ||
| PY02878 | Hypothetical protein | 1.61 | Gametocyte and schizonts | Ookinete | MAL7P1.74 | |
| PY01150 | ∈-adaptin, putative-related | 1.6 | PFI0200c | |||
| PY00977 | Exonuclease, putative | 1.55f | Gametocyte | MAL13P1.311 | ||
| PY01299 | Orthologue of AMY-1 | 1.34f | Gametocyte | PF07_0060 |
FC indicates fold change calculated as normalized signal intensity of the wild type P. yoelii YM divided by normalized signal intensity of the KO (two clones). Data are from two biological replicates hybridized to the oligonucleotide array as discussed under “Experimental Procedures” (eight hybridization signals per gene for wild type). Data from the two KO clones were combined (8 hybridization signals per gene). All genes that showed significant change were down-regulated in the KO.
Data are as reported by Le Roch et al. (2) (data publicly available on line).
Data are up-regulated in gametocytes as reported by Hall et al. (4).
Data are as reported by Hall et al. (4) (data publicly available on line).
P. falciparum homologue annotation number as reported on PlasmoDB.
Down-regulation of genes was verified by quantitative RT-PCR (Fig. 3).
The differential expression of 10 down-regulated genes was independently confirmed by real time quantitative RT-PCR (Fig. 3). We also confirmed, by real time quantitative RT-PCR, that the transcript coding for the PyHMGB1 (PY05184) protein was not differentially expressed, suggesting that compensation by the overexpression of another HMGB did not occur in the Δpyhmgb2 parasites. Among control genes analyzed by quantitative RT-PCR were those with ubiquitous expression (pyada (PY02076) or pytubI (PY01155); Fig. 3 and supplemental Fig. 1) and other known gametocyte-specific genes whose expression was not altered by microarray analysis (Fig. 3 and supplemental Fig. 1). These were expressed at similar levels in WT and Δpyhmgb2 parasites, confirming that the differences found by microarray profiling were not because of discrepancies in the number or maturity of asexual parasites and/or gametocytes present in the WT and Δpyhmgb2 samples.
FIGURE 3.
Real time quantitative RT-PCR verifies down-regulation of genes differentially expressed between WT and Δpyhmgb2 in microarray experiments. Real time quantitative RT-PCR data are represented by the black bars. The microarray data are represented by gray bars. The ratio of transcript expression in the WT parasites to transcript expression in KO is represented for the genes listed at the bottom of the graph. Differential expression was verified for nine genes listed in Table 3 along with genes with ubiquitous expression (Pypnp, PytubI, Pyhmgb1, and Pyada) or selected gametocyte-specific genes whose expression was not affected in the KO (PytubII, Pywarp, Pys36, PY06600, pys48/45, PY00415, Pynima). A thick black line delimits the ratio for which genes are down-regulated in the KO when compared with the WT.
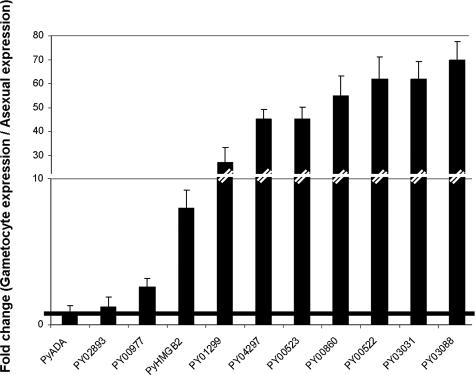
Down-regulated Genes in the Δpyhmgb2 Parasites Are Conserved and Expressed at the Gametocyte Stage—Among the genes down-regulated in the Δpyhmgb2 parasite, we found P. yoelii homologues of the P25 and P28 ookinete genes (PY00522 and PY00523, respectively) and of PfCCp2 (PY01580; also named lap4 in P. berghei) that have critical importance in oocyst development (27, 28). Of the 32 genes, all but three were conserved among all Plasmodium species (Table 3 and supplemental Table I). Over 92% of these genes are specifically expressed at the gametocyte stage or show enhanced expression at the gametocyte stage (Fig. 4). Transcripts of two P. falciparum homologues of down-regulated P. yoelii genes (PY06201 and PY02893) are exclusively expressed at the asexual stage (Fig. 4).
FIGURE 4.
Distribution of the of the expression peaks for the P. falciparum homologues of the down-regulated genes in the Δpyhmgb2 parasites. Expression data were available on PlasmoDB and from Ref. 2 for 27 down-regulated genes among the 29 genes conserved in at least P. falciparum and another Plasmodium species. 25 of the 27 genes (92%) show peak expression at the gametocyte stage (black bar). Genes expressed during the asexual stage exclusively or at the asexual and gametocyte stages (9 of 27; 33%) are represented by a white bar. 17 of the 27 genes (63%) show expression only at the gametocyte stage (dark gray bar). Genes expressed in only the asexual stages (2 of 27; 7%) are represented by a light gray bar.
We compared the steady state transcript levels in purified WT gametocytes and asexual erythrocytic forms for 10 of the genes down-regulated in the Δpyhmgb2 by real time quantitative RT-PCR (Fig. 5). As shown for its P. falciparum (29) and P. berghei homologues (4), the pyhmgb2 transcript is overexpressed (8-fold) in gametocytes compared with asexual stages (Fig. 5). We confirmed that eight of the down-regulated genes in the Δpyhmgb2 parasites were specifically expressed or overexpressed at the gametocyte stage in WT P. yoelii (Fig. 5). Analogous to its P. falciparum homologue, the PY02893 transcript (adenosylhomocysteinase) is not over-represented in gametocytes. Similarly, pyada transcript level was equivalent in gametocyte and asexual parasite extracts.
FIGURE 5.
Genes down-regulated in the KO are expressed specifically at the gametocyte stage of WT P. yoelii parasites. Quantification of the relative level of expression in gametocytes and asexual stages of WT P. yoelii parasites for 10 genes down-regulated in the KO was performed using real time quantitative RT-PCR. The normalized ratio of the measured transcript expression in purified gametocyte over asexual parasites in the WT parasites is represented for the genes listed at the bottom of the graph. A thick black line marks the ratio for which genes are overexpressed in gametocytes when compared with their expression in asexual parasites.
The Peak of Expression of the Pfhmgb2 Transcript Precedes or Overlaps Those of the Down-regulated Genes in the Δpyhmgb2 Parasites—We queried available expression profile data for gametocytogenesis of the P. falciparum 3D7 strain (3) for pfhmgb2 and other orthologues of genes down-regulated in the Δpyhmgb2 parasites (supplemental Fig. 2 and Table 3). The pfhmgb2 transcript expression peaks between day 6 and day 8 of the gametocytogenesis of 3D7 strain, whereas the genes down-regulated in the Δpyhmgb2 parasites peak at day 8 (supplemental Fig. 2 and Table 3). Interestingly, the signal yielded by the oligonucleotides representing the two genes expressed exclusively at the asexual stage (Table 3) does not follow this pattern (data not shown). These data are consistent with the hypothesized role of PyHMGB2 in the transcriptional regulation of the down-regulated genes.
Genes Down-regulated in the Δpyhmgb2 Parasites Encode Proteins Expressed at the Ookinete Stage—Translational repression during Plasmodium sexual development is described as a major mechanism of gene regulation. We examined the proteomic data (4, 30) available for the genes whose expression was down-regulated in the Δpyhmgb2 parasites. Of the 18 genes transcripts down-regulated in the Δpyhmgb2 parasites for which proteomic data are available, 16 (88%) had mRNA expressed at the gametocyte stage but only 5 had protein detectable in gametocytes (including 4 with both gametocyte and ookinete protein expression). Twelve of the 18 genes (67%; all with mRNA expressed in gametocytes stages) had gene products detected in only ookinetes or oocysts (Table 3). Interestingly, more than 80% of the genes down-regulated in the Δpyhmgb2 parasites had their expression reduced by at least 2-fold in the ΔDOZI parasites (7) (supplemental Table SI).
We performed an analysis of the expression of the PY00522 and PY00523 proteins, homologues of the P. falciparum Pfs25 and Pfs28 proteins. A similar number of in vitro cultured ookinetes from WT or Δpyhmgb2 parasites was loaded on a polyacrylamide gel, separated by electrophoresis, and subjected to Western blot. As shown in Fig. 6A, the expression of PyADA (upper panel) was comparable in WT and KO, whereas the Δpyhmgb2 parasites expressed ∼3–4-fold less Pys25 and Pys21 (also known as Pys28) protein as quantified by densitometry (Fig. 6B) in concordance with the transcriptomic data. An immunofluorescence analysis performed on the Δpyhmgb2 parasites in in vitro cultured ookinetes did not reveal a subpopulation with expression of these two proteins at the WT level (data not shown).
FIGURE 6.
Pys21 and Pys25 proteins are down-regulated in KO ookinetes. A, Western blot analysis of PyADA expression in WT and KO ookinetes (upper panel). Pys21 and Pys25 (lower panel) were detected in WT and KO cultured ookinetes. (Pys21 is also known as Pys28 and is a member of the P28 family that includes Pbs21, Pfs28, and Pvs28). B, densitometry quantification of the expression of Pys21 (or Pys28) and Pys25 proteins. Values for WT and KO protein levels were normalized using the PyADA loading control. Comparisons between WT and KO are presented in arbitrary units. WT values are represented with black bars. KO values are represented by gray bars.
DISCUSSION
In this study, we investigated the role of the PyHMGB2 during the complete life cycle of the malaria parasite P. yoelii. HMGB1 and HMGB2 are highly conserved among Plasmodium species (16). The P. falciparum homologue of the PyHMGB2 protein is expressed by schizonts at the end of the erythrocytic asexual cycle and exhibits enhanced expression during gametocytogenesis (29). P. yoelii homologues follow the same pattern of expression as confirmed by real time quantitative RT-PCR data (Fig. 5).
Loss of expression of the pyhmgb2 transcript during the erythrocytic asexual cycle in the mouse resulted in the delay of the onset of the parasitemia and time to death for the mice infected by the KO strains (Fig. 2). Although Δpyhmgb2 parasites are attenuated, PyHMGB2 is not essential during the asexual cycle of the parasite. The mild phenotype in asexual stages may be due to functional redundancy of PyHMGB2 with PyHMGB1 (16), but Δpyhmgb2 parasites did not compensate with overexpression of the pyhmgb1 transcript (Fig. 4).
Analysis of the Δpyhmgb2 parasite transcriptome (Table 3) revealed down-regulation of a subset of gametocyte transcripts. Two down-regulated genes, P. yoelii acid protease (PY06201) and adenosylhomocysteinase (PY02893), have P. falciparum orthologues exclusively expressed during the asexual cycle. The P. berghei homologue of the acid protease is expressed at the gametocyte stage indicating that this gene might be gametocyte-specific in rodent malarias (4). The adenosylhomocysteinase (PY02893) transcript expression profile was confirmed by real time quantitative PCR (Fig. 5). P. berghei adenosylhomocysteinase protein is expressed in both asexual parasites and ookinetes (4), indicating a potential role of this gene during both asexual and sexual development.
The majority of the genes that are differentially expressed in the KO have a homologue in all the Plasmodium species sequenced to date. In contrast, only 50% of the genes induced at the gametocyte stage for P. berghei have a homologue in P. falciparum (4). Thus the genes whose expression is regulated by HMGB2 probably play conserved roles in all Plasmodium species.
The process of gametogenesis that is triggered in the mosquito after the blood meal is controlled by kinases that are able to rapidly induce a cascade of events in response to an external stimuli (31). Interestingly, none of the genes for these kinases is differentially regulated in the Δpyhmgb2 parasites (Table 3 and supplemental Fig. 1).
The number and the maturity of the gametocytes were similar between Δpyhmgb2 and the WT (Table 1) suggesting that PyHMGB2 is not involved in the induction, commitment, and establishment of the gametocytemia in the blood. Gametocyte exflagellation was also comparable in the Δpyhmgb2 parasite when compared with the WT.
Ookinete formation was significantly decreased in Δpyhmgb2 and comparable with that found for a double knock-out of the P28 and P25 P. berghei genes (27, 32). In vivo, we observed a drastic impairment of oocyst formation (Table 2), although our microarray data indicate that PyHMGB2 regulates expression of genes transcribed in gametocytes.
We confirmed the gametocyte-specific expression of nine genes down-regulated in the Δpyhmgb2 parasites, verifying concordance of the expression profiles in P. falciparum, P. berghei, and P. yoelii. The P. berghei proteomic data suggest that many of these genes are translationally repressed until the ookinete stage (Table 3) (4). However, because of limitations in sensitivity of proteomic technology, we cannot rule out that some genes down-regulated in the KO are expressed in other stages of the life cycle.
Translational repression seems to be widespread during the sexual stages of Plasmodium (7) and has been well characterized for Pbs21 and Pbs25, two proteins whose transcript levels are down-regulated in the KO (33). In accordance with the proteomics data, 80% of the genes down-regulated in the Δpyhmgb2 parasites (supplemental Table SI) have their expression at least partially controlled through the action of DOZI as shown by the microarray data collected for the P. berghei ΔDOZI parasites (7).
Three genes down-regulated in the mutant parasites (Table 3) have been disrupted in different Plasmodium species. The Pbs21 (or Pbs28) and Pbs25 genes, which encode surface proteins of ookinetes, have been shown to have redundant function as only a double KO exhibited a clear phenotype (27). The P. berghei strain with double KO of Pbs21 and Pbs25 has modest reduction of ookinete production and a drastic impairment of oocyst production in the mosquito (27). The ookinetes produced in the double KO are not able to cross the midgut epithelium and interact with the basal lamina and consequently do not produce oocysts (27). Similarly, PfCCp2 (P. falciparum) and its orthologue pblap4 (P. berghei) have been disrupted with a phenotype in the oocyst stage despite protein expression at the gametocyte stage (28, 34). The observation that pys25, pys21, and pyCCP2 genes are down-regulated in our mutant could be sufficient to explain the phenotype of the Δpyhmgb2 parasite. Nevertheless, the other down-regulated genes could contribute to ookinete and oocyst formation given their protein expression profile and conservation across Plasmodium species. For example three gametocyte-specific articulins, a class of cytoskeletal proteins, are down-regulated in the Δpyhmgb2 parasites suggesting involvement of articulins in the maintenance of stage-specific cellular shapes (35).
The few oocysts generated by the Δpyhmgb2 were able to produce a reduced number of infective sporozoites (data not shown). The Pbs21 and Pbs25 double KO parasites were also able to produce infective sporozoites (27). Therefore, PyHMGB2 appears to play a crucial role for the full expression of these genes but is not absolutely required.
Because the HMGB box family does not bind to DNA in a sequence-specific manner, PyHMGB2 is likely to be recruited to DNA by other factors or by a specific chromatin context (17). Interestingly, yeast two-hybrid interaction data (36) show that HMGB2 is in the center of a broader network of proteins potentially involved in transcription regulation, including three AP2 domain plant-like transcription factors (supplemental Fig. 3). AP2 proteins regulate the transcription of a number of stress-induced pathways in plants (15). Genes for AP2 family members in P. falciparum are differentially expressed at specific developmental stages implicating them in transcriptional regulation of stage-specific gene expression (14). HMGB2 also interacts in yeast two-hybrid with chromatin remodeling and modification enzymes like the acetyltransferase GCN5 and a potential histone demethylase containing a Jumonji domain (36) (supplemental Fig. 3). PyHMGB2 may act cooperatively with AP2 sequence-specific transcription factors and the chromatin modification machinery to control gene expression.
The sexual cycle of Plasmodium is a critical process required for transmission of malaria via the mosquito. In this study, we demonstrate that PyHMGB2 is not required for asexual growth but is involved in controlling genes important for oocyst development in the mosquito. Analysis of gene expression and proteomic and protein interaction data indicate that PyHMGB2 plays a pivotal role in controlling the transcriptional expression of a set of genes that are crucial for completion of the sexual cycle of malaria.
Supplementary Material
Acknowledgments
We thank Alida Coppi for sharing her expertise of the mosquito life cycle of Plasmodium. We thank Sandra Gonzalez and Jean Nonon for their assistance with the rearing and infection of the mosquitoes. The design and construction of the P. yoelii microarray was supported by the National Institutes of Health (to L. W. B.).
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AI056840 (to P. S.). This work was also supported by a Philippe Foundation fellowship (to M. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental data, Figs. 1–3, and Table I.
Footnotes
The abbreviations used are: HMGB, high mobility group box; RT, reverse transcription; WT, wild type; KO, knock-out; GFP, green fluorescent protein; DIG, digoxigenin.
References
- 1.Alano, P. (2007) Mol. Microbiol. 66 291–302 [DOI] [PubMed] [Google Scholar]
- 2.Le Roch, K. G., Johnson, J. R., Florens, L., Zhou, Y., Santrosyan, A., Grainger, M., Yan, S. F., Williamson, K. C., Holder, A. A., Carucci, D. J., Yates, J. R., III, and Winzeler, E. A. (2004) Genome Res. 14 2308–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young, J. A., Fivelman, Q. L., Blair, P. L., de la Vega, P., Le Roch, K. G., Zhou, Y., Carucci, D. J., Baker, D. A., and Winzeler, E. A. (2005) Mol. Biochem. Parasitol. 143 67–79 [DOI] [PubMed] [Google Scholar]
- 4.Hall, N., Karras, M., Raine, J. D., Carlton, J. M., Kooij, T. W., Berriman, M., Florens, L., Janssen, C. S., Pain, A., Christophides, G. K., James, K., Rutherford, K., Harris, B., Harris, D., Churcher, C., Quail, M. A., Ormond, D., Doggett, J., Trueman, H. E., Mendoza, J., Bidwell, S. L., Rajandream, M. A., Carucci, D. J., Yates, J. R., III, Kafatos, F. C., Janse, C. J., Barrell, B., Turner, C. M., Waters, A. P., and Sinden, R. E. (2005) Science 307 82–86 [DOI] [PubMed] [Google Scholar]
- 5.Khan, S. M., Franke-Fayard, B., Mair, G. R., Lasonder, E., Janse, C. J., Mann, M., and Waters, A. P. (2005) Cell 121 675–687 [DOI] [PubMed] [Google Scholar]
- 6.Kumar, N., and Carter, R. (1985) Mol. Biochem. Parasitol. 14 127–139 [DOI] [PubMed] [Google Scholar]
- 7.Mair, G. R., Braks, J. A., Garver, L. S., Wiegant, J. C., Hall, N., Dirks, R. W., Khan, S. M., Dimopoulos, G., Janse, C. J., and Waters, A. P. (2006) Science 313 667–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aravind, L., Iyer, L. M., Wellems, T. E., and Miller, L. H. (2003) Cell 115 771–785 [DOI] [PubMed] [Google Scholar]
- 9.Callebaut, I., Prat, K., Meurice, E., Mornon, J. P., and Tomavo, S. (2005) BMC Genomics 6 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gissot, M., Kelly, K. A., Ajioka, J. W., Greally, J. M., and Kim, K. (2007) Plos Pathog. 3 e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voss, T. S., Kaestli, M., Vogel, D., Bopp, S., and Beck, H. P. (2003) Mol. Microbiol. 48 1593–1607 [DOI] [PubMed] [Google Scholar]
- 12.Dechering, K. J., Kaan, A. M., Mbacham, W., Wirth, D. F., Eling, W., Konings, R. N., and Stunnenberg, H. G. (1999) Mol. Cell. Biol. 19 967–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gissot, M., Briquet, S., Refour, P., Boschet, C., and Vaquero, C. (2005) J. Mol. Biol. 346 29–42 [DOI] [PubMed] [Google Scholar]
- 14.Balaji, S., Babu, M. M., Iyer, L. M., and Aravind, L. (2005) Nucleic Acids Res. 33 3994–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutterson, N., and Reuber, T. L. (2004) Curr. Opin. Plant Biol. 7 465–471 [DOI] [PubMed] [Google Scholar]
- 16.Briquet, S., Boschet, C., Gissot, M., Tissandie, E., Sevilla, E., Franetich, J. F., Thiery, I., Hamid, Z., Bourgouin, C., and Vaquero, C. (2006) Eukaryot. Cell 5 672–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agresti, A., and Bianchi, M. E. (2003) Curr. Opin. Genet. Dev. 13 170–178 [DOI] [PubMed] [Google Scholar]
- 18.Arai, M., Billker, O., Morris, H. R., Panico, M., Delcroix, M., Dixon, D., Ley, S. V., and Sinden, R. E. (2001) Mol. Biochem. Parasitol. 116 17–24 [DOI] [PubMed] [Google Scholar]
- 19.Carter, V., Nacer, A. M., Underhill, A., Sinden, R. E., and Hurd, H. (2007) Int. J. Parasitol. 37 1221–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jongco, A. M., Ting, L. M., Thathy, V., Mota, M. M., and Kim, K. (2006) Mol. Biochem. Parasitol. 146 242–250 [DOI] [PubMed] [Google Scholar]
- 21.Mota, M. M., Thathy, V., Nussenzweig, R. S., and Nussenzweig, V. (2001) Mol. Biochem. Parasitol. 113 271–278 [DOI] [PubMed] [Google Scholar]
- 22.Tarun, A. S., Peng, X., Dumpit, R. F., Ogata, Y., Silva-Rivera, H., Camargo, N., Daly, T. M., Bergman, L. W., and Kappe, S. H. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 305–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smyth, G. K. (2004) Stat. Appl. Genet Mol. Biol. 3 article 3 [DOI] [PubMed]
- 24.Smyth, G. K. (2005) in Bioinformatics and Computational Biology Solutions Using R and Bioconductor (Gentleman, R., ed) pp. 397–420, Springer-Verlag, New York
- 25.Tsuboi, T., Cao, Y. M., Hitsumoto, Y., Yanagi, T., Kanbara, H., and Torii, M. (1997) Infect. Immun. 65 2260–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi, Q., Cernetich, A., Daly, T. M., Galvan, G., Vaidya, A. B., Bergman, L. W., and Burns, J. M., Jr. (2005) Infect. Immun. 73 6363–6371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomas, A. M., Margos, G., Dimopoulos, G., van Lin, L. H., de Koning-Ward, T. F., Sinha, R., Lupetti, P., Beetsma, A. L., Rodriguez, M. C., Karras, M., Hager, A., Mendoza, J., Butcher, G. A., Kafatos, F., Janse, C. J., Waters, A. P., and Sinden, R. E. (2001) EMBO J. 20 3975–3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raine, J. D., Ecker, A., Mendoza, J., Tewari, R., Stanway, R. R., and Sinden, R. E. (2007) Plos Pathog. 3 e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Roch, K. G., Zhou, Y., Blair, P. L., Grainger, M., Moch, J. K., Haynes, J. D., De La Vega, P., Holder, A. A., Batalov, S., Carucci, D. J., and Winzeler, E. A. (2003) Science 301 1503–1508 [DOI] [PubMed] [Google Scholar]
- 30.Florens, L., Washburn, M. P., Raine, J. D., Anthony, R. M., Grainger, M., Haynes, J. D., Moch, J. K., Muster, N., Sacci, J. B., Tabb, D. L., Witney, A. A., Wolters, D., Wu, Y., Gardner, M. J., Holder, A. A., Sinden, R. E., Yates, J. R., and Carucci, D. J. (2002) Nature 419 520–526 [DOI] [PubMed] [Google Scholar]
- 31.Billker, O., Dechamps, S., Tewari, R., Wenig, G., Franke-Fayard, B., and Brinkmann, V. (2004) Cell 117 503–514 [DOI] [PubMed] [Google Scholar]
- 32.Siden-Kiamos, I., Vlachou, D., Margos, G., Beetsma, A., Waters, A. P., Sinden, R. E., and Louis, C. (2000) J. Cell Sci. 113 3419–3426 [DOI] [PubMed] [Google Scholar]
- 33.Paton, M. G., Barker, G. C., Matsuoka, H., Ramesar, J., Janse, C. J., Waters, A. P., and Sinden, R. E. (1993) Mol. Biochem. Parasitol. 59 263–275 [DOI] [PubMed] [Google Scholar]
- 34.Pradel, G., Hayton, K., Aravind, L., Iyer, L. M., Abrahamsen, M. S., Bonawitz, A., Mejia, C., and Templeton, T. J. (2004) J. Exp. Med. 199 1533–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Templeton, T. J., Iyer, L. M., Anantharaman, V., Enomoto, S., Abrahante, J. E., Subramanian, G. M., Hoffman, S. L., Abrahamsen, M. S., and Aravind, L. (2004) Genome Res. 14 1686–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.LaCount, D. J., Vignali, M., Chettier, R., Phansalkar, A., Bell, R., Hesselberth, J. R., Schoenfeld, L. W., Ota, I., Sahasrabudhe, S., Kurschner, C., Fields, S., and Hughes, R. E. (2005) Nature 438 103–107 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.